Sirt6 Regulates the Development of Medullary Thymic Epithelial Cells and Contributes to the Establishment of Central Immune Tolerance
- 1State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Department of Central Laboratory and Institute of Clinical Molecular Biology, Peking University People’s Hospital, Beijing, China
- 4Center of Organ Transplantation, Second Xiangya Hospital of Central South University, Changsha, China
- 5Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing, China
Although some advances have been made in understanding the molecular regulation of mTEC development, the role of epigenetic regulators in the development and maturation of mTEC is poorly understood. Here, using the TEC-specific Sirt6 knockout mice, we found the deacetylase Sirtuin 6 (Sirt6) is essential for the development of functionally competent mTECs. First of all, TEC-specific Sirt6 deletion dramatically reduces the mTEC compartment, which is caused by reduced DNA replication and subsequent impaired proliferation ability of Sirt6-deficient mTECs. Secondly, Sirt6 deficiency specifically accelerates the differentiation of mTECs from CD80–Aire– immature population to CD80+Aire– intermediate mature population by promoting the expression of Spib. Finally, Sirt6 ablation in TECs markedly interferes the proper expression of tissue-restricted antigens (TRAs) and impairs the development of thymocytes and nTreg cells. In addition, TEC conditional knockout of Sirt6 results in severe autoimmune disease manifested by reduced body weight, the infiltration of lymphocytes and the presence of autoantibodies. Collectively, this study reveals that the expression of epigenetic regulator Sirt6 in TECs is crucial for the development and differentiation of mTECs, which highlights the importance of Sirt6 in the establishment of central immune tolerance.
Introduction
As a primary lymphoid organ, thymus plays an indispensable role in the establishment of central immune tolerance (Anderson and Takahama, 2012; Abramson and Anderson, 2017). Among the thymic stromal cells, thymic epithelial cells (TECs) provide a special microenvironment for the survival, development and maturation of thymocytes to the development of immunological competent T lymphocytes which can recognize foreign antigens rather than self-antigens (Sun et al., 2014; Abramson and Anderson, 2017; Nitta and Takayanagi, 2020). TECs include cortical thymic epithelial cells (cTECs) and medullary thymic epithelial cells (mTECs) (Alawam et al., 2020). cTECs are essential for positive selection of T lymphocytes (Takada et al., 2017). mTECs mediate negative selection by eliminating the autoreactive T lymphocytes and promoting the generation of nTreg cells, which are critical to build central immune tolerance and prevent the occurrence of autoimmune diseases (Akiyama et al., 2005; Anderson et al., 2007; Kadouri et al., 2020). The development of mTECs is mainly regulated by CD40, TNF receptor family (TNFRF) protein receptor activator of NF-κB (RANK), and lymphotoxin β receptor (LTβR) (Rossi et al., 2007; Zhu et al., 2007; Akiyama et al., 2008, 2012; Hikosaka et al., 2008; Seach et al., 2008). The cooperation of CD40 and RANK in mTECs is crucial for the establishment of self-tolerance (Akiyama et al., 2008; Hikosaka et al., 2008), while LTβR-mediated mTECs development is not involved in the establishment of self-tolerance (Cosway et al., 2017). Among the NF-κB signaling pathways, RelB subunit of the NF-κB complex (Burkly et al., 1995; Weih et al., 1995; Riemann et al., 2017; Jin and Zhu, 2018), NF-κB inducing kinase (NIK) (Kajiura et al., 2004), TNF receptor-activated factor 6 (TRAF6) (Akiyama et al., 2005), IκB-kinase (IKK) (Kinoshita et al., 2006), and NF-κB2 (Zhang et al., 2006; Zhu et al., 2006) have been widely recognized to regulate the development of mTEC and establish of self-tolerance. Compared with immature mTECs, functional mature mTECs act as the antigen present cell characterized by high expression of CD80, MHC class II (MHC II), and Aire (Kyewski and Klein, 2006; Klein et al., 2009; Anderson and Su, 2016). The expression of these molecules by mTECs is indispensable for thymocytes negative selection (Kadouri et al., 2020). Mature mTECs mediate the deletion of autoreactive T cells and promote the development of natural regulatory T cell (nTreg) by expressing tissue-restricted antigens (TRAs), which is essential for the establishment of central immune tolerance (Aschenbrenner et al., 2007; Cowan et al., 2013; Malhotra et al., 2016; Lebel et al., 2020). As so far, the molecular mechanisms regulating the development and differentiation of mTECs are still elusive. A recent report shows that SIRT1, a member of sirtuins family, could interact with Aire and induces its deacetylation, which is essential for Aire-driven TRAs expression and subsequent establishment of central immune tolerance (Chuprin et al., 2015). The epigenetic regulation of other Sirtuins family members in TEC development is not clear.
As a NAD+ dependent histone deacetylase, SIRT6 plays key roles in the regulation of metabolism, inflammation, longevity, genome stability and cancer (Kugel and Mostoslavsky, 2014; Chang et al., 2020). The phenotype of Sirt6 knockout mice was early reported by Mostoslavsky et al. (2006). They found that Sirt6 knockout mice died at about 4 weeks, the thymus of the mice diminished drastically and displayed a profound lymphopenia. The defects of lymphocytes in Sirt6 knockout mice was non-cell-autonomous (Mostoslavsky et al., 2006), which implies the defect of thymus in Sirt6 knockout mice may be caused by Sirt6 ablation in thymic stromal cells. We therefore investigated the role of Sirt6 in TECs using the TEC-specific Sirt6 knockout mice. We found that Sirt6 deficient in TECs caused severe thymic atrophy and that the proliferation, maturation and function of mTECs were drastically affected by Sirt6 deletion. Impressively, TEC-specific Sirt6 knockout mice spontaneously developed autoimmune disease. Our study unveils the indispensable role of Sirt6 in the development and maturation of mTECs and in the establishment of central immune tolerance.
Results
TEC-Specific Sirt6 Ablation Leads to Severe Thymic Atrophy
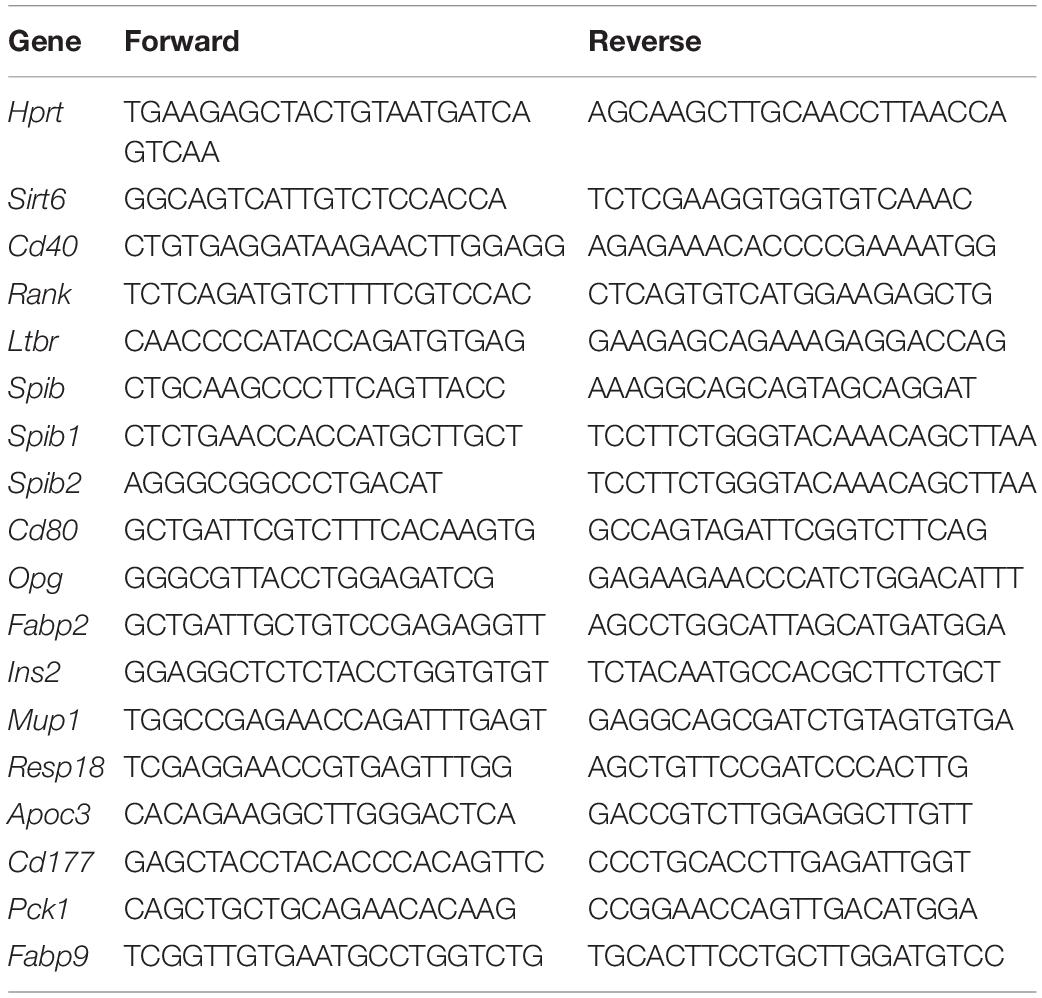
To investigate the role of Sirt6 in TECs, we crossed Foxn1-cre mice (Soza-Ried et al., 2008; Liu et al., 2013) and Sirt6flox/flox mice (Kim et al., 2010; Wang et al., 2016) to generate Sirt6 conditional knock out mice (designated as Sirt6 cKO mice henceforth) and we further confirmed Sirt6 was indeed inactivated (Supplementary Figures 1A–C). First, we checked the effect of TEC-specific Sirt6 knockout on the thymus of the 4-week-old mice. Compared with wild-type mice, the specific deletion of Sirt6 in TECs led to severe thymic atrophy (Figures 1A,B). The morphological analysis showed that thymic medullary region decreased significantly while the cortical region had no obvious change after Sirt6 ablation (Figures 1C,D).
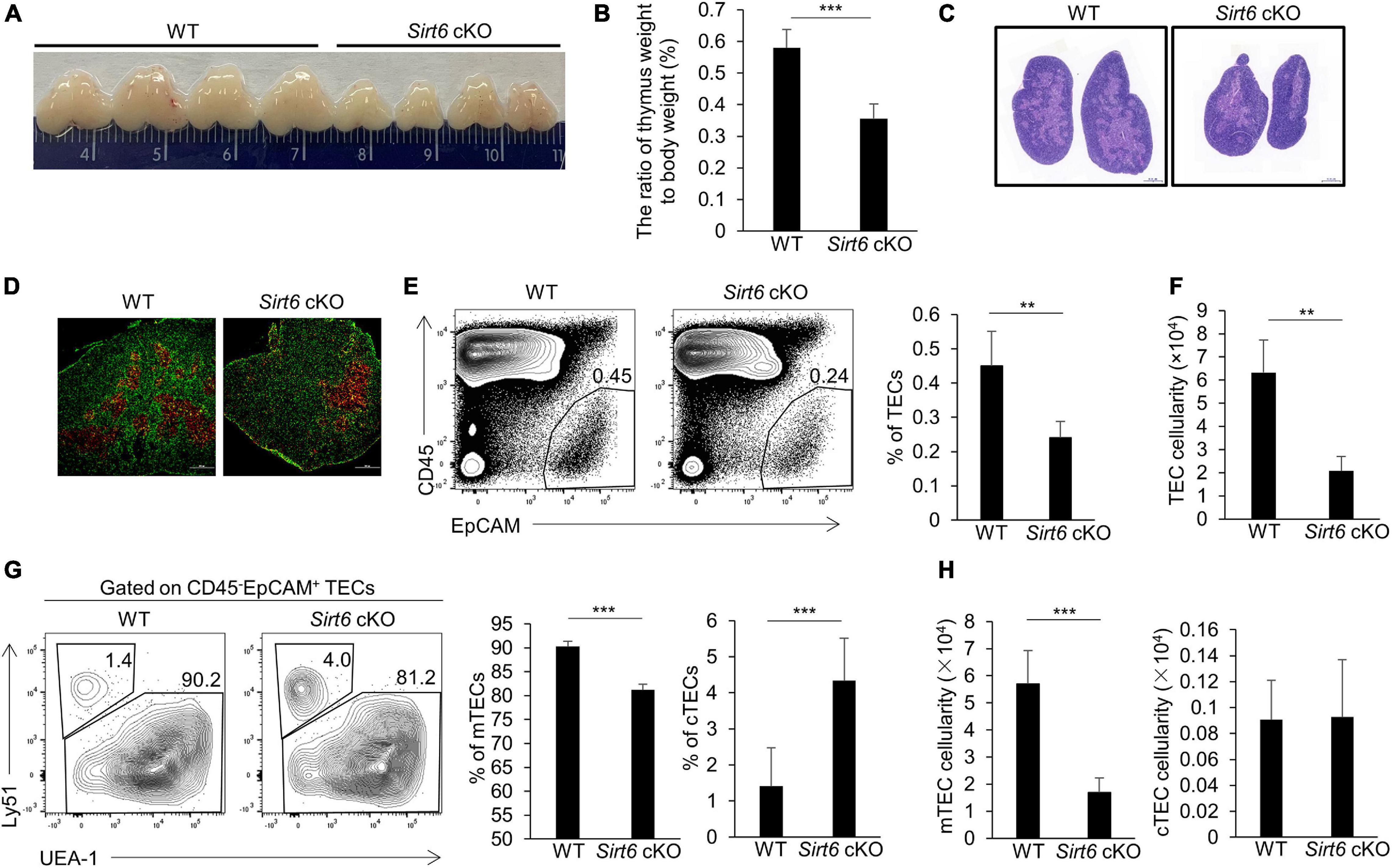
Figure 1. TEC-specific ablation of Sirt6 causes thymus atrophy and reduces mTEC compartment. (A) Representatives of thymi isolated from 4-week-old Sirt6 cKO mice and wild-type littermates. (B) Thymic weight normalized to body weight in 4-week-old wild-type and Sirt6 cKO mice. (C) Hematoxylin and eosin (H&E) stained thymic sections of 4-week-old wild-type and Sirt6 cKO mice. Scale bars: 1,000 μm. (D) Representative staining of frozen thymic sections of wild-type and Sirt6 cKO mice. KRT8 (green) and KRT5 (red) highlights cortical regions and medullary regions, respectively. Scale bars: 500 μm. (E) Representative flow cytometric profiles and frequency of CD45–EpCAM+ TECs obtained from 4-week-old Sirt6 cKO mice and littermates. (F) Total cell numbers of TECs of 4-week-old Sirt6 cKO mice and littermates. (G) Representative flow cytometric profiles showing frequencies of mTECs (UEA-1+Ly51–) and cTECs (UEA-1–Ly51+) were first gated on TECs. (H) Total cell numbers of mTECs (left) and cTECs (right) of 4-week-old wild-type and Sirt6 cKO mice. N = 4 per group. **p < 0.01 and ***p < 0.001 (Student’s t-test).
