- 1Indian Council of Agricultural Research-Central Soil Salinity Research Institute, Regional Research Station, Lucknow, India
- 2Indian Council of Agricultural Research-Central Institute for Subtropical Horticulture, Lucknow, India
Fusarium wilt in bananas is one of the most devastating diseases that poses a serious threat to the banana industry globally. With no effective control measures available to date, biological control has been explored to restrict the spread and manage the outbreak. We studied the effective biological control potential of different Trichoderma spp. in the management of Fusarium oxysporum f. sp. cubense tropical race 4 (Foc TR4). Expression of the defense related genes and metabolites in banana plants inoculated with Foc TR4 and treated with effective Trichoderma sp interactions were also studied. The in vitro growth inhibition of Foc TR4 by Trichoderma reesei isolate CSR-T-3 was 85.19% indicating a higher antagonistic potential than other Trichoderma isolates used in the study. Further, in in vivo assays, the banana plants treated with the isolate CSR-T-3 T. reesei had a significant reduction in the disease severity index (0.75) and also had increased phenological indices with respect to Foc TR4 treated plants. Enhanced activity of defense enzymes, such as β-1, 3-glucanase, peroxidase, chitinase, polyphenol oxidase, and phenylalanine ammonia lyase with higher phenol contents were found in the Trichoderma isolate CSR-T-3 treated banana plants challenge-inoculated with Foc TR4. Fusarium toxins, such as fusaristatin A, fusarin C, chlamydosporal, and beauveric acid were identified by LC-MS in Foc TR4-infected banana plants while high intensity production of antifungal compounds, such as ß-caryophyllene, catechin-o-gallate, soyasapogenol rhamnosyl glucoronide, peptaibols, fenigycin, iturin C19, anthocyanin, and gallocatechin-o-gallate were detected in T. reesei isolate CSR-T-3 treated plants previously inoculated with Foc TR4. Gene expression analysis indicated the upregulation of TrCBH1/TrCBH2, TrXYL1, TrEGL1, TrTMK1, TrTGA1, and TrVEL1 genes in CSR-T-3 treatment. LC-MS and gene expression analysis could ascertain the upregulation of genes involved in mycoparasitism and the signal transduction pathway leading to secondary metabolite production under CSR-T-3 treatment. The plants in the field study showed a reduced disease severity index (1.14) with high phenological growth and yield indices when treated with T. reesei isolate CSR-T-3 formulation. We report here an effective biocontrol-based management technological transformation from lab to the field for successful control of Fusarium wilt disease caused by Foc TR4 in bananas.
Introduction
Fusarium wilt caused by Fusarium oxysporum f. sp. cubense tropical race 4 (Foc TR4) is considered to be one of the most devastating diseases of bananas limiting banana production worldwide (Ploetz and Pegg, 2000; Dita et al., 2018). The pathogen was first reported in South East Asia during the early 1990s and toward the end of the century, endemic outbreak of Foc TR4 was realized over large areas in the northern territory of Australia, China, Indonesia, Malaysia, and Taiwan (Ploetz, 2006a,b). Most recently, severe outbreak of Foc TR4 in the northern region in India covering the states of Uttar Pradesh and Bihar threatening banana cultivation in the region were reported, which is the largest producer of bananas (Damodaran, 2018). The polycyclic nature of the disease suggests that its multiple cycle of infection in bananas is causing devastating losses over a period of time (Buddenhagen, 2009). Different types of toxins are produced by Fusarium which are involved in pathogenesis and disease development. The major toxins that are reported from Fusarium species include; nivalenol, zearalenone, fumonisins, trichothecene, fusarins, and Fusaric acid (Niehaus et al., 2014; Dong et al., 2017; López-Díaz et al., 2017; Perincherry et al., 2019; Qiu et al., 2019). Differential colonization patterns of Foc race 4 from race 1 were identified and explained by upregulation of pathogenicity-associated genes, such as Fga1, Fhk1, Fow2, and Ste12 that allowed faster colonization in the xylem with higher virulence (Guo et al., 2015). The discrimination of Foc TR4 from Foc race 1 proved very efficient with the PCR-based detection employed for secreted in xylem (SIX) genes (Carvalhais et al., 2019) and comparative analysis of various specific primers has also ascertained the efficiency of SIX gene-specific primers in the molecular detection of Foc TR4 (Magdama et al., 2019). These advances have now paved the way for a reliable dependable confirmatory assay for the early detection/diagnosis of Foc TR4.
Management and control of Foc TR4 is a matter of serious concern and different approaches, such as chemical and biological control, manipulation of cultural practices, and resistant breeding have been attempted by different research groups. The ability of Foc TR4 to survive in the absence of its host is a significant factor limiting the successful management of this disease through agronomic practices like crop rotation (Brandes, 1919). Besides this, use of chemical control measures involving corm injection and soil drenching of carbendazim fungicide failed to provide sustainable management of the disease but their repeated use has raised environmental concerns (Getha and Vikineswary, 2002). Application of soil fumigants, field sanitation, and flood fallowing provided limited control over a large area (Ploetz et al., 1990). In another approach using resistance breeding, some advances were reported for evolving resistant varieties and hybrids against Foc race 1 (Damodaran et al., 2009) but to date no successful reports are available for Foc TR4 (de Matos et al., 2011; Amorim et al., 2013; Garcez et al., 2016; Rebouças et al., 2018; Ribeiro et al., 2018; Gonçalves et al., 2019). Moreover, the selection of resistant cultivars seems to be an effective control strategy against the disease, however, the existence of poor fertility in the Cavendish group (AAA) limited its improvement programs through conventional breeding (Aguilar Moran, 2013). Thus, the only alternative approach for management is by exploring antagonistic and growth promoting fungi or bacteria which have earlier been successfully demonstrated in biological control of soil borne pathogens in different crop plants (Tan et al., 2015, Khan et al., 2018). Rhizospheric existence and endophytic colonization of the potential antagonist are known to not only suppress the pathogen but also incite systemic-induced host plant resistance (Ting et al., 2009, Sood et al., 2020).