As a longevity gene, whether Sirt6 deletion leads to thymus senescence attracts our attention. Interestingly, although thymic atrophy still could be seen in 8-month-old Sirt6 cKO mice, the extent of reduction in thymus size was smaller than that in 4-week-old Sirt6 cKO mice (Supplementary Figure 2A). The rate of thymus weight loss (normalized for total body weight) and the rate of thymocytes number reduction did not increase in 8-month-old Sirt6 cKO mice compared with wild-type control littermates (Supplementary Figure 2B). The morphology of thymus in 8-month-old Sirt6 cKO mice showed no obvious signs of aging and the cortico-medullary junctions was integrated as assessed by H&E staining and Masson staining (Supplementary Figures 2C,D). Loss of naïve T cells in spleen is another indicator of age-related thymic degeneration (Aw and Palmer, 2012). However, from the 4 weeks to 8 months of age, the decline of naïve T cells in the spleen of Sirt6 cKO mice did not worsen compared with control mice (Supplementary Figures 2E,F). Thus, we focused our studies on the role of Sirt6 in the development and differentiation of TECs but not on the aging of TECs. Unless otherwise indicated, further experiments were carried out in 4-week-old Sirt6 cKO mice and their littermate controls.
To assess the endogenous role of Sirt6 in TECs, we checked the frequency and total number of TECs (CD45–EpCAM+) in Sirt6 cKO mice and their littermate controls. Flow cytometry analysis revealed that Sirt6 deficiency resulted in the reduction of TECs, both in proportion and in absolute cell number (Figures 1E,F). These results suggested the thymic hypoplasia in Sirt6 cKO mice was caused by impaired TEC development. As we mentioned above, TECs are composed of cTECs and mTECs, we further scrutinized the effect of Sirt6 ablation on cTECs and mTECs. We found the percentage and cell number of mTECs reduced remarkable and absolute number of cTECs was obviously unchanged although the proportion of cTECs was relatively increased in Sirt6 cKO mice (Figures 1G,H). These results showed that Sirt6 deficiency predominately influence the development of mTECs rather than cTECs.
Sirt6 Deficiency Impairs the Proliferation of mTECs by Reducing DNA Replication
Due to the decreased thymus size and the cell number of mTECs in Sirt6 cKO mice, we examined the proliferation and apoptosis of mTECs. The results showed that the percentage of Ki67-positive cells in total mTECs reduced by nearly half in Sirt6 cKO mice (Figures 2A,B). Whereas, the expression of active caspase 3, an indicator of intrinsic apoptosis, did not increase in mTECs of Sirt6 cKO mice compared with littermate controls (Supplementary Figure 3).
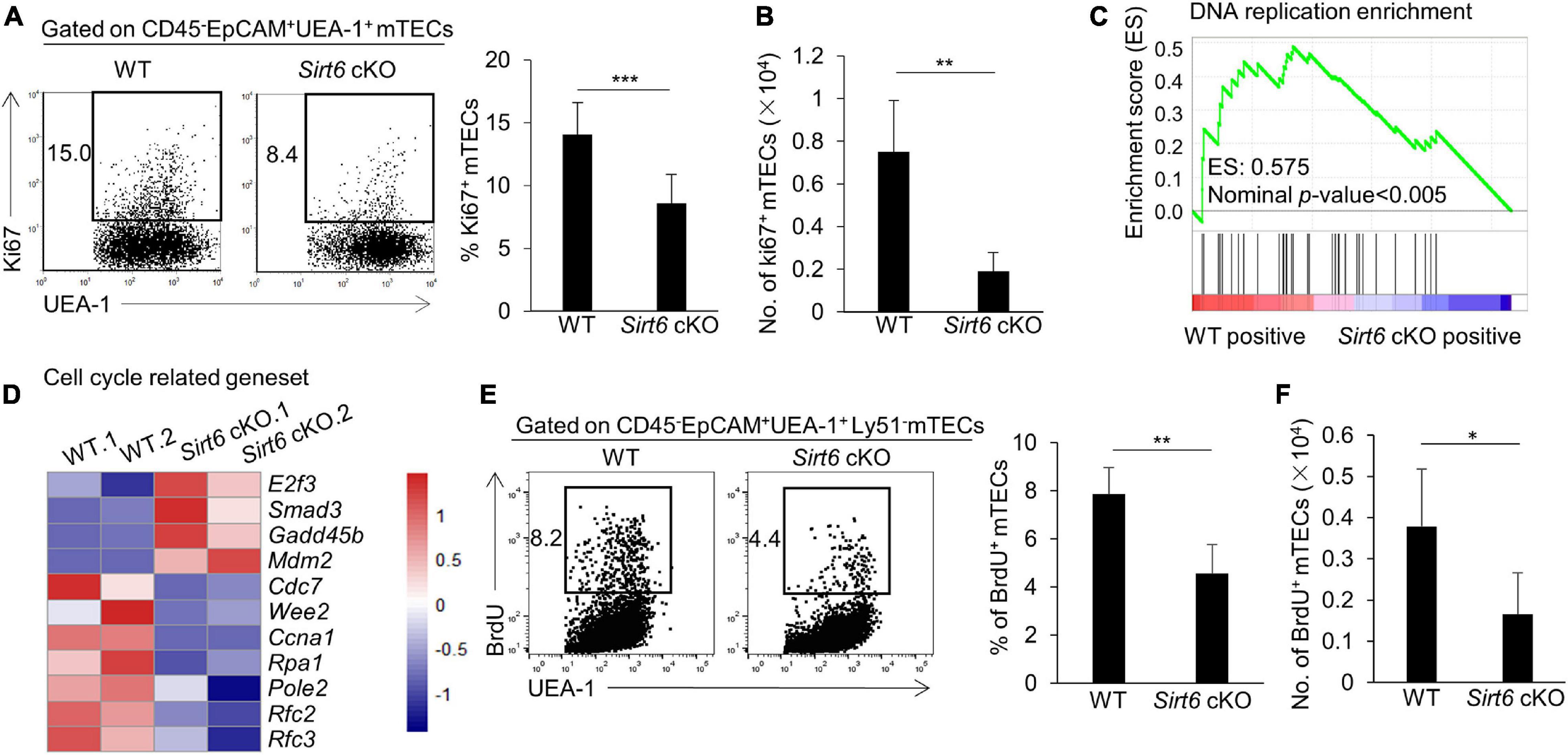
Figure 2. Sirt6 deficiency impairs mTEC proliferation capacity by reducing DNA replication in cell cycle. (A) Flow cytometry plots and frequency for the staining of Ki67 in mTECs. (B) Cell numbers of Ki67+ mTECs of 4-week-old Sirt6 cKO mice and littermates. (C) GSEA analysis reveals that DNA replication process had a less positive expression in Sirt6 deficient mTECs compared with wild-type mTECs defined by the criterion of p < 0.005. (D) Heatmap of the significantly changed genes (p < 0.05) associated with cell cycle and proliferation. (E) BrdU staining was used for flow cytometry analysis of mTECs of 2-week-old Sirt6 cKO mice and littermates 24 h after intraperitoneal injection of BrdU (1 mg per mice). (F) Cell numbers of BrdU+ mTECs of 2-week-old Sirt6 cKO mice and littermates. N ≥ 4 per group. *p < 0.05, **p < 0.01, and ***p < 0.001 (Student’s t-test).
To better understand how Sirt6 regulates cell proliferation, we performed RNA-sequencing analysis (RNA-seq) on mTECs (CD45–EpCAM+UEA-1+ly51–) sorted from 4-week-old wild-type or Sirt6 cKO mice, with two parallel samples in each group (named WT.1 and WT.2, Sirt6 cKO. 1 and Sirt6 cKO. 2). Gene Set Enrichment Analysis (GSEA) showed that DNA replication process was significantly reduced in Sirt6-deficient mTECs (Figure 2C), indicating that Sirt6 ablation may affect the cell cycle progression of mTECs. The expressions of many proliferation-related genes changed in Sirt6 deficient mTECs (Figure 2D). The expressions of genes promoting cell cycle and cell proliferation such as Cdc7, Ccna1, Rpa1, Pole2, Rfc2, and Rfc3 (Chen and Wold, 2014; Klajic et al., 2014; Tudzarova et al., 2016; Hu et al., 2020; Rogers et al., 2020) were down-regulated in Sirt6 deficient mTECs compared with wild-type TECs, whereas genes inhibiting cell cycle and proliferation such as E2f3, Smad3, and Gadd45b (Salvador et al., 2013; Park et al., 2016; Clijsters et al., 2019) were significantly up-regulated in Sirt6 deficient mTECs (Figure 2D). The modulated expressions of these genes would collectively contribute to the poor mTECs proliferation in Sirt6 cKO mice. 5-bromo-2’-deoxyuridine (BrdU) incorporation assay was used to detect newly synthesized DNA in S phase. The portion and the absolute number of proliferating cell (BrdU-positive cells) decreased markedly in mTECs of 2-week-old Sirt6 cKO mice in comparison with their littermate controls (Figures 2E,F). In contrast, Sirt6 deficiency did not affect the proliferation of cTECs as showed by the similar BrdU incorporation (Supplementary Figure 4A).
Sirt6 Deficiency Promotes CD80 Expression on mTECs
We further investigated the effect of Sirt6 deficiency on the differentiation and maturation of mTECs. Compared with immature mTECs, functional mature mTECs highly expressed CD80, MHC II and Aire, which were considered to play an critical role in negative selection (Kadouri et al., 2020). Flow cytometry analysis showed that the expression of MHC II and Aire on mTECs was similar between wild-type and Sirt6 cKO mice, whereas the proportion of CD80+ mTECs increased significantly in Sirt6 cKO mice (Figure 3A). Due to the overall reduction of mTECs, the absolute number of CD80+ mTECs, MHC IIhigh mTECs and Aire+ mTECs all decreased dramatically (Figure 3B). Specifically, Aire expressed on CD80+MHC IIhigh mTECs (mTEChigh) and then regulated the expression of thousands of TRAs (Heino et al., 2000; Gray et al., 2007; Wells et al., 2020). Sirt6 deletion accelerated the maturation of CD80–Aire– mTECs to the differentiation of CD80+Aire–mTEC, but did not affect its further differentiation into CD80+Aire+ mTECs (Figures 3C,D). As shown by similar expression of MHC II and CD40, Sirt6 did not affect the maturation of cTECs (Supplementary Figures 4B,C).
Figure 3. The proportion of CD80+ mTECs specifically increases in Sirt6 cKO mice. (A) Representative flow cytometric profiles and frequencies of MHC II, CD80 and Aire expressed on mTECs of WT and Sirt6 cKO mice. (B) Cell numbers of MHC IIhigh, CD80+ and Aire+ mTECs of Sirt6 cKO mice and littermates. (C) Flow cytometry plots showed the mature stage by detecting the expression of CD80 and Aire. (D) The ratio statistics of different stages in the maturation of mTECs have been shown. N ≥ 4 per group. **p < 0.01 and ***p < 0.001 (Student’s t-test).
TNFR family members CD40, RANK, and LTβR play a critical role in promoting the development of mTECs (Rossi et al., 2007; Zhu et al., 2007; Akiyama et al., 2008, 2012; Hikosaka et al., 2008; Seach et al., 2008; Cosway et al., 2017). There was no significant difference in Cd40 and Rank expression in mTECs between Sirt6 cKO mice and wild-type mice, but the expression of Ltbr in Sirt6 deficient mTECs decreased slightly (Supplementary Figure 5).
Sirt6 Regulates the Development and Maturation of mTECs Through SPIB-OPG Feedback Mechanism
In order to figure out the regulation of mTEC maturation in molecular basis, we performed KEGG analysis for the RNA-seq results. We found that the endocytosis, NF-κB signaling pathway and focal adhesion were upregulated in Sirt6-deficient mTECs (Figure 4A). Among them, NF-κB signaling pathway has been reported to be essential in the development and maturation of mTECs (Burkly et al., 1995; Weih et al., 1995; Kajiura et al., 2004; Akiyama et al., 2005; Kinoshita et al., 2006; Zhang et al., 2006; Zhu et al., 2006; Riemann et al., 2017; Jin and Zhu, 2018). NF-κB target geneset was also enriched in Sirt6 absent mTECs, indicating that NF-κB signaling pathway was indeed activated after Sirt6 deletion (Figures 4B,C).
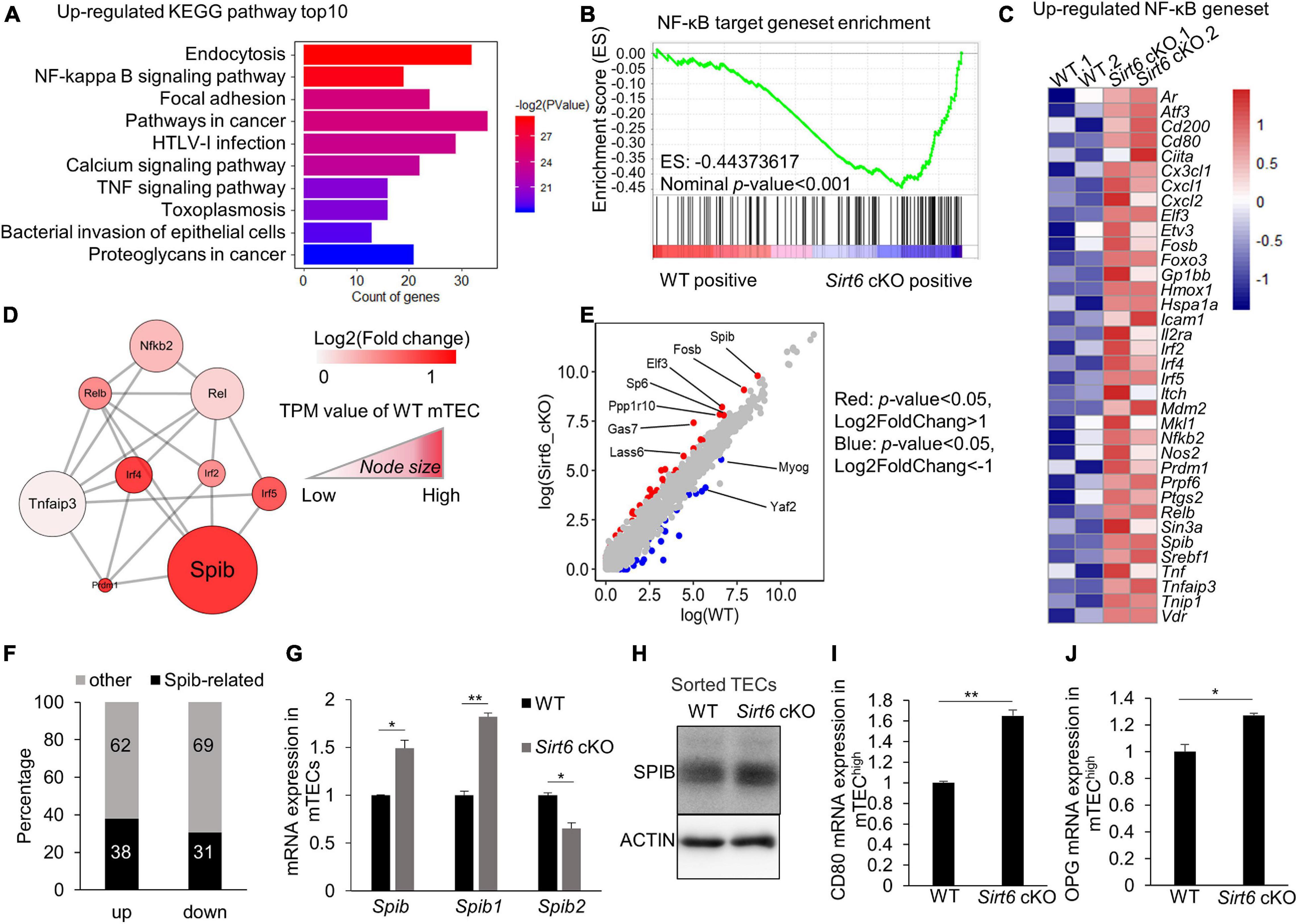
Figure 4. Sirt6 deficiency leads to the activation of NF-κB pathway which in turn upregulates the expression of Spib. (A) Upregulated genes in Sirt6 deficient mTECs were enriched in KEGG pathways, top10 pathways were ordered by p-value. All pathways were selected under the standard of p < 0.05. (B) GSEA analysis reveals that NF-κB target geneset had a more positive expression in Sirt6 deficient mTECs defined by the criterion of p < 0.001. (C) The upregulated genes (p < 0.05) involved in NF-κB target geneset were performed by heatmap. (D) The molecular network between NF-κB and its downstream associated upregulated genes was constructed by STRING. All genes belong to transcription factors, color indicated the change of log2FoldChange and Node size indicated the TPM value of wild-type mTECs. (E) The scatter plot showed the difference of TPM values between wild-type and Sirt6 deficient mTECs. The transcription factors with significant changes were color-coded in the plot, red indicated that genes under the criterion of p < 0.05 and log2foldchange > 1 and blue indicated that genes under the criterion of p < 0.05 and log2foldchange < -1. (F) The bar graph shows the changes of SPIB target genes in Sirt6 deficient mTECs relative to wild-type mTECs. Black indicates the proportion of SPIB target genes, which was obtained from the results of ChIP-seq. (G) Expression of Spib and its two different promoters (Spib1 and Spib2) in mTECs (CD45–EpCAM+UEA-1+Ly51–) sorted from wild-type or Sirt6 cKO mice were measured by quantitative Real-Time PCR analysis. Data were normalized to Hprt mRNA levels. (H) Western blot result for SPIB expression in TECs (CD45–EpCAM+) sorted from wild-type or Sirt6 cKO mice. (I,J) Quantitative Real-Time PCR analysis of Opg (I) and Cd80 (J) mRNA expression in mTEChigh (CD45–EpCAM+UEA-1+CD80+MHC IIhigh) sorted from wild-type and Sirt6 cKO mice. Data were normalized to Hprt mRNA levels. *p < 0.05 and **p < 0.01 (Student’s t-test).
Next, we focused on the NF-κB signaling pathway to understand why Sirt6 specifically restricts the development of CD80+ mTECs. We analyzed the network of NF-κB related transcription factor and found that Spib, as an important transcription factor related to the development and differentiation of mTEC (Akiyama et al., 2014), was notably increased in Sirt6-deficient mTECs (Figure 4D). Among the known transcription factors involved in the development and differentiation of mTEC, Spib showed the greatest difference between WT and Sirt6 deficient mTECs (Supplementary Figure 6A). Furthermore, Spib was the highest expressed compared with other transcription factors that changed (p < 0.05) between Sirt6 deficient mTECs and wild-type control (Figure 4E).
In order to verify the contribution of upregulated Spib in Sirt6 deficient mTECs, we compared the genes directly regulated by Spib (Christie et al., 2015) and the genes changed in Sirt6 deficient mTECs. Our RNA-seq results showed that 920 genes was down-regulated (p < 0.05), and 1,217 genes was up-regulated (p < 0.05) in the Sirt6 deficient mTECs. Among the genes regulated by Spib, 461 genes were up-regulated (38% of 1,217 upregulated genes) and 281 genes were down-regulated (31% of 920 down-regulated genes) (Figure 4F). The genes directly regulated by Spib accounted for 35% of those with statistical difference between wild-type and Sirt6 deficient mTECs.
Previous studies showed that Spib1 rather than Spib2 is highly expressed in mTECs (Akiyama et al., 2014). To verify the expression of Spib in mTECs, we isolated mTECs (CD45–EpCAM+UEA-1+ly51–) from wild-type or Sirt6 cKO mice. Quantitative PCR analysis showed that Spib1, not Spib2, was upregulated after Sirt6 deletion (Figure 4G). In addition, western blot result of sorted TECs (CD45–EpCAM+) confirmed that SPIB was indeed up-regulated after Sirt6 conditional knockout (Figure 4H and Supplementary Figure 6B). It has been reported that Spib promotes osteoprotegerin (Opg) expression and participates in the mTEC developmental stage–specific negative feedback regulation (Akiyama et al., 2014). The development of mTECs was limited in Sirt6 cKO neonates but not in E16.5 (Supplementary Figures 7A,B), which was consistent with the previous report that Spib regulated the development of mTECs in neonates rather than embryos (Akiyama et al., 2014). mTEChi population (CD45–EpCAM+UEA-1+CD80+MHC IIhigh) were sorted from wild-type or Sirt6 cKO mice (Supplementary Figure 6C), and the expression of Cd80 and Opg were evaluated by quantitative PCR analysis. After Sirt6 deletion, the expressions of Cd80 and Opg were up-regulated (Figures 4I,J).
The Development of Thymocytes Is Abnormal in Sirt6 cKO Mice
Positive and negative selection T cells in thymus is mainly orchestrated by TECs (Anderson et al., 2000; Rodewald, 2008). We examined the development of thymocytes in Sirt6 cKO mice to determine whether the impaired mTEC development affected its function in orchestrating thymocytes development. Although the proportion of DN, DP, CD4 SP, and CD8 SP was unaffected by Sirt6 deletion, their cellularity decreased significantly in Sirt6 cKO mice compared with wild-type control (Figures 5A,B). The further maturation of post-selected thymocytes downregulates the expression of CD24, CCR7 and upregulates the expression of CD62L (Zuklys et al., 2016; Liang et al., 2018). We found the frequency of CD24lowCD62LhighCD4+CD8–TCRβ+ and CD24lowCD62LhighCD4–CD8+TCRβ+ reduced in Sirt6 cKO mice (Figure 5C and Supplementary Figure 8A). The percentage of CD24–CCR7loCD4+CD8–TCRβ+CD5+Foxp3– thymocytes and CD24–CCR7loCD4–CD8+TCRβ+CD5+ thymocytes decreased in Sirt6 cKO mice (Figures 5D,E and Supplementary Figure 8B), implying the further maturation of thymocytes was restrained by dysfunctional mTECs in Sirt6 cKO mice. The number of CD4+ T cells or CD8+ T cells and the portion of naïve CD4+ T cells or naïve CD8+ T cells reduced in spleen, indicating that T cells output from thymus decreased after Sirt6 deletion in TECs (Figures 5F,G and Supplementary Figure 8C).