Trichoderma (Teleomorph: Hypocrea) is a fast-growing soil inhabitant fungus known for the production of a large numbers of spores and has been reported to have biocontrol potentials (Monfil and Casas-Flores, 2014). The antagonism of Trichoderma against phytopathogens is achieved by mycoparasitism (Papavizas, 1985) through the production of enzymes (chitinases, glucanases, and proteases) that degrade the fungal cell wall which is mainly composed of chitin (Qian et al., 2019) as well as antibiosis (Harman and Kubicek, 1998); through production of secondary metabolites and antimicrobial compounds active against a wide array of phytopathogens (Speckbacher and Zeilinger, 2018). Additionally, when the plant encounters the pathogen it induces a host defense mechanism while on interaction with a non-pathogenic organism the mechanism for induced systemic resistance (ISR) gets activated (Hermosa et al., 2013; Mukherjee et al., 2013). A plethora of studies suggest that Trichoderma spp. induce systemic resistance to the subsequent attack by the pathogens in the host plants (Martínez-Medina et al., 2013). Elevated production of antioxidants like polyphenols, cell wall degrading enzymes like glucanases, chitinases, etc. by Trichoderma probably induced the host defense mechanism against the invading pathogens including Fusarium (Kavino et al., 2009). Though, long term efficacy of biocontrol agents in field studies have seldom come up with repeatability at a field level (Ploetz, 2004), yet, repeated application of biocontrol agents has resulted in the suppression of soil borne pathogens. These discoveries showed promise for exploring antagonistic biocontrol agents and developing a package for the management of the Foc TR4 pathogen causing Fusarium wilt in bananas. The prime focus of the study was to identify an elite antagonistic fungus by screening different Trichoderma isolates from diverse biotic and abiotic stressed environments that has the potential to inhibit the Foc TR4 pathogen, assess its efficacy in the management of the disease under pot culture and sick field experiments, and to decipher its antifungal mode of action/mechanism in disease suppression through gene expression and LC-MS analysis. We report here a systematic development of a technological innovation involving a Trichoderma isolate and its field transformation as a biocontrol agent toward successful management of banana wilt caused by Fusarium oxysporum f. sp. cubense tropical race 4.
Materials and Methods
Collections of Trichoderma spp. and Fusarium oxysporum f. sp. cubense Tropical Race 4
The virulent strain of the Fusarium oxysporum f. sp. cubense tropical race 4 was collected from the culture collections of the Soil Microbiology Laboratory, ICAR-Central Soil Salinity Research Institute, Regional Research Station, Lucknow, India that had submitted the first report on the outbreak of the disease in India (Damodaran et al., 2018). The three Trichoderma isolates used in the study are described as follows; (i) Trichoderma reesei (CSR-T-3, MK050013) isolated from the rhizosphere soil of banana cultivar G-9 grown in salt affected soil, (ii) Trichoderma asperellum (CSR-T-4, MN227242) isolated from the disease suppressive rhizosphere of banana cultivar G-9 grown in the Foc TR4 affected region of Uttar Pradesh, India, and (iii) Trichoderma koningiopsis (CSR-T-2, KJ812401) obtained from the rhizosphere of Saccharam spontaneum in the barren sodic soil of Uttar Pradesh, India. These isolates were identified and submitted to the culture collections (NBIMCC-SF-0030) at the ICAR-National Bureau of Agriculturally Important Microorganisms (ICAR-NBAIM), Mau, India and have been reported in our earlier studies (Damodaran et al., 2019c; Yadav et al., 2020).
In vitro Antagonistic Assays
The three isolates of the Trichoderma species were evaluated for their in vitro antagonistic potential against CSR-F-1 (the virulent strain of Foc TR4) through a dual culture test as described by Tian et al. (2016). Mycelial disks of Foc TR4 and Trichoderma spp. from actively growing colonies were placed on a 9 cm diameter petri dish filled with potato dextrose medium (PDA) in combinations. The isolates were placed at 5.5 cm apart on the same plate and were incubated for 72 h at 22°C. Foc TR4 isolate CSR-F-1 was also placed without disks of Trichoderma isolates as control. The growth of the isolates was monitored and observations were recorded at an interval of every 24 h from which the percentage of pathogen growth inhibition was calculated. The inhibition efficacy of Trichoderma isolates on the mycelia growth of Foc TR4 and the radius of the Foc colony in each plate were measured, and a standard growth curve was derived as described by Matarese et al. (2012). The distance between the Foc TR4 inoculating point and furthest point of the colony in control plates (r1) and the distance between the Foc TR4 sowing point and the edge of the colony where the mycelia came in contact with Trichoderma spp. in the treatment plates was taken at an interval of 24 h for calculating the inhibition percentage (IP) using the formula; IP = ((r1 – r2)/r1)*100. This in vitro dual culture assay was carried out in five replicates and the data was statistically analyzed by analysis of variance (ANOVA) using Duncan's multiple range tests using the SAS 9.0 software.
SEM Analysis of Dual Culture Interaction to Ascertain Antagonism of the Best Performing Isolate
Scanning electron microscopy of the dual culture regions was carried out for establishing and observing the CSR-T-3 antagonism against Foc TR4 (CSR-F-1) as described in protocol given by Kumar et al. (2018). Pieces of agar from the interaction zones of the dual culture plates at their early stage of interaction were fixed in 3% glutaraldehyde dissolved in 0.1 M phosphate buffer (pH 7.0). The specimens were dehydrated using acetone at a series of concentration (30, 50, 60, 70, 80, 90, and 95%) for 20 min. The samples were dried for 30 min and mounted on a steel stub with double sided carbon tape. The samples were finally coated with a film of gold-palladium alloy under vacuum and observed with a scanning electron microscope (SEM, Fei Quanta 200) at Bhaba Bhimrao Ambedkar University, Lucknow, India. The control plates involving the individual plates of T. reesei (CSR-T-3) and Foc TR4 (CSR-F-1) were also maintained for comparison.
In vivo Assay of Antifungal Potential Using Pot Culture
The in vivo assays were performed with the best performing Trichoderma isolate from the in vitro dual culture assay. A systematic pot culture experiment was carried out in June-July 2017 and June-July 2018 at the ICAR-Central Soil Salinity Research Institute, Regional Research Station, Lucknow, India by following a completely randomized design (CRD) with three replicates. The temperature of the green house during the experimental period ranged from 29 to 32°C with 70% humidity. The fungal culture was prepared by inoculating the 5-days-old culture of the best performing Trichoderma isolate in potato dextrose broth (PDB) with subsequent incubation for 5 days at 28°C in an incubated shaker. The spore suspension was obtained by filtering the broth through double-layered cheese cloth. The filtrate thus obtained was diluted 50 times and the spore number was adjusted from 107 to 108 using a hemocytometer. About 100 ml of the diluted filtrate was added in sterile soil. Likewise, the pathogen inoculum was prepared using the same procedure and the spore count was adjusted from 105 to 106. For sterilizing the soil, normal clayey loam soil (pH 7.45, E.C. 0.32 dS/m, organic carbon content of 3.0 g/kg) was collected from the research farm of the Institute, passed through a 20 mm mesh sieve and sterilized in an autoclave at 121°C for 30 min for 3 consecutive days.
Secondary hardened tissue cultured banana plantlets of the Grand Naine variety (susceptible) of uniform height were selected to conduct the experiment. The plants were inoculated with the pathogen by dipping them in the pathogen spore suspension containing 106 conidia/ml for 30 min as described by Pérez-Vicente et al. (2014) before planting. Three treatments viz, TC: uninoculated plants, TF: plants inoculated with CSR-F-1 (Foc TR4) at the time of planting and TFTR: plants inoculated with CSR-F-1 (Foc TR4) along with soil drenching with suspension of the best performing Trichoderma isolate were included in the pot experiment with five replicates for each treatment. The experiment was repeated three times.
The phenological indices of banana seedlings like plant height (cm), girth (cm), and number of leaves was noted at 0, 30, 60, and 90 days after planting. The morphological growth-related parameters like root biomass and shoot biomass were recorded at the end of the experiment. The wilt disease was scored by adopting the following disease scale as followed by Pérez-Vicente et al. (2014).