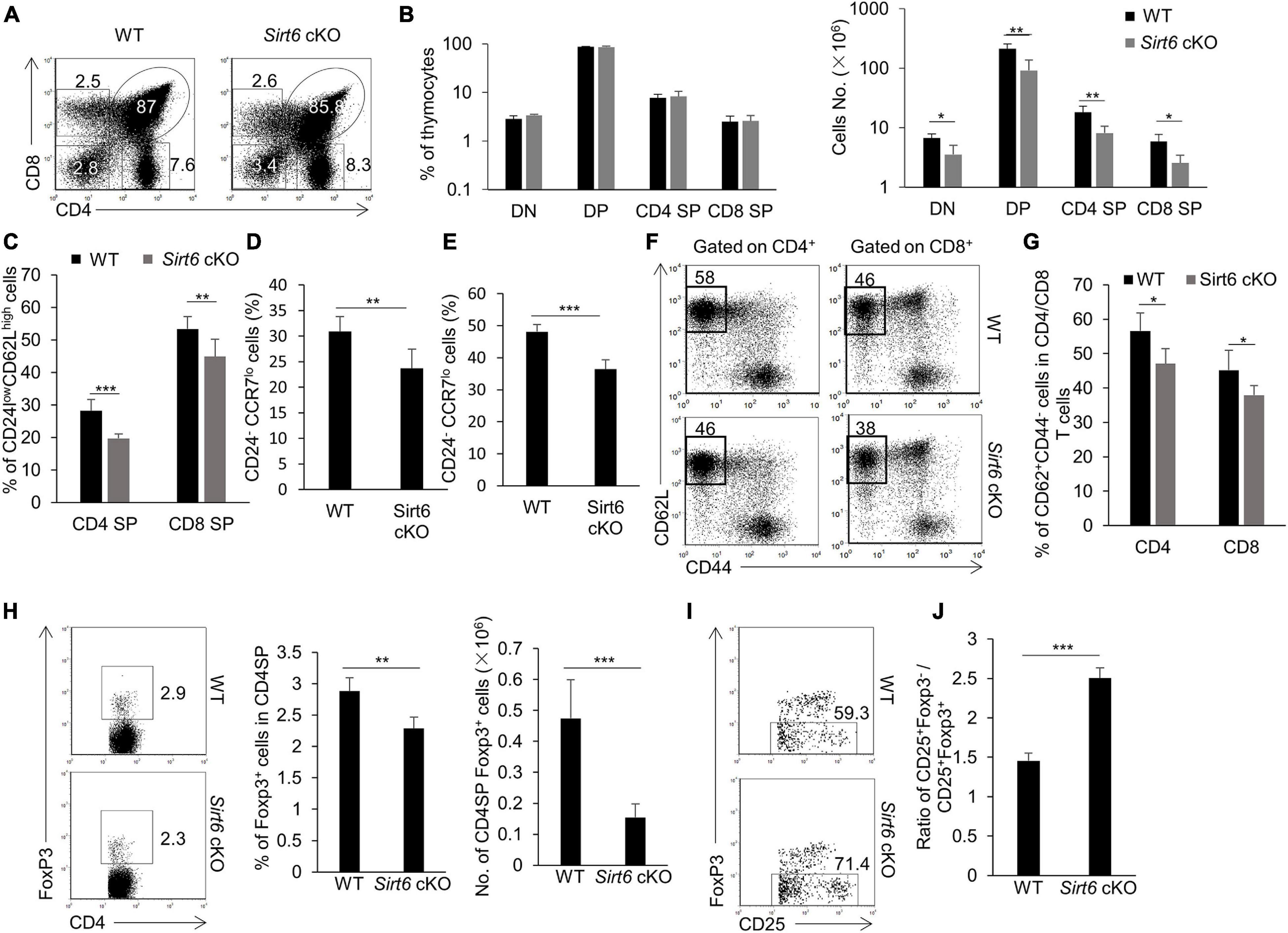
Figure 5. The development of thymocytes is impaired after Sirt6 deletion in TECs. (A) Representative flow cytometry plots of CD4 and CD8 expressed on thymocytes derived from wild-type and Sirt6 cKO mice. (B) Frequencies (left) and absolute cell numbers (right) of DN (CD4–CD8–), DP (CD4+CD8+), CD4SP (CD4+CD8–), and CD8 SP (CD4–CD8+) thymocytes of Sirt6 cKO mice and littermate controls. (C) Frequencies of CD24lowCD62LhighTCRβ+ CD4SP and CD24lowCD62LhighTCRβ+ CD8SP mature thymocytes of wild-type and Sirt6 cKO mice. (D) Frequencies of CD24–CCR7loCD4+CD8–TCR+CD5+Foxp3– thymocytes of wild-type and Sirt6 cKO mice. (E) Frequencies of CD24–CCR7loCD4–CD8+TCR+CD5+ thymocytes of wild-type and Sirt6 cKO mice. (F,G) Representative flow cytometry plots (F) and frequency (G) of naïve (CD62L+CD44–) T cells in CD4+ or CD8+ splenocytes of wild-type and Sirt6 cKO mice. (H) Flow cytometry plots (left) and frequency (middle) and absolute cell numbers (right) of Foxp3+nTreg of Sirt6 cKO mice and littermate controls. (I) Flow cytometry plots show the maturation of nTreg from precursors (CD4+CD8–CD25+Foxp3–) to mature (CD4+CD8–CD25+Foxp3+) in wild-type and Sirt6 cKO mice. (J) Ratio of precursor to mature nTreg between wild-type and Sirt6 cKO mice. N ≥ 4 per group. *p < 0.05, **p < 0.01, and ***p < 0.001 (Student’s t-test).
In addition to the elimination of self-reactive T cells, mTECs play an important role in promoting the diversion of nTreg cells lineage (Cowan et al., 2013; Kadouri et al., 2020). The results showed that the percentage and the cell number of mature nTreg cells (Foxp3+CD4 SP) decreased obviously in Sirt6 cKO mice compared to their littermate controls (Figure 5H). By detecting the maturation process of nTreg cells (Lio and Hsieh, 2008), we found the ratio of immature nTreg cells (Foxp3–CD25+CD4 SP) to mature nTreg cells (Foxp3+CD25+CD4 SP) increased notably in Sirt6 cKO mice, implying that the maturation of nTreg cells was blocked during the differentiation of Foxp3–CD25+CD4 SP to Foxp3+CD25+CD4 SP (Figures 5I,J and Supplementary Figure 8D).
Central Immune Tolerance Is Disrupted in the Sirt6 cKO Mice
Due to the developmental and functional defects of mTECs in Sirt6 cKO mice, we next determined whether such defects affected the establishment of central immune tolerance. We compared the body weight of Sirt6 cKO mice with their age-matched littermate controls and found the body weight of Sirt6 cKO mice was lower obviously than their age-matched littermate controls 20 weeks after birth (Figure 6A). Lymphocytic infiltrates in multiple organs is an important index to judge the occurrence of autoimmune disease (Chuprin et al., 2015). H&E staining results showed that many organs in the 8-month-old Sirt6 cKO mice had more severe lymphocytes infiltration than wild-type control mice, including the salivary gland, kidney, lung, and liver (Figure 6B). We further examined the presence of autoantibodies in serum of 8-month-old Sirt6 cKO. Indeed, there were high levels of antinuclear antibodies in the serum of Sirt6 cKO mice (Figure 6C). What’s more, compared with wild-type control mice, Sirt6 cKO mice had more autoantibodies against many organs of Rag2 KO mice (Shinkai et al., 1992), such as liver, colon, and salivary gland (Figure 6D).
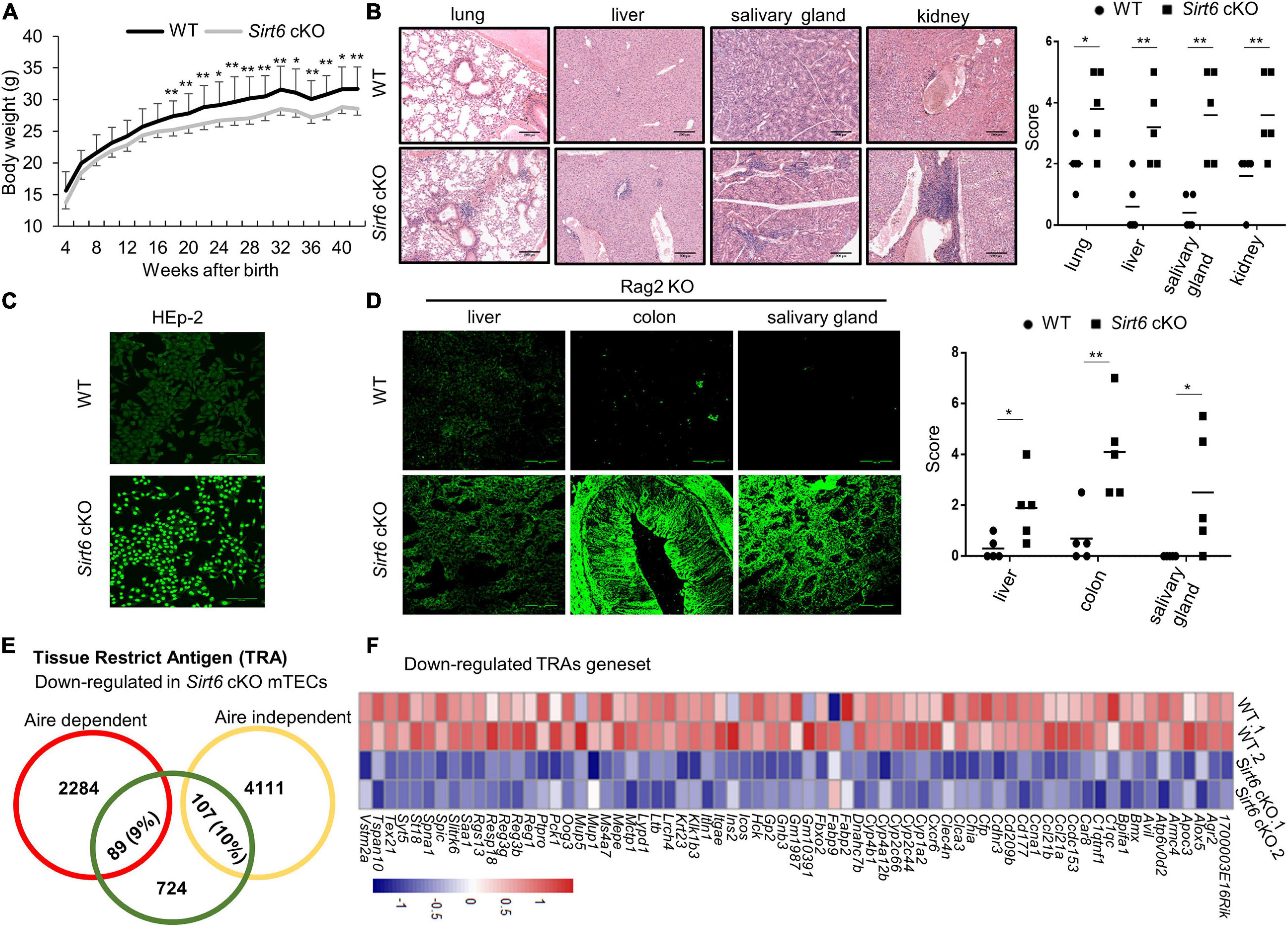
Figure 6. Sirt6 cKO mice spontaneously develop severe autoimmune disorder. (A) Curve of body weight with age of wild-type and Sirt6 cKO mice. (B) Hematoxylin and eosin (H&E) stained paraffin embedded sections of lung, liver, salivary gland, and kidney of 8-month-old wild-type and Sirt6 cKO mice. Infiltration scores and means are indicated. Scale bars: 200 μm. (C) Antinuclear antibodies from the serum of 8-month-old wild-type and Sirt6 cKO mice combined with HEp-2 cell line then detected by anti-mice IgG-AF488 antibody. Scale bars: 100 μm. (D) Tissue sections (liver, colon, and salivary gland) of Rag2 knockout mice were incubated with the serum of 8-month-old wild-type and Sirt6 cKO mice to detect autoantibodies. Scores and means are indicated. Scale bars: 100 μm. (E) The downregulated of Aire-dependent and Aire-independent TRA genes in the comparison of wild-type and Sirt6 deficient mTEC were calculated in venn diagram. Aire-dependent genes were circled in red and Aire-independent genes were circled in yellow. (F) Heatmap of the downregulated TRAs genes (p < 0.05) in Sirt6 deleted mTEC. N ≥ 5 per group. *p < 0.05 and **p < 0.01 (Student’s t-test).
The expression of TRAs on the mature mTECs is essential for eliminating auto-reactive T cells and promoting the development of nTreg cells, which is essential for the establishment of central immune tolerance (Klein et al., 2000). We analyzed the expression of Aire-dependent and Aire-independent TRAs (Sansom et al., 2014; Rodrigues et al., 2017) and found that the expression of both Aire-dependent and Aire-independent TRAs was affected in the mTECs after Sirt6 deletion (Figure 6E). Compared with Sirt1 which only specifically regulates Aire-dependent TRAs (Chuprin et al., 2015), Sirt6 affect a wider range of TRAs expression. Most of the down-regulated TRAs in Sirt6 deficient mTECs were shown in Figure 6F. The decreased TRAs expression indicated that the function of mTECs in establishing central immune tolerance was impaired after Sirt6 deletion.
Discussion
We define Sirt6 as an important regulator in mTEC development. As Sirt6 ablation inhibits the proliferation of mTECs, the percentage and absolute number of mTECs decrease significantly. These results suggest that, in contrast to Sirt1, Sirt6 is involved in the regulation of mTEC development. Previous reports have shown that Sirt6 deficiency has linked to many diseases, such as cancer, neurodegeneration and aging (Khan et al., 2018). Because Sirt6 knockout mice died several weeks after birth (Mostoslavsky et al., 2006), the specific function of Sirt6 in different organs are unclear. Age-related thymic involution occurs at the early stage of organisms (Taub and Longo, 2005; Chinn et al., 2012) and TEC-intrinsic molecules are considered to be an important and sufficient to initiate thymus involution (Lynch et al., 2009; Rezzani et al., 2014; Cheng and Anderson, 2018; Wang et al., 2019; Baran-Gale et al., 2020). Although a recent study showed that Sirt6 deficiency in cynomolgus monkey causes developmental retardation, the main effect of Sirt6 deficiency in rodents was showed to be associated with accelerated aging (Mostoslavsky et al., 2006; Kawahara et al., 2009; Kanfi et al., 2012; Zhang et al., 2018). Compared with thymus weight loss and naïve T cell levels in spleens of wild-type and Sirt6 cKO mice during 4 weeks to 8 months after birth, Sirt6 cKO mice do not show any detectable increased thymus weight loss and the reduced naïve T cells in spleen. These results suggest that Sirt6 ablation in TECs do not accelerate thymic involution.
On the other hand, we have found that 8-month-old Sirt6 cKO mice, rather than age-matched wild-type mice, have substantial lymphocytic infiltrates in multiple organs and obvious autoantibodies in the serum, indicating that Sirt6 deficient in mTECs leads to autoimmune disorders. These pathological changes can be supported by the decrease of Aire-dependent and independent TRA expressions and the blockage of nTreg cell maturation in the thymus of Sirt6 cKO mice. The impaired TRA expression and the poor nTreg cell maturation in Sirt6 cKO mice may lead to the failure of establishing central immune tolerance, which subsequently contribute to the development of autoimmune disease in Sirt6 cKO mice. On the other hand, the frequency of Helios expressing thymocytes during T cell maturation (Daley et al., 2013; Zuklys et al., 2016) in Sirt6 cKO mice did not change compared with those in WT mice, indicating that the negative selection of these cells in Sirt6 cKO mice seems not remarkably affected (Data not shown). However, the development of CD24+CCR7+ to CD24–CCR7lo CD4SP and CD8 SP cell in Sirt6 cKO mice was blocked, and the frequency of TCRβhighCD24lowCD62Lhigh CD4 SP and TCRβhighCD24lowCD62Lhigh CD8 SP in Sirt6 cKO mice were decreased, implying the further maturation of T cells may be restrained by dysfunctional mTECs in Sirt6 cKO mice. It is reported that chemokines CCL19 and CCL21, as functional ligands of CCR7, play an important role in the accumulation of positively selected thymocytes in thymic medulla, which is crucial for the elimination of auto-reactive T cells (Ueno et al., 2002, 2004; Anderson and Takahama, 2012; Kozai et al., 2017). RNA-seq data showed that the expression of Ccl19 and Ccl21a in Sirt6 deficient mTECs decreased significantly, which indicated that the ability of mTEC to eliminate self-reactive T cells might be impaired. Previous reports show that different GFP levels in Rag2p GFP transgenic mice can be used to evaluate thymocyte medullary dwell time, “thymic age” of thymocytes and identify recent thymic emigrants directly (Yu et al., 1999; Boursalian et al., 2004; McCaughtry et al., 2007; Hauri-Hohl et al., 2014; Cowan et al., 2016; White et al., 2017). The medullary residency time is closely related to the establishment of central immune tolerance (McCaughtry et al., 2007). By using the OT2/RIP-OVA system, self-reactivity and TCR affinity for self-antigen are proved to be related to the negative selection and the development of nTreg cells (Lee et al., 2012; Wyss et al., 2016; Santamaria et al., 2021). Thus, we should employ these mouse models to directly and systemically address whether thymic negative selection and Treg development are impaired by Sirt6 deficiency in TECs in the future.
The results showed that the proportion and the cell number of Ki67+ mTECs and BrdU+ mTECs decreased after Sirt6 deletion. Together with the analysis of RNA-seq assays, our data indicate that the reduced proliferation ability of Sirt6-deficient mTECs is likely due to the decrease of DNA replication. The differentiation from CD80– mTECs to CD80+ mTECs has been accelerated specifically in Sirt6 cKO mice, implying the differentiation of mTECs is affected in the absence of Sirt6. SIRT6 is originally thought to be a mono-ADP-ribosyltransferase, which has later been found to be involved in DNA repair (Liszt et al., 2005; Mao et al., 2011). SIRT6, on the other hand, is widely known as a deacetylase (Michishita et al., 2008; Kawahara et al., 2009; Michishita et al., 2009; Yang et al., 2009; Zhong et al., 2010; Tasselli et al., 2016). In sorted mTECs, acetylation do not increase after Sirt6 ablation, indicating that SIRT6 may not act as a key deacetylase in the development of mTEC. This observation is supported by the studies showing that deacetylase activity of SIRT6 is 1,000 times slower than other sirtuin family members (Pan et al., 2011). Our RNA-seq analysis data show that NF-κB signaling pathway is remarkably up-regulated in the Sirt6-deficient mTECs, which is nicely in line with the observation showing that Sirt6 deletion up-regulated the expression NF-κB target genes (Kawahara et al., 2009). It is well-known that the development and differentiation of mTEC depends on NF-κB signaling pathway (Burkly et al., 1995; Weih et al., 1995; Kajiura et al., 2004; Akiyama et al., 2005; Kinoshita et al., 2006; Zhang et al., 2006; Zhu et al., 2006; Riemann et al., 2017; Jin and Zhu, 2018). The enhanced NF-κB signal pathway up-regulated the expression of Spib in mTECs after Sirt6 deletion as indicated by RNA-seq, real-time PCR and western blot assays, as previously reported (Akiyama et al., 2014). It is nicely demonstrated by Akiyama et al. that Spib remarkably limits the development and maturation of mTECs but promotes CD80 expression in mature mTECs (Akiyama et al., 2014). Importantly, RANKL stimulation consecutively induces Spib expression in TECs and Spib facilitates expression of OPG protein, which competitively inhibits RANKL–RANK interactions as a decoy receptor of RANKL, in mTECs by maintenance of the hypomethylated states in Opg (Akiyama et al., 2014; Tsukasaki et al., 2020). Thus, SPIB–mediated negative feedback regulation of RANKL signaling limits mTEC development in neonates but not in embryos by forming RANKL-NF-κB-SPIB-OPG regulating loop in mTECs (Akiyama et al., 2014). Our results suggest that Sirt6 negatively regulates the expression of NF-κB-SPIB-OPG pathway, which is crucial for the postnatal development and maturation of mTECs. However, it should be noted that mTECs are heterogenous and some atypical types of terminally differentiated mTECs exist in the thymus (Bornstein et al., 2018; Miller et al., 2018; Park et al., 2020). A newly defined IL25+ thymic tuft cells are regulated by Pou2f3 and affect the development of thymus-resident type-2 innate lymphoid cells and thymic invariant natural killer T cells (Bornstein et al., 2018; Miller et al., 2018; Lucas et al., 2020). The latest research shows that there are two distinct groups in human TECs, TEC(myo)s and TEC(neuro)s (Park et al., 2020). Considering the diversity and heterogeneity of TECs, we may speculate another possibility that the increased Spib expression in Sirt6 cKO mTECs may be caused by the enhanced Spib expression in some unidentified mTEC subpopulation rather than that the expression level of Spib was simply increased in whole mTECs. Unfortunately, our present results could not exclude this possibility and more detailed studies on Sirt6-mediated regulation in the development of mTEC subpopulations should be performed in the future.
In summary, our study reveals that Sirt6 is involved in the regulation of the development, maturation and function of mTECs and is critical for establishment of central immune tolerance. These results support the non-redundant role of different epigenetic molecules in maintaining the functional integrity of mTEC and preventing autoimmune disorders.
Materials and Methods
Mice
Sirt6loxp/loxp mice (Kim et al., 2010; Wang et al., 2016) were crossed with Foxn1-Cre mice (Soza-Ried et al., 2008; Liu et al., 2013) to generate Foxn1-Cre Sirt6loxp/loxp mice. Littermates or age-matched wild-type mice (Foxn1-Cre negative) were used as controls. We obtained Sirt6loxp/loxp mice from Dr. Zhenyu Ju of Hangzhou Normal University, Hangzhou, China, and obtained Foxn1-Cre mice from Dr. Yu Zhang of Peking University Health Science Center, Beijing, China. Rag2 KO mice (Shinkai et al., 1992) was purchased from HFK Bioscience, Beijing, China. All mice were maintained under specific pathogen–free conditions and treated in accordance with Animal Experiments Guidelines of the animal Ethics Committee of Institute of Zoology, Beijing, China.
Thymic Stromal Cell Isolation
Thymic stromal cells was isolated from whole thymus by using the previous thymic stromal cell separation method (Sun et al., 2013; Liang et al., 2018). In brief, fresh thymus tissue was cut into pieces and suspended the thymus fragments in DMEM (Hyclone Laboratories, SH30022.01B) medium with 2% fetal bovine serum (FBS; Gibco, 16000-044). Most thymocytes in the supernatant were removed, and the remaining thymus fragments were incubated at 37°C for 15 min in 2ml solution of 1 mg/ml collagenase/dispase (Sigma-Aldrich, 11097113001) with 20 U/ml DNAse I (Sigma-Aldrich, D5025). Repeat the above digestion three times, gently shaking to make digestion more thorough. The digested cell suspension was terminated with PBS containing 1% FBS and 5 mM EDTA until all fragments disappeared. After centrifugation, cells were suspended in DMEM (containing 2% FBS). Cell suspension was gently blown and filtered with a 200 mesh filter to remove clumps and form a single cell suspension.
Flow Cytometric Analysis and Antibodies
Fc-receptor was blocked by 2.4G2 before staining with indicated antibodies. All type of cell were stained with the appropriate fluorophore-labeled antibodies at 4°C for 30 min. For intracellular staining, the fixation buffer (eBioscience, 00-5123-43 and 00-5223-56) and permeabilization buffer (eBioscience, 00-8333-56) was used according to the supplier’s protocol. The flow cytometry was performed with Gallios Flow Cytometer (BeckMan Coulter, United States) or BD LSRFortessa X-20 Flow Cytometer (BD Biosciences, United States).
The Fluorescein labeled Ulex Europaeus Agglutinin I (UEA I) (Vector Laboratories, FL-1061) was obtained from Vector Laboratories. The CD45-PerCP/Cy5.5 (Biolegend, clone 30-F11, 103132), CD326-PE/Cy7 (Biolegend, clone G8.8, 188216), CD326-FITC (Biolegend, clone G8.8, 118207), Ly51-AF647 (Biolegend, clone 6C3, 108312), CD40-PE (Biolegend, clone 3/23, 124610), I-A/I-E-Brilliant Violet 421 (Biolegend, clone M5/114.15.2, 107632), CD80-PE (Biolegend, clone 16-10A1, 104708), CD80-BV650 (Biolegend, clone 16-10A1, 104732), CD4-FITC (Biolegend, clone GK1.5, 100405), CD4-PE (Biolegend, clone GK1.5, 100408), CD4-PE/Cy5 (Biolegend, clone GK1.5, 100410), CD4-APC (Biolegend, clone GK1.5, 100412), CD4-APC/Cy7 (Biolegend, clone GK1.5, 100412), CD8a-PE (Biolegend, clone 53-6.7, 100725), CD8a-PE/Cy5 (Biolegend, clone 53-6.7, 100710), CD8a-Brilliant Violet 421 (Biolegend, clone 53-6.7, 100738), TCR-β-PE-Cy7 (Biolegend, clone H57-597, 109222), CD24-FITC (Biolegend, clone M1/69, 101805), CD24-PE/Cy5 (Biolegend, clone M1/69, 101812), CD44-FITC (Biolegend, clone IM7, 103006), CCR7-PE (Biolegend, clone 4B12, 120106), CD5-APC (Biolegend, clone 53-7.3, 100626) were purchased from Biolegend. The Aire (eBioscience, clone 5H12, 50-5934-80), Fixable Viability Dye eFluorTM 506 (eBioscience, 65-0866-18), CD62L-PE (eBioscience, clone MEL-14, 12-0621-82), CD25-PE-Cy5 (eBioscience, clone PC61.5, 12-0251-82), Foxp3-FITC (eBioscience, clone FJK-16s, 11-5773-82), were purchased from eBioscience. CD45-BUV395 (BD Biosciences, clone 30-F11, 564279), Ly51-BV786 (BD Biosciences, clone BP-1, 740882), PE Mice Anti-Ki-67 Set (BD Biosciences, 556027), APC BrdU Flow Kit (BD Biosciences, 552598), and PE Active Caspase-3 Apoptosis Kit (BD Biosciences, 550914) were purchased from BD Biosciences.
BrdU Incorporation Assays
For Brdu incorporation, 2-week-old Sirt6 cKO and littermate control mice were intraperitoneally injected with BrdU (BD Biosciences), 1 mg per mice. Twenty-four hours after injection, thymic stromal cells were isolated according to the above-mentioned method before flow cytometric analysis. After surface antigens staining, according to the supplier’s protocol (BD PharmingenTM BrdU Flow Kits Instruction Manual), BrdU was detected by using APC-BrdU Flow Kit (BD Biosciences, 552598) and was analyzed by flow cytometry.