Grade 1 No symptoms
Grade 2 10–20% initial corm discoloration
Grade 3 20–40% slight discoloration of the corm
Grade 4 40–60% discoloration of the corm
Grade 5 60–80% complete discoloration of the corm
Besides this, the chlorophyll content was also estimated at the end of the experiment. Chlorophyll a, b, and total chlorophyll contents were estimated by following the method of Sadasivam and Manickam (1996) using 80% acetone and magnesium carbonate. The data were statistically analyzed by subjecting them to analysis of variance (ANOVA) and means were compared by a Duncan's multiple range test (p < 0.05) (Rangaswamy, 1995).
Liquid Chromatography Coupled With Mass Spectrometric (LC-MS) Analysis
LC-MS analysis was performed in the plant samples of treatments TC (treatment negative control), TF (treatment Foc-TR4), and TFTR (treatment Foc-TR4 and Trichoderma reesei isolate CSR-T-3) of the pot experiment to identify important organic compounds and their role in inducing host tolerance. The whole plant was uprooted, washed with sterile distilled water, and then shade-dried for 30 days. Further the protocol for sample preparation was as prescribed by the Sophisticated Analytical Instrumentation Facility (SAIF), Central Drug Research Institute (CDRI), Lucknow, Uttar Pradesh, India.
A total of 250 g of dry weight plant material was used for the preparation of plant extracts consisting of root, leaves, and stem. Ethanol crude extract was obtained by crushing the plants in 70% ethanol which was further purified by using ethanol acetate. A small fraction of the solvent free plant extract was collected in Eppendorf for further analysis. The extract residue was stored at 4°C until used for the antimicrobial assay.
The ESI-LC–MS of the TLC purified sample was analyzed using a MICROMASS QUATTRO II triple quadrupole mass spectrometer with a JASCO PU-980 HPLC pump at the Sophisticated Analytical Instrumentation Facility, CSIR-Central Drug Research Institute, Lucknow, India. The WATER SPHERISORB ODS 2 (250 _ 4.6 mm _ 5 l) column was used with acetonitrile: water +0.1% formic acid solvent system. Gradient elution was performed at 1.0 ml/min. The photodiode array was monitored at 200–650 nm and recorded at 220 nm. The mass spectra were scanned in the range 80–1,000 DA in 2.5 s. The ESI capillary was set at 3.5 kV and the cone voltage at 40 V. The m/z spectral chromatographs were analyzed, key metabolites were predicted based on already published reports and tabulated for comparative analysis between the TC, TF, and TFTR samples.
Biocontrol Mechanism of Trichoderma reesei Against Foc Through Gene Expression Analysis
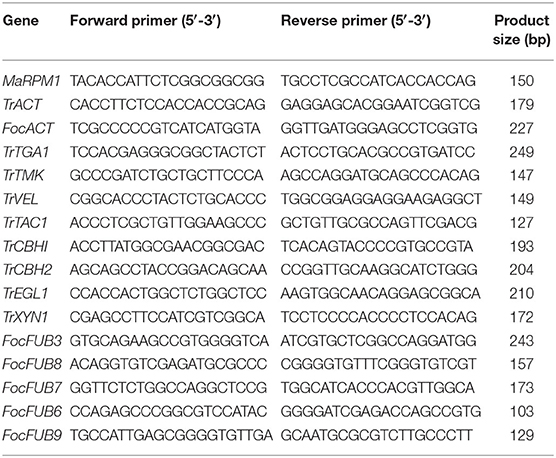
To study the basis of resistance achieved in banana plants against Foc TR4 through drenching the soil with the best performing Trichoderma isolate at the time of planting, the expression levels of the genes of T. reesei and Foc TR4 were studied using a real time-polymerase chain reaction (RT-PCR) assay. Genomic sequence information for the genes involved in mycoparasitism viz., TrCBH1/TrCBH2, TrXYL1, and TrEGL1 encoding cellobiohydrolase, xylanase, and endoglucanase, respectively and few genes involved in the signaling transduction pathway leading to secondary metabolite production in T. reesei, i.e., TrTMK1, TrTGA1, and TrVEL1 genes encoding mitogen activated protein kinase (MAPK), G-protein alpha subunit, and VELVET1 protein, respectively were mined from the whole genome sequence of T. reesei QM6a as reported by Martínez-Hidalgo et al. (2015) (Bioproject: PRJNA15571) available at NCBI database. Similarly, the genes involved in Fusaric acid biosynthesis were mined from the whole genome information of F. oxysporum f. sp. cubense (Foc TR4) in the NCBI database (Bioproject: PRJNA529756). The selection of these genes was based on the well-established antagonistic biocontrol potential evincing molecular mechanism of Trichoderma sp. against Fusarium species earlier reported by Druzhinina et al. (2012) and Zeilinger et al. (2016). The primers for the genes used in RT-PCR analysis were designed for exonic segments contributing to the functional domains based on protein structural domain analysis and exon predictions (phylogenetic analysis of genes and structural predictions are presented in Supplementary Figures 1, 2) using the primer3 software and were synthesized from Xcelris Labs Pvt. Ltd, Ahmedabad, India (Table 1). The root samples of healthy banana plants (negative control), banana plants inoculated with CSR-F-1 (Foc TR4), banana plants challenged with CSR-F-1 (Foc TR4) followed by subsequent treatment with the best performing Trichoderma isolate (Foc-TR4 + CSR-T-3), and banana plants treated with the best performing Trichoderma isolate alone were collected at 30 days after treatment. Total RNA was isolated from the root samples of all four treatments using the Spectrum™ Plant Total RNA Kit (Sigma USA) following the manufacturer's instructions. The quality of total RNA was checked on 1% agarose gel and quantified using a QIAxpert spectrophotometer (Qiagen Germany). The RNA (3 μg) was subjected to cDNA synthesis using a Maxima First Strand cDNA Synthesis Kit. RT-PCR was performed using 10 ng cDNA as the template, forward and reverse primers (5 picomoles each), and the SYBR master mix (ABI, NY, USA) in Applied Biosystems™ 7500 Real-Time PCR Systems (ABI, NY, USA). PCR amplification was programmed with a pre PCR read at 60°C for 30 s, a holding stage at 95°C for 10 min, and a cycling stage at 95°C for 15 s and at 60°C for 1 min. Data were normalized twice, once with banana housekeeping gene encoding disease resistance protein RPM (MaRPM) and then with actin genes of Fusarium and Trichoderma (FocACT, TrACT) for assessing the relative abundance of the transcripts related with Fusaric acid biosynthesis and Trichoderma related genes, respectively. The differential expression of the genes was expressed in terms of 2-fold change using the formula, Fold Change (FC) = 2(−ΔΔCt).