Immunofluorescence Staining
Tissues were embedded in optimum cutting temperature compound (Sakura, 4583) and frozen in liquid nitrogen. Sections (6 μm in thickness) and/or cells were fixed for 20 min with 4% polyoxymethylene (Solarbio, P1110) and blocked in PSB containing 1% BSA. Then, sections and/or cells were incubated with primary and secondary antibodies for 1 h at room temperature. Samples were stained with DAPI (1:1,000) after secondary staining. The following antibodies were used for staining: rabbit anti-KRT5 (Covance, PRB-160P; clone AF 138) diluted by 1:400 and rat anti-KRT8 (DSHB, AB 531826; Troma-I) diluted by 1:200. Sera of 8-month-old wild-type and Sirt6 cKO mice (diluted by 1:30) were used as primary antibodies for the detection of the autoantibodies. The secondary antibodies were used for staining: Alexa Fluor 594-conjugated donkey anti-rabbit IgG (H + L) (Jackson ImmunoResearch Laboratories, 711-586-152) diluted by 1:400, Alexa Fluor 488-conjugated donkey anti-rat IgG (H + L) (Jackson ImmunoResearch Laboratories, 712-546-150) diluted by 1:400, and Alexa Fluor 488-conjugated donkey anti-mice IgG (H + L) antibodies (Jackson ImmunoResearch Laboratories, 715-546-150) diluted by 1:300. All antibodies were diluted in 0.5% BSA in PBS. Laser scanning confocal microscope (Zeiss LSM710, Oberkochen, Germany) were used to acquire images.
RNA-Seq Data Processing
As mentioned above, thymic stromal cells were isolated from 4-week-old wild-type and Sirt6 cKO mice, with two independent parallel samples in each group. The next RNA sequencing step of sorted mTECs (CD45–EPCAM+UEA-1+Ly51–) was performed according to the previous method (Liang et al., 2020).
Low quality reads (Q < 20) were assessment by FastQC and the adaptor sequence was filtered by Trimgalore. Processed reads were aligned to mice genome (mm10) via HISAT2 (Pertea et al., 2016). StringTie was applied to assemble and quantify the transcripts in each sample to obtain the number of exon, transcription initiation/stop site, count and TPM (Transcripts Per Kilobase of exon model per Million mapped reads) values. The identification of differential expression genes (DEGs) was performed by using the DEseq2 (Love et al., 2014) R-packages with count value. The threshold is under the condition of adjusted p < 0.05 and | Foldchange| > 2. The normalized gene expression only retained the gene with TPM > 0.1 in all samples.
KEGG Pathway Enrichment Analysis
The KEGG pathway analysis of the differentially expressed genes were performed by KOBAS 3.0 on the web1 (Ai and Kong, 2018). In addition, GSEA was carried out by searching KEGG Database (Subramanian et al., 2007). All analyses were selected with p < 0.05 as the cut-off criterion.
Network Creation and Customization
Functional protein association network was created by STRING (Szklarczyk et al., 2015) (Available online: https://string-db.org/) which was constructed by the transcriptional factors obtain by website (Available online: http://bioinfo.life.hust.edu.cn/AnimalTFDB/). Networks were all visualized by Cytoscape (Demchak et al., 2014).
Culture of TECs
Thymi of wild-type and Sirt6 cKO neonatal mice were cut into small pieces and suspended with DMEM containing 2% FBS. After the small pieces settled at the bottom of the tube, the cell suspension was discarded. The remaining small pieces were suspended in TyEpiCM (ScienCell Research Laboratories, Catalog #3911) and incubator at 37°C with 5% CO2 for 7 days, exchanging the medium every other day.
Western Blot Assay
Cultured wild-type and Sirt6 cKO TECs were used to detect SIRT6 expression. TECs (CD45–EPCAM+) isolated from wild-type and Sirt6 cKO mice were used to detect the expression of SPIB. After being washed with cold PBS, TEC were lysed in RIPA buffer (140 mM NaCl, 10 mM Tris-Cl (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate and 0.1% SDS) complemented with a proteinase inhibitor cocktail (Sigma-Aldrich, P8340). Protein concentration was detected with Bradford assay. Proteins were analyzed by 10% SDS-PAGE and transferred onto PVDF membranes (Merck Millipore, IPFL00010). Each PVDF membrane was blocked with 5% non-fat dried milk (OXOID, LP0031) for 60–90 min at room temperature and incubated with each primary antibodies overnight on the shaking table at 4°C. After cleaning PVDF membrane with TBST solution for four times, the corresponding secondary antibodies were added for 45–60 min at room temperature. Protein bands were detected by chemiluminescence (Merck Millipore, WBKLS0500). ACTIN is used as internal reference for protein standardization. The primary antibodies used for western blot are as follows: Anti-SIRT6 (Cell Signaling Technology, 12486) diluted by 1:1,000; anti-SPIB (Cell Signaling Technology, 14337S) diluted by 1:1,000; anti-ACTIN (Sigma-Aldrich, A5441) diluted by 1:20,000.
Quantitative RT-PCR
Thymic stromal cell was isolated from 4-week-old wild-type or Sirt6 cKO mice and then sorted with MoFlo XDP cell sorter (Beckman Coulter, Brea, CA, United States), characterized as TEC (CD45–EPCAM+), mTEC (CD45–EPCAM+UEA-1+Ly51–), cTEC (CD45–EPCAM+UEA-1–Ly51+), mTEClow (CD45–EPCAM+UEA-1+CD80–MHCIIlow) and mTEChigh (CD45–EPCAM+UEA-1+CD80+MHCIIhigh). Total RNA was isolated with MicroElute Total RNA Kits (Omega Bio-tek, R6831) and reverse transcription was performed with Super-Script III Reverse Transcription (Invitrogen, 18080–093) according to manufacturer’s instructions.
Real-time PCR was performed with multiple kits (SYBR Premix Ex Taq, Takara Bio, DRR041A) on CFX96 (Bio-Rad Laboratories, Hercules, CA, United States). All primers used in this article were listed in Table 1.
Statistical Analysis
All data are presented as the means ± SD. The statistical significance of differences between two experimental groups was tested by Student’s t-test. A p-value < 0.05 was considered statistically significant.
Data Availability Statement
The data presented in the study are deposited in the NCBI Gene Expression Omnibus public repository, accession number GSE166840.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee of Institute of Zoology, Beijing, China.
Author Contributions
QZ, ZL, and TL designed and carried out the experiments, analyzed data, and wrote the manuscript. JZ performed the bioinformatics analyses and wrote the manuscript. XD, HS, and YC performed the experiments. ZZ performed the fluorescence staining. LT genotyped the genetically modified mice. YZ designed the experiments, analyzed the data, wrote the manuscript, and provided overall supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2017YFA0105002 and 2017YFA0104402, YZ), the National Natural Science Foundation for General and Key Programs (31930041, YZ and 31800754, ZL), and the Knowledge Innovation Program of Chinese Academy of Sciences (XDA16030301, YZ).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Zhenyu Ju (Hangzhou Normal University) for kindly providing the Sirt6loxp/loxp mice and Yu Zhang (Peking University Health Science Center) for kindly providing Foxn1-Cre mice. We thank Qing Meng and Xia Yang for the technical assistance in cell sorting. We thank Ling Li for her excellent laboratory management.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.655552/full#supplementary-material
Supplementary Figure 1 | SIRT6 was successfully knockout in TECs. (A) Identification method of WT (Foxn1.cre–) and Sirt6 cKO (Foxn1.cre+, Sirt6loxp/loxp) mice. (B) Western blot result for Foxn1.Cre-mediated recombination efficacy of Sirt6loxp/loxp between wild-type TECs and Sirt6 cKO TECs cultured in vitro. (C) Quantitative Real-Time PCR analysis of Sirt6 mRNA expression in mTEC (CD45–EpCAM+UEA-1+Ly51–) and cTEC (CD45–EpCAM+UEA-1–Ly51+) sorted from wild-type and Sirt6 cKO mice. Data were normalized to Hprt mRNA levels.
Supplementary Figure 2 | Aging-like phenotype did not observed in Sirt6 cKO mice. (A) Representative figure of thymi isolated from 8-month-old WT and Sirt6 cKO mice. (B) Thymic weight normalized to body weight (left) and the thymocytes cellularity (right) in 4-week-old WT and Sirt6 cKO mice and 8-month-old WT and Sirt6 cKO mice. WT (4-week-old: n = 4; 8-month old: n = 5) and Sirt6 cKO mice (4-week-old: n = 4; 8-month-old: n = 9). (C,D) Hematoxylin and eosin (H&E) staining (C) and Masson’s Trichrome staining (D) stained sections of thymi from 8-month-old WT and Sirt6 cKO mice. Scale bars: 500 μm. (E,F) Representative flow cytometric profiles (E) and frequency (F) of naïve T cells (CD62L+CD44–) in CD4+ (left) or CD8+ (right) splenocytes from 8-month-old or 4-week-old WT and Sirt6 cKO mice (4-week-old: n = 14 per group; 8-month-old: n = 10 per group).
Supplementary Figure 3 | Decreased mTECs in Sirt6 cKO mice is not caused by apoptosis. Flow cytometry plots and frequency for the staining of Active Caspase3 in WT and Sirt6 deficient mTECs. N = 4 per group. ∗∗p < 0.01 (Student’s t-test).
Supplementary Figure 4 | The maturation and proliferation of cTECs were not influenced in Sirt6 cKO mice. (A) Flow cytometry plots and frequency for BrdU+ cTECs from 2-week-old Sirt6 cKO mice and littermates. (B) Representative flow cytometry plots and frequencies of CD40 (upper) and MHC II (lower) expressed on cTECs. (C) Cell numbers of CD40+ cTECs (upper) and MHC IIhigh cTECs (lower) of 4-week-old Sirt6 cKO mice and littermates. N ≥ 4 per group. ∗p < 0.05 (Student’s t-test).
Supplementary Figure 5 | The expression of TNFRF in mTECs. Quantitative Real-Time PCR analysis of Cd40, Rank, and Ltbr mRNA expression in mTECs (CD45–EpCAM+UEA-1+Ly51–) sorted from wild-type and Sirt6 cKO mice. Data were normalized to Hprt mRNA levels. ∗p < 0.05 (Student’s t-test).
Supplementary Figure 6 | SPIB expression was up-regulated in Sirt6 cKO TECs. (A) The scatter plot showed the difference of transcription factors between wild-type and Sirt6 deficient mTECs. Genes with significant changes were color-coded in the plot. Red indicated that genes up-regulated in Sirt6 cKO mTECs, while blue indicated that genes down-regulated in Sirt6 cKO mTECs. The color of genes was under the criterion of p < 0.05. (B) The original picture of western blot result for Figure 4H. (C) Sorting strategies for Figures 4I,J.
Supplementary Figure 7 | Sirt6 deficient mTECs began to decrease in neonates, but not in E16.5. (A) Frequencies and absolute numbers of TECs (CD45–EpCAM+), cTECs (CD45–EpCAM+UEA-1–Ly51+) and mTECs (CD45–EpCAM+UEA-1+Ly51–) obtained from E16.5 Sirt6 cKO mice and littermates. (B) Frequencies and absolute numbers of TECs (CD45–EpCAM+), cTECs (CD45–EpCAM+UEA-1–Ly51+), mTECs (CD45–EpCAM+UEA-1+Ly51–), CD80+ mTECs (CD45–EpCAM+UEA-1+Ly51–CD80+) obtained from WT and Sirt6 cKO neonatals. N ≥ 4 per group. ∗p < 0.05 and ∗∗p < 0.01 (Student’s t-test).
Supplementary Figure 8 | The development of thymocytes in WT and Sirt6 cKO mice. (A) Gating strategies for Figure 5C. (B) FACS plots for Figures 5D,E. (C) Absolute numbers of CD4+ splenocytes and CD8+ splenocytes of wild-type and Sirt6 cKO mice. (D) Gating strategies for Figure 5I.
Abbreviations
BrdU, 5-bromo-2′-deoxyuridine; cTEC, cortical thymic epithelial cells; DN, double negative; DP, double positive; ETPs, early thymic precursors; GSEA, Gene Set Enrichment Analysis; IKK, I κ B-kinase; LT β R, lymphotoxin β receptor; mTECs, medullary thymic epithelial cells; MHC II, major histocompatibility complex class II; NIK, NF- κ B inducing kinase; nTreg, thymic regulatory T cell; OPG, osteoprotegerin; RANK, receptor activator of NF- κ B; SP, single positive; TECs, thymic epithelial cells; TRAs, tissue restricted antigens; TRAF6, TNF receptor-activated factor 6; TNFRF, TNF receptor family; UEA1, Ulex Europaeus Lectin 1.
Footnotes
References
Ai, C., and Kong, L. (2018). CGPS: a machine learning-based approach integrating multiple gene set analysis tools for better prioritization of biologically relevant pathways. J. Genet. Genomics 45, 489–504. doi: 10.1016/j.jgg.2018.08.002
Akiyama, N., Shinzawa, M., Miyauchi, M., Yanai, H., Tateishi, R., Shimo, Y., et al. (2014). Limitation of immune tolerance-inducing thymic epithelial cell development by Spi-B-mediated negative feedback regulation. J. Exp. Med. 211, 2425–2438. doi: 10.1084/jem.20141207
Akiyama, T., Maeda, S., Yamane, S., Ogino, K., Kasai, M., Kajiura, F., et al. (2005). Dependence of self-tolerance on TRAF6-directed development of thymic stroma. Science 308, 248–251. doi: 10.1126/science.1105677
Akiyama, T., Shimo, Y., Yanai, H., Qin, J., Ohshima, D., Maruyama, Y., et al. (2008). The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity 29, 423–437. doi: 10.1016/j.immuni.2008.06.015
Akiyama, T., Shinzawa, M., and Akiyama, N. (2012). TNF receptor family signaling in the development and functions of medullary thymic epithelial cells. Front. Immunol. 3:278. doi: 10.3389/fimmu.2012.00278
Alawam, A. S., Anderson, G., and Lucas, B. (2020). Generation and regeneration of thymic epithelial cells. Front. Immunol. 11:858. doi: 10.3389/fimmu.2020.00858
Anderson, G., Lane, P. J., and Jenkinson, E. J. (2007). Generating intrathymic microenvironments to establish T-cell tolerance. Nat. Rev. Immunol. 7, 954–963. doi: 10.1038/nri2187
Anderson, G., and Takahama, Y. (2012). Thymic epithelial cells: working class heroes for T cell development and repertoire selection. Trends Immunol. 33, 256–263. doi: 10.1016/j.it.2012.03.005
Anderson, M., Anderson, S. K., and Farr, A. G. (2000). Thymic vasculature: organizer of the medullary epithelial compartment? Int. Immunol. 12, 1105–1110. doi: 10.1093/intimm/12.7.1105
Anderson, M. S., and Su, M. A. (2016). AIRE expands: new roles in immune tolerance and beyond. Nat. Rev. Immunol. 16, 247–258. doi: 10.1038/nri.2016.9
Aschenbrenner, K., D’Cruz, L. M., Vollmann, E. H., Hinterberger, M., Emmerich, J., Swee, L. K., et al. (2007). Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat. Immunol. 8, 351–358. doi: 10.1038/ni1444
Aw, D., and Palmer, D. B. (2012). It’s not all equal: a multiphasic theory of thymic involution. Biogerontology 13, 77–81. doi: 10.1007/s10522-011-9349-0
Baran-Gale, J., Morgan, M. D., Maio, S., Dhalla, F., Calvo-Asensio, I., Deadman, M. E., et al. (2020). Ageing compromises mouse thymus function and remodels epithelial cell differentiation. eLife 9:e56221.
Bornstein, C., Nevo, S., Giladi, A., Kadouri, N., Pouzolles, M., Gerbe, F., et al. (2018). Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature 559, 622–626. doi: 10.1038/s41586-018-0346-1
Boursalian, T. E., Golob, J., Soper, D. M., Cooper, C. J., and Fink, P. J. (2004). Continued maturation of thymic emigrants in the periphery. Nat. Immunol. 5, 418–425. doi: 10.1038/ni1049
Burkly, L., Hession, C., Ogata, L., Reilly, C., Marconi, L. A., Olson, D., et al. (1995). Expression of relB is required for the development of thymic medulla and dendritic cells. Nature 373, 531–536. doi: 10.1038/373531a0
Chang, A. R., Ferrer, C. M., and Mostoslavsky, R. (2020). SIRT6, a mammalian deacylase with multitasking abilities. Physiol. Rev. 100, 145–169. doi: 10.1152/physrev.00030.2018
Chen, R., and Wold, M. S. (2014). Replication protein A: single-stranded DNA’s first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays 36, 1156–1161. doi: 10.1002/bies.201400107
Cheng, M., and Anderson, M. S. (2018). Thymic tolerance as a key brake on autoimmunity. Nat. Immunol. 19, 659–664. doi: 10.1038/s41590-018-0128-9
Chinn, I. K., Blackburn, C. C., Manley, N. R., and Sempowski, G. D. (2012). Changes in primary lymphoid organs with aging. Semin. Immunol. 24, 309–320. doi: 10.1016/j.smim.2012.04.005
Christie, D. A., Xu, L. S., Turkistany, S. A., Solomon, L. A., Li, S. K., Yim, E., et al. (2015). PU.1 opposes IL-7-dependent proliferation of developing B cells with involvement of the direct target gene bruton tyrosine kinase. J. Immunol. 194, 595–605. doi: 10.4049/jimmunol.1401569
Chuprin, A., Avin, A., Goldfarb, Y., Herzig, Y., Levi, B., Jacob, A., et al. (2015). The deacetylase Sirt1 is an essential regulator of Aire-mediated induction of central immunological tolerance. Nat. Immunol. 16, 737–745. doi: 10.1038/ni.3194
Clijsters, L., Hoencamp, C., Calis, J. J. A., Marzio, A., Handgraaf, S. M., Cuitino, M. C., et al. (2019). Cyclin F controls cell-cycle transcriptional outputs by directing the degradation of the three activator E2Fs. Mol. Cell 74, 1264.e7–1277.e7.
Cosway, E. J., Lucas, B., James, K. D., Parnell, S. M., Carvalho-Gaspar, M., White, A. J., et al. (2017). Redefining thymus medulla specialization for central tolerance. J. Exp. Med. 214, 3183–3195. doi: 10.1084/jem.20171000
Cowan, J. E., McCarthy, N. I., and Anderson, G. (2016). CCR7 controls thymus recirculation, but not production and emigration, of Foxp3(+) T Cells. Cell Rep. 14, 1041–1048. doi: 10.1016/j.celrep.2016.01.003
Cowan, J. E., Parnell, S. M., Nakamura, K., Caamano, J. H., Lane, P. J., Jenkinson, E. J., et al. (2013). The thymic medulla is required for Foxp3+ regulatory but not conventional CD4+ thymocyte development. J. Exp. Med. 210, 675–681. doi: 10.1084/jem.20122070
Daley, S. R., Hu, D. Y., and Goodnow, C. C. (2013). Helios marks strongly autoreactive CD4+ T cells in two major waves of thymic deletion distinguished by induction of PD-1 or NF-kappaB. J. Exp. Med. 210, 269–285. doi: 10.1084/jem.20121458
Demchak, B., Hull, T., Reich, M., Liefeld, T., Smoot, M., Ideker, T., et al. (2014). Cytoscape: the network visualization tool for GenomeSpace workflows. F1000Res 3:151. doi: 10.12688/f1000research.4492.2
Gray, D., Abramson, J., Benoist, C., and Mathis, D. (2007). Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J. Exp. Med. 204, 2521–2528. doi: 10.1084/jem.20070795
Hauri-Hohl, M., Zuklys, S., Hollander, G. A., and Ziegler, S. F. (2014). A regulatory role for TGF-beta signaling in the establishment and function of the thymic medulla. Nat. Immunol. 15, 554–561. doi: 10.1038/ni.2869
Heino, M., Peterson, P., Sillanpaa, N., Guerin, S., Wu, L., Anderson, G., et al. (2000). RNA and protein expression of the murine autoimmune regulator gene (Aire) in normal, RelB-deficient and in NOD mouse. Eur. J. Immunol. 30, 1884–1893. doi: 10.1002/1521-4141(200007)30:7<1884::aid-immu1884>3.0.co;2-p
Hikosaka, Y., Nitta, T., Ohigashi, I., Yano, K., Ishimaru, N., Hayashi, Y., et al. (2008). The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity 29, 438–450. doi: 10.1016/j.immuni.2008.06.018
Hu, T., Shen, H., Li, J., Yang, P., Gu, Q., and Fu, Z. (2020). RFC2, a direct target of miR-744, modulates the cell cycle and promotes the proliferation of CRC cells. J. Cell Physiol. 235, 8319-8333
Jin, C., and Zhu, M. (2018). RelB intrinsically regulates the development and function of medullary thymic epithelial cells. Sci. China Life Sci. 61, 1039–1048. doi: 10.1007/s11427-017-9298-3
Kadouri, N., Nevo, S., Goldfarb, Y., and Abramson, J. (2020). Thymic epithelial cell heterogeneity: TEC by TEC. Nat. Rev. Immunol. 20, 239–253. doi: 10.1038/s41577-019-0238-0
Kajiura, F., Sun, S., Nomura, T., Izumi, K., Ueno, T., Bando, Y., et al. (2004). NF-kappa B-inducing kinase establishes self-tolerance in a thymic stroma-dependent manner. J. Immunol. 172, 2067–2075. doi: 10.4049/jimmunol.172.4.2067
Kanfi, Y., Naiman, S., Amir, G., Peshti, V., Zinman, G., Nahum, L., et al. (2012). The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221. doi: 10.1038/nature10815
Kawahara, T. L., Michishita, E., Adler, A. S., Damian, M., Berber, E., Lin, M., et al. (2009). SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell 136, 62–74. doi: 10.1016/j.cell.2008.10.052
Khan, R. I., Nirzhor, S. S. R., and Akter, R. (2018). A review of the recent advances made with SIRT6 and its implications on aging related processes, major human diseases, and possible therapeutic targets. Biomolecules 8:44. doi: 10.3390/biom8030044
Kim, H. S., Xiao, C., Wang, R. H., Lahusen, T., Xu, X., Vassilopoulos, A., et al. (2010). Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 12, 224–236. doi: 10.1016/j.cmet.2010.06.009
Kinoshita, D., Hirota, F., Kaisho, T., Kasai, M., Izumi, K., Bando, Y., et al. (2006). Essential role of IkappaB kinase alpha in thymic organogenesis required for the establishment of self-tolerance. J. Immunol. 176, 3995–4002. doi: 10.4049/jimmunol.176.7.3995
Klajic, J., Busato, F., Edvardsen, H., Touleimat, N., Fleischer, T., Bukholm, I., et al. (2014). DNA methylation status of key cell-cycle regulators such as CDKNA2/p16 and CCNA1 correlates with treatment response to doxorubicin and 5-fluorouracil in locally advanced breast tumors. Clin. Cancer Res. 20, 6357–6366. doi: 10.1158/1078-0432.ccr-14-0297
Klein, L., Hinterberger, M., Wirnsberger, G., and Kyewski, B. (2009). Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 9, 833–844. doi: 10.1038/nri2669
Klein, L., Klugmann, M., Nave, K. A., Tuohy, V. K., and Kyewski, B. (2000). Shaping of the autoreactive T-cell repertoire by a splice variant of self protein expressed in thymic epithelial cells. Nat. Med. 6, 56–61. doi: 10.1038/71540
Kozai, M., Kubo, Y., Katakai, T., Kondo, H., Kiyonari, H., Schaeuble, K., et al. (2017). Essential role of CCL21 in establishment of central self-tolerance in T cells. J. Exp. Med. 214, 1925–1935. doi: 10.1084/jem.20161864
Kugel, S., and Mostoslavsky, R. (2014). Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem. Sci. 39, 72–81. doi: 10.1016/j.tibs.2013.12.002
Kyewski, B., and Klein, L. (2006). A central role for central tolerance. Annu. Rev. Immunol. 24, 571–606. doi: 10.1146/annurev.immunol.23.021704.115601
Lebel, M. E., Coutelier, M., Galipeau, M., Kleinman, C. L., Moon, J. J., and Melichar, H. J. (2020). Differential expression of tissue-restricted antigens among mTEC is associated with distinct autoreactive T cell fates. Nat. Commun. 11:3734.
Lee, H. M., Bautista, J. L., Scott-Browne, J., Mohan, J. F., and Hsieh, C. S. (2012). A broad range of self-reactivity drives thymic regulatory T cell selection to limit responses to self. Immunity 37, 475–486. doi: 10.1016/j.immuni.2012.07.009
Liang, Z., Zhang, L., Su, H., Luan, R., Na, N., Sun, L., et al. (2018). MTOR signaling is essential for the development of thymic epithelial cells and the induction of central immune tolerance. Autophagy 14, 505–517. doi: 10.1080/15548627.2017.1376161
Liang, Z., Zhang, Q., Dong, X., Zhang, Z., Wang, H., Zhang, J., et al. (2020). mTORC2 negatively controls the maturation process of medullary thymic epithelial cells by inhibiting the LTbetaR/RANK-NF-kappaB axis. J. Cell Physiol. [Epub ahead of print]. doi: 10.1002/jcp.30192
Lio, C. W., and Hsieh, C. S. (2008). A two-step process for thymic regulatory T cell development. Immunity 28, 100–111. doi: 10.1016/j.immuni.2007.11.021
Liszt, G., Ford, E., Kurtev, M., and Guarente, L. (2005). Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 280, 21313–21320. doi: 10.1074/jbc.m413296200
Liu, B., Liu, Y. F., Du, Y. R., Mardaryev, A. N., Yang, W., Chen, H., et al. (2013). Cbx4 regulates the proliferation of thymic epithelial cells and thymus function. Development 140, 780–788. doi: 10.1242/dev.085035
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550.
Lucas, B., White, A. J., Cosway, E. J., Parnell, S. M., James, K. D., Jones, N. D., et al. (2020). Diversity in medullary thymic epithelial cells controls the activity and availability of iNKT cells. Nat. Commun. 11:2198.