Biochemical Analysis of Defense-Related Enzymes and Phenol Content
Leaf samples of 1 g (three samples from each replicate) were homogenized with 2 ml of 0.1 M sodium citrate buffer (pH 5.0) at 48°C and centrifuged for 20 min at 10,000 rpm. The supernatant crude enzyme extract was used for assaying 1, 3-glucanase (Pan et al., 1991) and chitinase (Boller and Mauch, 1988) activity. Enzyme extracted in 0.1 M sodium phosphate buffer (pH 7.0) was used for the estimation of peroxidase (PO) (Hammerschmidt et al., 1982), polyphenol oxidase (PPO) (Mayer et al., 1965), and phenylalanine ammonia lyase (PAL) (Ross and Sederoff, 1992). The phenol content was estimated following the procedure of Zieslin and Ben-Zaken (1993) and expressed as catechol equivalents g−1 of protein. The assays were carried out for the TC, TF, and TFTR samples and the data were statistically analyzed using the SAS 9.2 statistical software.
Preparation of Bioformulation of CSR-T-3 and Application Techniques
A 2 mm disc of the best performing Trichoderma isolate was inoculated into the CSR patent protected culture media (Rai et al., 2012) and incubated in a rotary shaker at 150 rpm for 72 h at room temperature (28 ± 2°C). After 72 h of incubation, the formulation containing 6 × 107 spores ml−l was used for drenching the plants. The plants were drenched with 500 ml/plant with 1% of the formulation prepared using the best performing Trichoderma isolate CSR-T-3 at 2, 4, 6, and 8 months after planting (Damodaran et al., 2019a). The untreated control plants were drenched with the plain culture at the same quantity used in treatment. The incidence of wilt disease in the treatments was scored based on a 1–5 disease scale (Ploetz et al., 1999). Besides disease incidence, observations of growth parameters, such as the girth and height of the pseudo stem, leaf area, yield, and number of hands were recorded according to the guidelines of the International Network for the Improvement of Banana and Plantain (INIBAP).
Field Evaluation of CSR-T-3 on Disease Affected Hotspot of Uttar Pradesh (India)
A field experiment was performed in the Sohawal block (26° 46′ 3.3852″ N latitude; 81° 59′ 42.7992″ E longitude) of the Faizabad district (Uttar Pradesh, India) during 2018–2019 wherein there was an extensive outbreak reported in 2017 (Damodaran et al., 2018) to evaluate the effectiveness of bioformulation in Fusarium wilt management at field level. The fields (clayey loam soil, pH-7.2) that were reported to have 60–80% Fusarium wilt incidence over the past 2 years (2017 and 2018) were selected. Average annual rainfall at the experimentation fields was in the range of 800–900 mm. Disease free banana plantlets of cv. Grand Naine (G-9) produced in the tissue culture laboratory of the ICAR-Central Soil Salinity Research Institute, Regional Research Station, Lucknow, Uttar Pradesh were planted and 50 plants per replication were used for each treatment. The experiment was laid out in a completely randomized block design with two treatments (one treatment included G-9 banana plantlets planted in a Foc infested field while the other treatment included banana G-9 plantlets planted in a Foc infested field and treated with the CSR-T-3 Trichoderma isolate formulation). The plants under the treatment were drenched with 500 ml/plant with 3% of the formulation at 2, 4, 6, and 8 months after planting replicated three times with 50 plants in each replication. The fertilizers and farm yard manure (FYM with nitrogen 0.5–1.5, phosphorus 0.4–0.8, and potassium 0.5–1.9) were applied as per the recommended dosage (Crop Production Guide, 2017). The data on growth and yield were recorded and the data on field experiment were subjected to a paired t-test (p < 0.05) for each parameter and were statistically analyzed using the SAS 9.2 statistical software. The percentage values of the disease index were arcsine transformed (Rangaswamy, 1995).
Results
In vitro Bioassay of the Trichoderma Isolates for Selecting an Elite Antagonistic Isolate Against Foc TR4
Three Trichoderma isolates viz., T. reesei (CSR-T-3), T. koningiopsis (CSR-T-2), and T. asperellum (CSR-T-4) were assessed for their in vitro efficacy by a dual co-culture technique on PDA medium. All of the three isolates utilized the nutrient source and showed a significant increase in the inhibition percentage between 48 and 72 h after inoculation by inhibiting the mycelia growth of Foc TR4 (CSR-F-1). The maximum reduction of the mycelia growth of the pathogen was elicited by the isolate CSR-T-3 that showed the highest inhibition percentage of 85.19% (Figure 1) at 120 h after inoculation, whereas CSR-T-1 showed the least inhibition (50.00%). The inhibition percentage of CSR-T-2 (T. koningiopsis) was 62.65%. Based on these results, it is evident that CSR-T-3 (T. reesei) is the most effective Trichoderma isolate with a tremendous antagonistic ability against Foc TR4.
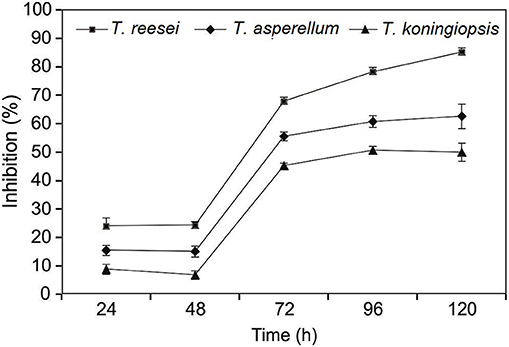
Figure 1. Inhibitory effect of CSR-T-3 (Trichoderma reesei) against pathogenic isolate CSR-F-1 of Foc TR4. Percentage inhibition of Foc TR4 was found to be highest in CSR-T-3 (Trichoderma reesei) over a period of time in comparison with T. asperellum and T. koningiopsis.
SEM Analysis of Dual Culture Interaction to Ascertain CSR-T-3 Antagonism
SEM analysis of the dual culture interaction zones between the CSR-T-3 (T. reesei isolate) and CSR-F-1 Foc TR4 isolates revealed considerable changes in the morphology including collapse, deformation, and broken cells (Figures 2A–C) leading to the loss of integrity of the Foc TR4 hyphae while the spores and mycelium in the control plates were intact and undisturbed leading to normal spore formation. This morphological microscopic identification indicates the mycoparasitism exhibited by CSR-T-3 which is identified as T. reesei (NAIMCC-SF-0030; NCBI No. MH997668).

Figure 2. Characterization and evaluation of CSR-T-3 as a biocontrol agent for the management of banana wilt caused by Fusarium oxysporum f. sp. cubense tropical race 4 (Foc TR4). (A) in vitro dual culture assay showing inhibition of Foc TR4 (F) by the CSR-T-3 Trichoderma reesei isolate (T). Scanning electron micrograph showing (B,C) mycoparasitism of the Trichoderma reesei CSR-T-3 isolate on Foc TR-4 mycelium. Pot culture experiment of banana plants for testing the biocontrol potential of the formulation of CSR-T-3 showing (D) TF (treatment with Foc TR4), (E) TFTR (treatment with Foc TR4 + CSR-T-3), and (F) TC (treatment negative control). Characteristic wilting symptoms manifested in TF (D) where the pathogen was challenge-inoculated whereas, the plants treated with CSR-T-3 and challenged with Foc TR4 showed healthy growth indicating the protection offered by CSR-T-3 as a biocontrol agent.
Green House Evaluation of CSR-T-3 (Trichoderma reesei) on Fusarium Disease Control
From the in vitro antifungal assay of the Trichoderma isolates against CSR-F-1 (Foc TR4), Trichoderma isolate CSR-T-3 with a significantly high inhibition percentage was used for in vivo pot culture studies. The disease spread was rapid and the symptoms of yellowing of older leaves started appearing in the Foc TR4 inoculated plants from 24 days after planting in control plants (Figure 2D; additional root morphological differences in Supplementary Figure 3). As time went on the plants in the Foc TR4 alone treatment began to show mortality between 60 and 90 days. At 90 days after planting when the experiment was completed, the plants that had been treated with Foc TR4 showed a significantly high mean disease severity index of 3.75 while the plants treated with TFTR recorded significantly lower DSI (0.75) that was similar to the plants of control that were not inoculated with the pathogen (Table 2). None of the plants treated with TFTR showed mortality until the end of the experiment (Figure 2E).
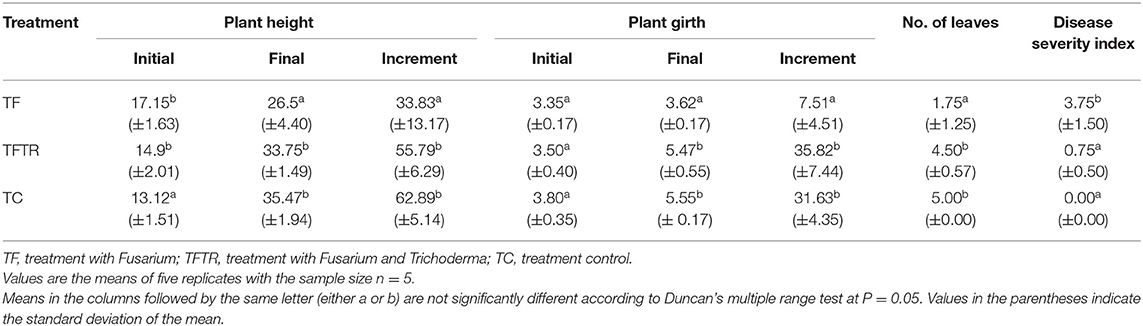
Table 2. Effect of Trichoderma reesei (CSR-T-3) on Fusarium wilt incidence and growth of banana plantlets under greenhouse pot experiment.
Plant height and girth in the TFTR-treated plants were recorded to be 33.75 and 5.47 cm, respectively which were significantly higher than the TF-treated plants (Foc TR4 alone treated banana plants). Interestingly, there were no significant difference in height and girth observed in the TFTR-treated plants (T. reesei isolate CSR-T-3 treated banana plants with challenge inoculation of Foc TR4 strain) as well as in the case of TC (untreated negative control). About a 55.79% increase in plant height was noticed in plants with TFTR treatment which also exhibited increased stem girth by 35.82% in comparison with plants with Foc TR4 alone treatment (TF) (Figures 2E,F). The results suggest that the treatment of the fungi T. reesei (CSR-T-3) significantly reduced the incidence of the disease and also promoted the growth of the seedlings on par with that of healthy plants.
Effect of Trichoderma reesei CSR-T-3 on Biomass and Chlorophyll Content of Bananas
Although, there was no improvement in plant height and pseudo stem girth in CSR-T-3 treatment, significant differences were recorded for growth parameters like weight of the pseudo stem and root. The mean fresh (Figure 3A) and dry (Figure 3B) weight of the plantlets treated with the T. reseei CSR-T-3 isolate were higher compared to those in the other treatments including control without any microbial inoculations (CSR-T-3, antagonistic biocontrol agent and CSR-F-1, Foc TR4 pathogen). The fresh root and shoot weight for the TFTR treatment (14.20 and 56.30 g, respectively) inoculated with CSR-T-3 T. reesei isolate along with Foc TR4 was significantly higher than the TC control and TF treatment. However, there was no significant difference between root and shoot dry weights in the TFTR treatment (1.11 and 2.60 g, respectively) and TC which were significantly higher than the TF treatment (0.96 and 2.60, respectively). Thus, treatment with the T. reesei CSR-T-3 isolate had a growth promoting effect on banana plants. Similarly, total chlorophyll content 0.60 mg/g was recorded in the TFTR treatment, which was significantly higher than the TF treatment (value). The total chlorophyll content in the untreated control TC treatment (0.72 mg/g) and TFTR treatment showed non-significant differences indicating the effect of the treatment in reducing the stress of the plants due to the inoculation of the pathogen (Figure 3C). Overall, it was observed that there was additional growth improvement noticed in the CSR-T-3 treatment apart from disease suppression, despite the stress imposed by the inoculation of Foc TR4. The growth attributes were found to be on par with the healthy plants.
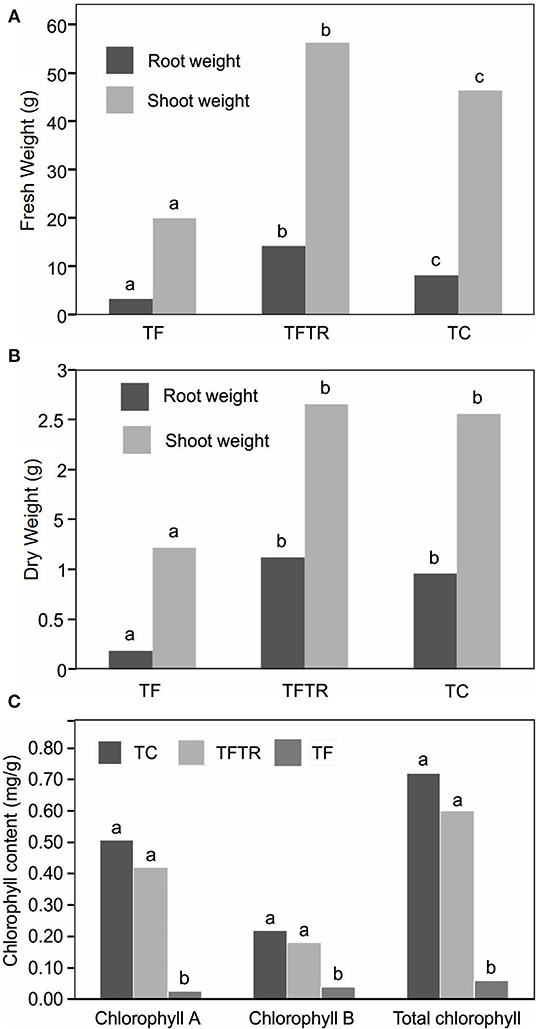
Figure 3. Effect of CSR-T-3 (Trichoderma reesei) on banana root and shoot tissues, (A) fresh weight in “g” and (B) dry weight in “g” under greenhouse pot experiment while (C) shows variations in chlorophyll content. Both fresh and dry weights of shoot and root tissues were found to be enhanced in TFTR indicating growth promotion aided by the treatment in comparison with control. Quite contrarily, the TF plants showed reduced shoot and root weights (fresh and dry) which could be attributed to the wilting and senescence of the tissues. Chlorophyll a and b as well as total chlorophyll content reduced in TFTR and TF over the control TC.