Lynch, H. E., Goldberg, G. L., Chidgey, A., Van den Brink, M. R., Boyd, R., and Sempowski, G. D. (2009). Thymic involution and immune reconstitution. Trends Immunol. 30, 366–373. doi: 10.1016/j.it.2009.04.003
Malhotra, D., Linehan, J. L., Dileepan, T., Lee, Y. J., Purtha, W. E., Lu, J. V., et al. (2016). Tolerance is established in polyclonal CD4(+) T cells by distinct mechanisms, according to self-peptide expression patterns. Nat. Immunol. 17, 187–195. doi: 10.1038/ni.3327
Mao, Z., Hine, C., Tian, X., Van Meter, M., Au, M., Vaidya, A., et al. (2011). SIRT6 promotes DNA repair under stress by activating PARP1. Science 332, 1443–1446. doi: 10.1126/science.1202723
McCaughtry, T. M., Wilken, M. S., and Hogquist, K. A. (2007). Thymic emigration revisited. J. Exp. Med. 204, 2513–2520. doi: 10.1084/jem.20070601
Michishita, E., McCord, R. A., Berber, E., Kioi, M., Padilla-Nash, H., Damian, M., et al. (2008). SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496. doi: 10.1038/nature06736
Michishita, E., McCord, R. A., Boxer, L. D., Barber, M. F., Hong, T., Gozani, O., et al. (2009). Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8, 2664–2666. doi: 10.4161/cc.8.16.9367
Miller, C. N., Proekt, I., von Moltke, J., Wells, K. L., Rajpurkar, A. R., Wang, H., et al. (2018). Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature 559, 627–631. doi: 10.1038/s41586-018-0345-2
Mostoslavsky, R., Chua, K. F., Lombard, D. B., Pang, W. W., Fischer, M. R., Gellon, L., et al. (2006). Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315–329. doi: 10.1016/j.cell.2005.11.044
Nitta, T., and Takayanagi, H. (2020). Non-epithelial thymic stromal cells: unsung heroes in thymus organogenesis and T Cell development. Front. Immunol. 11:620894. doi: 10.3389/fimmu.2020.620894
Pan, P. W., Feldman, J. L., Devries, M. K., Dong, A., Edwards, A. M., and Denu, J. M. (2011). Structure and biochemical functions of SIRT6. J. Biol. Chem. 286, 14575–14587. doi: 10.1074/jbc.m111.218990
Park, J. E., Botting, R. A., Dominguez Conde, C., Popescu, D. M., Lavaert, M., Kunz, D. J., et al. (2020). A cell atlas of human thymic development defines T cell repertoire formation. Science 367:eaay3224.
Park, S. J., Yang, S. W., and Kim, B. C. (2016). Transforming growth factor-beta1 induces cell cycle arrest by activating atypical cyclin-dependent kinase 5 through up-regulation of Smad3-dependent p35 expression in human MCF10A mammary epithelial cells. Biochem. Biophys. Res. Commun. 472, 502–507. doi: 10.1016/j.bbrc.2016.02.121
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Rezzani, R., Nardo, L., Favero, G., Peroni, M., and Rodella, L. F. (2014). Thymus and aging: morphological, radiological, and functional overview. Age 36, 313–351. doi: 10.1007/s11357-013-9564-5
Riemann, M., Andreas, N., Fedoseeva, M., Meier, E., Weih, D., Freytag, H., et al. (2017). Central immune tolerance depends on crosstalk between the classical and alternative NF-kappaB pathways in medullary thymic epithelial cells. J. Autoimmun. 81, 56–67. doi: 10.1016/j.jaut.2017.03.007
Rodewald, H. R. (2008). Thymus organogenesis. Annu. Rev. Immunol. 26, 355–388. doi: 10.1146/annurev.immunol.26.021607.090408
Rodrigues, P. M., Ribeiro, A. R., Perrod, C., Landry, J. J. M., Araujo, L., Pereira-Castro, I., et al. (2017). Thymic epithelial cells require p53 to support their long-term function in thymopoiesis in mice. Blood 130, 478–488. doi: 10.1182/blood-2016-12-758961
Rogers, R. F., Walton, M. I., Cherry, D. L., Collins, I., Clarke, P. A., Garrett, M. D., et al. (2020). CHK1 inhibition is synthetically lethal with loss of B-Family DNA polymerase function in human lung and colorectal cancer cells. Cancer Res. 80, 1735–1747. doi: 10.1158/0008-5472.can-19-1372
Rossi, S. W., Kim, M. Y., Leibbrandt, A., Parnell, S. M., Jenkinson, W. E., Glanville, S. H., et al. (2007). RANK signals from CD4(+)3(-) inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J. Exp. Med. 204, 1267–1272. doi: 10.1084/jem.20062497
Salvador, J. M., Brown-Clay, J. D., and Fornace, A. J. Jr. (2013). Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv. Exp. Med. Biol. 793, 1–19. doi: 10.1007/978-1-4614-8289-5_1
Sansom, S. N., Shikama-Dorn, N., Zhanybekova, S., Nusspaumer, G., Macaulay, I. C., Deadman, M. E., et al. (2014). Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 24, 1918–1931. doi: 10.1101/gr.171645.113
Santamaria, J. C., Borelli, A., and Irla, M. (2021). Regulatory T Cell heterogeneity in the thymus: impact on their functional activities. Front. Immunol. 12:643153. doi: 10.3389/fimmu.2021.643153
Seach, N., Ueno, T., Fletcher, A. L., Lowen, T., Mattesich, M., Engwerda, C. R., et al. (2008). The lymphotoxin pathway regulates Aire-independent expression of ectopic genes and chemokines in thymic stromal cells. J. Immunol. 180, 5384–5392. doi: 10.4049/jimmunol.180.8.5384
Shinkai, Y., Rathbun, G., Lam, K. P., Oltz, E. M., Stewart, V., Mendelsohn, M., et al. (1992). RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68, 855–867. doi: 10.1016/0092-8674(92)90029-c
Soza-Ried, C., Bleul, C. C., Schorpp, M., and Boehm, T. (2008). Maintenance of thymic epithelial phenotype requires extrinsic signals in mouse and zebrafish. J. Immunol. 181, 5272–5277. doi: 10.4049/jimmunol.181.8.5272
Subramanian, A., Kuehn, H., Gould, J., Tamayo, P., and Mesirov, J. P. (2007). GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics 23, 3251–3253. doi: 10.1093/bioinformatics/btm369
Sun, L., Li, H., Luo, H., Zhang, L., Hu, X., Yang, T., et al. (2013). Phosphatase Wip1 is essential for the maturation and homeostasis of medullary thymic epithelial cells in mice. J. Immunol. 191, 3210–3220. doi: 10.4049/jimmunol.1300363
Sun, L., Li, H., Luo, H., and Zhao, Y. (2014). Thymic epithelial cell development and its dysfunction in human diseases. Biomed. Res. Int. 2014:206929.
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452.
Takada, K., Kondo, K., and Takahama, Y. (2017). Generation of peptides that promote positive selection in the thymus. J. Immunol. 198, 2215–2222. doi: 10.4049/jimmunol.1601862
Tasselli, L., Xi, Y., Zheng, W., Tennen, R. I., Odrowaz, Z., Simeoni, F., et al. (2016). SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 23, 434–440. doi: 10.1038/nsmb.3202
Taub, D. D., and Longo, D. L. (2005). Insights into thymic aging and regeneration. Immunol. Rev. 205, 72–93. doi: 10.1111/j.0105-2896.2005.00275.x
Tsukasaki, M., Asano, T., Muro, R., Huynh, N. C., Komatsu, N., Okamoto, K., et al. (2020). OPG production matters where it happened. Cell Rep. 32:108124. doi: 10.1016/j.celrep.2020.108124
Tudzarova, S., Mulholland, P., Dey, A., Stoeber, K., Okorokov, A. L., and Williams, G. H. (2016). p53 controls CDC7 levels to reinforce G1 cell cycle arrest upon genotoxic stress. Cell Cycle 15, 2958–2972. doi: 10.1080/15384101.2016.1231281
Ueno, T., Hara, K., Willis, M. S., Malin, M. A., Hopken, U. E., Gray, D. H., et al. (2002). Role for CCR7 ligands in the emigration of newly generated T lymphocytes from the neonatal thymus. Immunity 16, 205–218. doi: 10.1016/s1074-7613(02)00267-4
Ueno, T., Saito, F., Gray, D. H., Kuse, S., Hieshima, K., Nakano, H., et al. (2004). CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J. Exp. Med. 200, 493–505. doi: 10.1084/jem.20040643
Wang, H., Diao, D., Shi, Z., Zhu, X., Gao, Y., Gao, S., et al. (2016). SIRT6 controls hematopoietic stem cell homeostasis through epigenetic regulation of Wnt signaling. Cell Stem Cell 18, 495–507. doi: 10.1016/j.stem.2016.03.005
Wang, H. X., Pan, W., Zheng, L., Zhong, X. P., Tan, L., Liang, Z., et al. (2019). Thymic epithelial cells contribute to thymopoiesis and T Cell development. Front. Immunol. 10:3099. doi: 10.3389/fimmu.2019.03099
Weih, F., Carrasco, D., Durham, S. K., Barton, D. S., Rizzo, C. A., Ryseck, R. P., et al. (1995). Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-kappa B/Rel family. Cell 80, 331–340. doi: 10.1016/0092-8674(95)90416-6
Wells, K. L., Miller, C. N., Gschwind, A. R., Wei, W., Phipps, J. D., Anderson, M. S., et al. (2020). Combined transient ablation and single-cell RNA-sequencing reveals the development of medullary thymic epithelial cells. eLife 9:e60188.
White, A. J., Baik, S., Parnell, S. M., Holland, A. M., Brombacher, F., Jenkinson, W. E., et al. (2017). A type 2 cytokine axis for thymus emigration. J. Exp. Med. 214, 2205–2216. doi: 10.1084/jem.20170271
Wyss, L., Stadinski, B. D., King, C. G., Schallenberg, S., McCarthy, N. I., Lee, J. Y., et al. (2016). Affinity for self antigen selects Treg cells with distinct functional properties. Nat. Immunol. 17, 1093–1101. doi: 10.1038/ni.3522
Yang, B., Zwaans, B. M., Eckersdorff, M., and Lombard, D. B. (2009). The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 8, 2662–2663. doi: 10.4161/cc.8.16.9329
Yu, W., Nagaoka, H., Jankovic, M., Misulovin, Z., Suh, H., Rolink, A., et al. (1999). Continued RAG expression in late stages of B cell development and no apparent re-induction after immunization. Nature 400, 682–687. doi: 10.1038/23287
Zhang, B., Wang, Z., Ding, J., Peterson, P., Gunning, W. T., and Ding, H. F. (2006). NF-kappaB2 is required for the control of autoimmunity by regulating the development of medullary thymic epithelial cells. J. Biol. Chem. 281, 38617–38624. doi: 10.1074/jbc.m606705200
Zhang, W., Wan, H., Feng, G., Qu, J., Wang, J., Jing, Y., et al. (2018). SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature 560, 661–665. doi: 10.1038/s41586-018-0437-z
Zhong, L., D’Urso, A., Toiber, D., Sebastian, C., Henry, R. E., Vadysirisack, D. D., et al. (2010). The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 140, 280–293. doi: 10.1016/j.cell.2009.12.041
Zhu, M., Chin, R. K., Christiansen, P. A., Lo, J. C., Liu, X., Ware, C., et al. (2006). NF-kappaB2 is required for the establishment of central tolerance through an Aire-dependent pathway. J. Clin. Invest. 116, 2964–2971. doi: 10.1172/jci28326
Zhu, M., Chin, R. K., Tumanov, A. V., Liu, X., and Fu, Y. X. (2007). Lymphotoxin beta receptor is required for the migration and selection of autoreactive T cells in thymic medulla. J. Immunol. 179, 8069–8075. doi: 10.4049/jimmunol.179.12.8069
Keywords: autoimmune disease, immune tolerance, Sirt6, Spib, thymic epithelial cells (TECs), thymus
Citation: Zhang Q, Liang Z, Zhang J, Lei T, Dong X, Su H, Chen Y, Zhang Z, Tan L and Zhao Y (2021) Sirt6 Regulates the Development of Medullary Thymic Epithelial Cells and Contributes to the Establishment of Central Immune Tolerance. Front. Cell Dev. Biol. 9:655552. doi: 10.3389/fcell.2021.655552
Received: 19 January 2021; Accepted: 09 March 2021;
Published: 29 March 2021.
Edited by:
Ivan Lilyanov Dzhagalov, National Yang-Ming University, TaiwanReviewed by:
Takeshi Nitta, The University of Tokyo, JapanMathias Hauri-Hohl, University Children’s Hospital Zurich, Switzerland
Copyright © 2021 Zhang, Liang, Zhang, Lei, Dong, Su, Chen, Zhang, Tan and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Zhao, zhaoy@ioz.ac.cn
†These authors have contributed equally to this work
 Qian Zhang
Qian Zhang Zhanfeng Liang
Zhanfeng Liang Jiayu Zhang
Jiayu Zhang Tong Lei2†
Tong Lei2†  Xue Dong
Xue Dong Zhaoqi Zhang
Zhaoqi Zhang Liang Tan
Liang Tan Yong Zhao
Yong Zhao