Metabolite Profiling of CSR-T-3 Efficacy Using LC-MS Analysis
The crude metabolites of plants treated with Foc TR4 (TF), Foc TR4, and CSR-T-3 (TFTR), and untreated negative control (TC) were analyzed by LC-MS to detect the production of antimicrobial compounds by CSR-T-3 as well as Fusarium toxins. LC-MS spectral profiles with different retention times and relative intensity (%) facilitated the identification of the compound showing distinct peak values by comparing their identity using similar molecular mass (m/z) values reported for distinct compounds (Figure 4). The compounds identified from LC-MS analysis could be broadly categorized as phenolic esters, antioxidants, fatty acids, fungal toxins, and compounds with antifungal and antibacterial principles. The extracts from the treatment control (TC) showed distinct peaks for fatty acid compound 20-methyl spirolide G (MeG) and soyasapogenol e-3-o-rhamnosyl glucosyl glucuronide (Table 3). The compounds in the crude extract of TFTR treatment with relative abundance was identified as ß-caryophyllene (1,429.9), catechin-o-gallate, soyasapogenol rhamnosyl glucoronide (729), peptaibols (1,788.2), fenigycin (1,462.2), iturin C19 (1,134.9), anthocyanin (1,191.7), and gallocatechin-o-gallate (913.5) (Figures 4C–F). Fungal toxins and metabolites produced by Fusarium spp. like enniatin A, fusarin C, chlamydosporal, etc. were also identified in TFTR treatment with low peak intensity (Figures 4C,D). The extracts of treatment Foc TR4 showed distinct compounds like fusaristatin A, fusarin C, chlamydosporal, and beauveric acid with high peak intensity (Figures 4A,B).
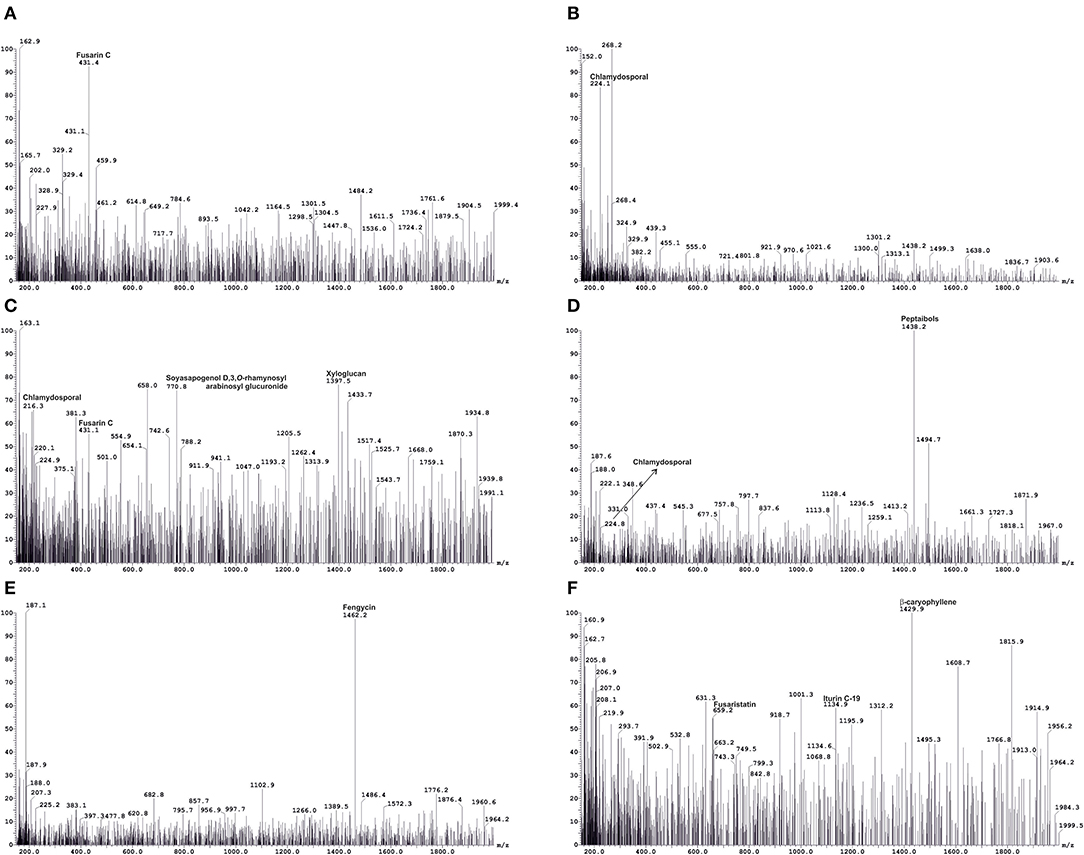
Figure 4. A representation of liquid chromatography-mass spectrometry (LC-MS) analysis used for identification of key metabolites produced by host-pathogen interactions. Metabolite profiling using LC-MS indicating the production of pathogen-related toxins, such as fusarin C and chlamydosporal (A,B) in Foc TR4-infected wilted plants (TF) and detection of antifungal compounds (C–F), such as ß caryophyllene, soyasapogenol D, 3-O-rhamnosyl arabinosyl glucuronide, peptaibols, and fengycinin CSR-T-3 treated banana plants challenged with Foc TR4 (TFTR). Simultaneous detection of pathogen-related toxins at low intensity (C,D) indicating a reduction of toxins and increase in antifungal metabolites under CSR-T-3 TFTR treatment.
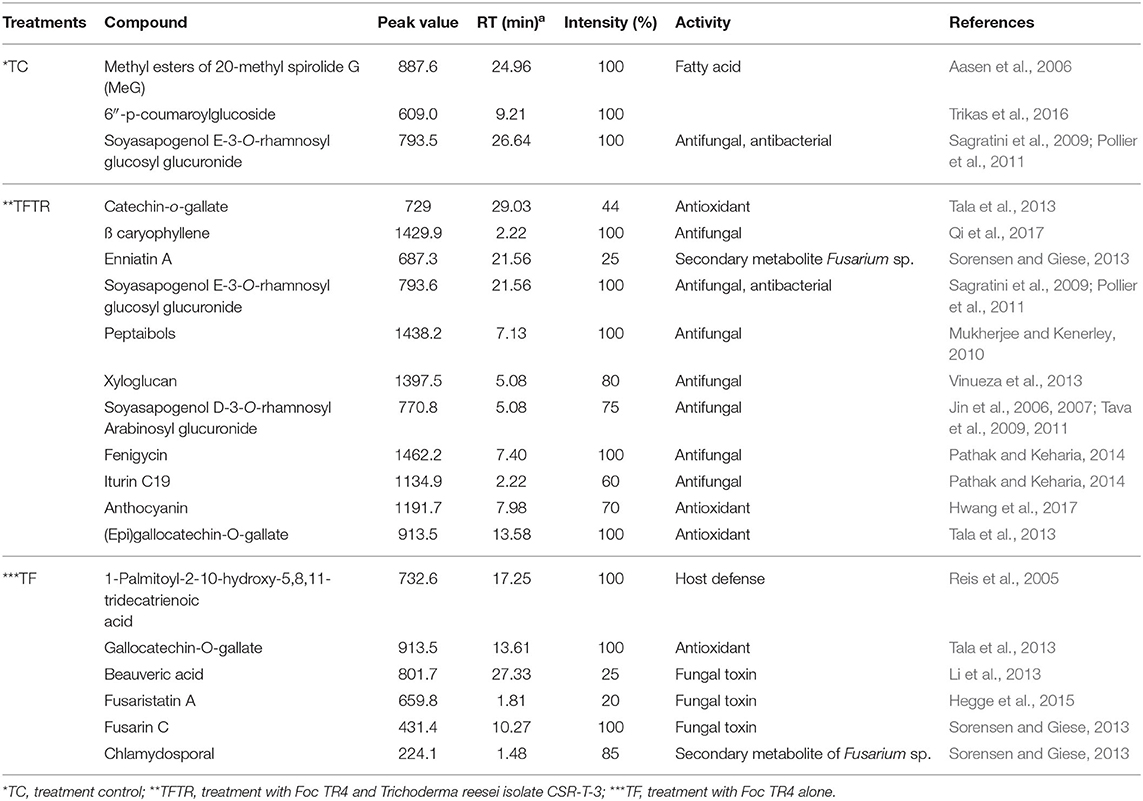
Table 3. Key metabolites identified from LC-MS analysis of crude extracts of the banana root samples under various treatments.
Gene Expression Profiling of Banana Plants With CSR-T-3 Treatment
Gene expression profiling revealed that the expression levels of Fusarium-related genes indicated a clear pattern of upregulation of the Fusaric acid biosynthesis genes in only Fusarium treatments (TF and TFTR) evident from the heat map presented in Figure 5. In the case of Trichoderma mycoparasitism genes (TrCBH1, TrCBH2, TrEGL1, TrXYN1) and signal transduction pathway genes for secondary metabolite production (TrTGA1, TrTMK1, and TrVEL1) a distinct pattern of upregulation was recorded in only the Trichoderma treatments (TFTR and TTR) (Figure 5). The significant upregulation of the Fusaric acid biosynthetic genes in TF and their corresponding downregulation in TFTR confirmed the suppression of Fusarium toxin production by T. reesei. An exception was recorded in the expression levels of the FUB3 gene which showed a trend toward no change in TFTR (Z score-0). Contrarily, the significant downregulation in TTR (i.e., almost no expression) also strongly affirms the absence of the pathogen Foc TR4 and thereby toxin pathway gene expressions are being curtailed. On the other hand, the mycoparasitism-related genes, such as TrCBH1, TrCBH2, TrEGL1, TrXYN1 were found to be significantly upregulated in TFTR and these genes were involved in the breakdown of the Foc cell wall and their components and are being produced by CSR-T-3. Interestingly, the TrCBH1 gene seems to be an inducible gene upon pathogen invasion as seen in the change from Z-score 0 (no change) in TTR toward +1 (significant upregulation) in TFTR. But other genes seemed to exhibit constitutive expression as both TFTR and TTR showed similar levels of upregulation. Some signal transduction pathway genes, such as TrVEL1 and TrTMK1 were found to be significantly upregulated in TFTR which was similar to the pattern exhibited by TTR even though an upstream gene TrTGA1 showed downregulation in TFTR which was observed to be upregulated in TTR.
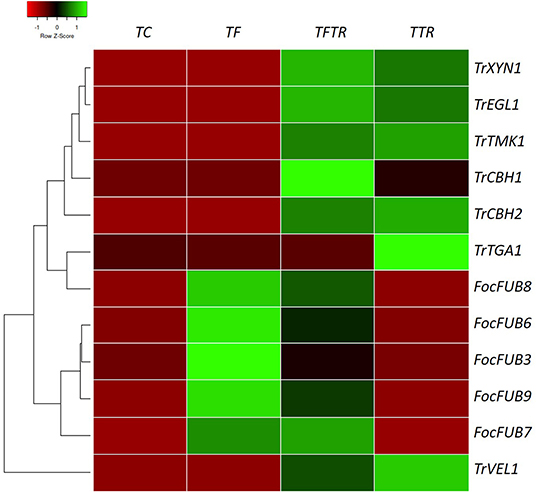
Figure 5. Heat map showing gene expression patterns of key genes involved in fusaric acid biosynthesis by Foc TR4, mycoparasitism, and secondary metabolite production inducing signal transduction pathway genes of Trichoderma reesei in various treatments for establishing the biocontrol mechanism of CSR-T-3. Fusaric acid biosynthesis involves a series of genes, such as FUB3, FUB6, FUB7, FUB8, and FUB9. Fusaric acid is one of the major toxins produced by Fusarium oxysporum f. sp. cubense tropical race 4 and the upregulation of these genes in banana plants challenge-inoculated with Foc TR4 (TF) while downregulation of the same was recorded in TFTR (CSR-T-3-treated banana plants challenge-inoculated with Foc TR4) and TTR (CSR-T-3-treated banana plants without pathogen). Similarly, the genes involved in mycoparasitism (TrCBH1, TrCBH2, TrEGL1, TrXYN1) and those involved in the signal transduction pathway inducing secondary metabolite production (TrVEL1, TrTGA1, TrTMK1) were found to be significantly upregulated in TTR and TFTR. Green color code represents upregulation and red color code indicates downregulation. TC indicates negative control (no pathogen, no biocontrol), TF represents Foc TR4-challenged banana plants, TFTR indicates CSR-T-3-treated banana plants challenged with Foc TR4 and TTR indicates positive control (banana plants with CSR-T-3 (Trichoderma reesei isolate).
Possible Biocontrol Mechanism Elucidated From Gene Expression and LC-MS Analysis
LC-MS and gene expression studies have provided some insights on the probable mode of action which is proposed as a probable mechanism of biocontrol exhibited by the CSR-T-3 Trichoderma reesei isolate against Foc TR4 (Figure 6). The role of Fusaric acid was confirmed from gene expression analysis but was not evident from LC-MS analysis. Fusaric acid is the major toxin reported in many Fusarium species. Fusaric acid biosynthesis involves several enzymes polyketide synthase (FUB1), aminoacid kinase (FUB3), hydrolase (FUB4), acetyl transferase (FUB5), dehydrogenase (FUB8), and proteins of unknown function (FUB2, FUB6s, and FUB9). But in the present study, from the LC-MS analysis, the role of other toxins, such as fusaristatin A, fusarin C, chlamydosporal, and beauveric acid could be established to be present in infected plants. Based on the time and location of the sampling for LC-MS analysis, it is evident that Fusarium-related toxins are accumulated in the Foc TR4-infected plants which are responsible for the discoloration (Figure 6A). The mycelial growth of the pathogen and establishment in the plant tissues caused vascular clogging thereby blocking nutrient-water movement and eventually resulting in the sudden wilting of the plants. The counteracting mechanism of biocontrol agent CSR-T-3 by mycoparasitism and production of antifungal metabolites was established by the results observed in gene expression and LC-MS analysis. The typical signal transduction pathway, especially the genes upstream to the production of secondary metabolites induction, as well as those associated with the antifungal properties were upregulated in both TFTR and TTR. This was supported by the downregulation of Fusarium-related toxins and the upregulation of antifungal compounds in the LC-MS data. Thus, a possible mechanism of biocontrol potential of the CSR-T-3 T. reesei isolate against Foc TR4 has mycoparasitism and antifungal metabolites as key contributors (Figure 6B).
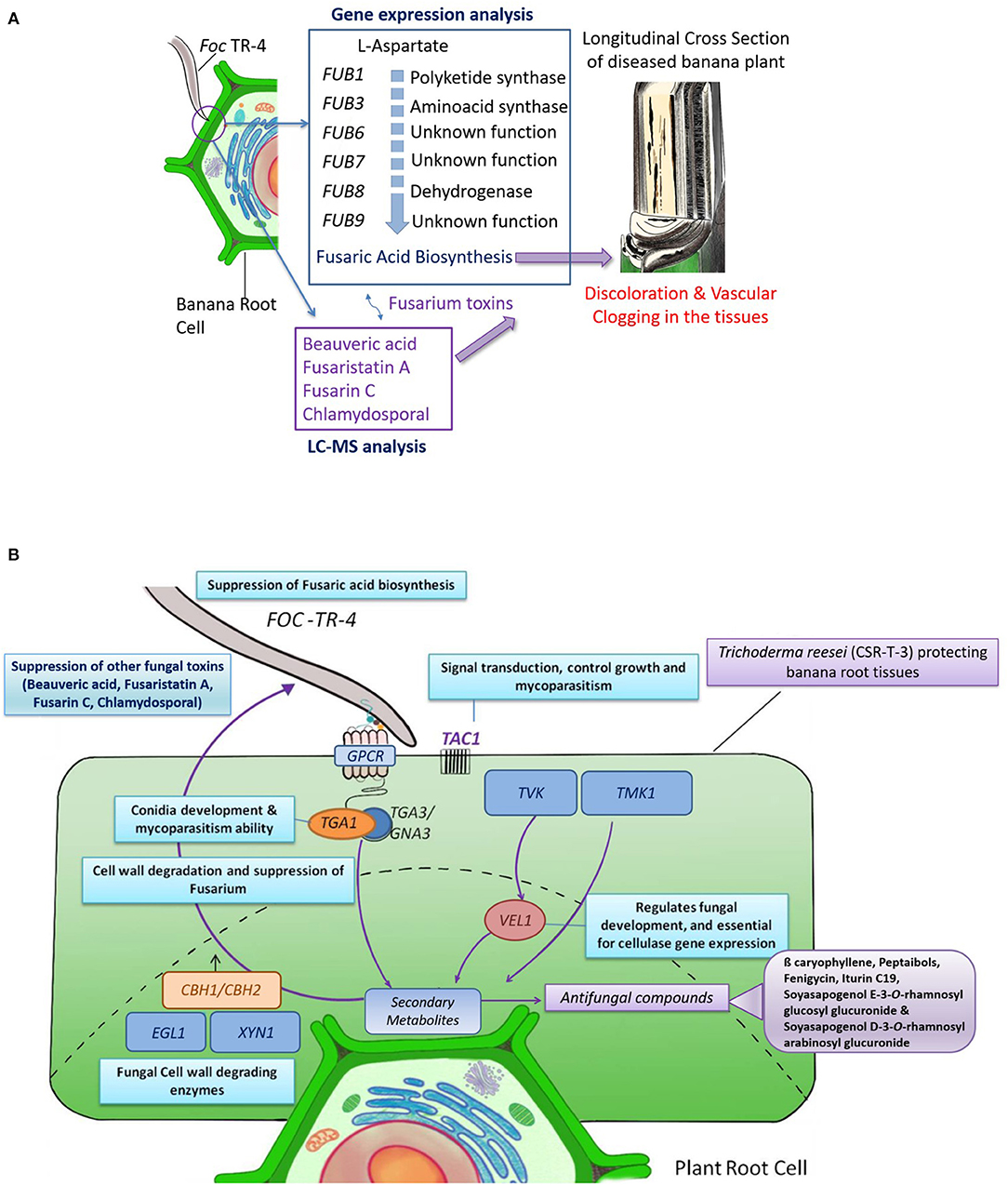
Figure 6. Possible mechanism of CSR-T-3 (Trichoderma reesei) in providing protection against Foc TR4 pathogen in bananas. (A) Mode of infection in the banana plants are induced by the toxins produced by Fusarium oxysporum f. sp. cubense tropical race 4 (Foc TR4) which was evident from the gene expression analysis and LC-MS profiling. The toxins and the mycelial mats results in discoloration and vascular clogging of the plant tissues, the most characteristic symptom of Foc TR4. (B) Mechanism of CSR-T-3 through the expression of mycoparasitism-related genes cellobiohydrolase (CBH1/CBH2), endoglucanse (EGL1), and xylanase (XYL1) genes and the signal pathway that are induced by TMK1 and VEL1 genes are evident from the gene expression studies. Expression of these genes also triggers the production of secondary metabolites especially antifungal compounds, such as ß-caryophyllene, catechin-o-gallate, soya sapogenol rhamnosyl glucoronide, peptaibols, fenigycin, iturin C19, anthocyanin, and gallocatechin-o-gallate. Co-suppression of fusaric acid and other toxins, such as beauveric acid, fusarastatin A, fusarin C, and chlamydosporal during the same time results in conferring tolerance in the banana plants.
Biochemical Analysis of Defense-Related Enzymes and Phenol Content
Stress conditions are known to induce reactive oxygen species (ROS) production thereby resulting in oxidative damage in the cells. This effect can be recorded by assaying and quantifying the ROS scavenging enzymes which may be produced by the host banana plants upon infection with Foc TR4. Sometimes, the biocontrol agents also facilitate production and induction of host defense enzymes. Assaying the defense enzymes in different treatments was completed with the aim of assessing the host response in combination with the CSR-T-3 treatment (Table 4). The accumulation of defense enzymes and phenols was significantly higher in the TFTR treatment (where plants were treated with the pathogenic isolate of Foc TR4 and the T. reesei CSR-T-3 isolate) in comparison with TF (Foc TR4-treated) and TC (negative control with no treatments). The treatments involving T. reesei isolate CSR-T-3 invariably had a higher induction of peroxidase (POD), polyphenol oxidase (PPO), phenylalanine ammonia lyase (PAL), and ß-1, 3-glucanase activity than the TF and TC treatments. Significant induction of chitinase activity (3.160-fold increase in nmol of GlcNAc g−1fw min−1) was observed in T. reesei isolate CSR-T-3 inoculated TFTR treatment while Foc TR4 alone inoculated TF treatment recorded induction but at low levels. The treatments involving T. reesei isolate CSR-T-3 had nearly 1.382 times higher phenolic content than the control plants.
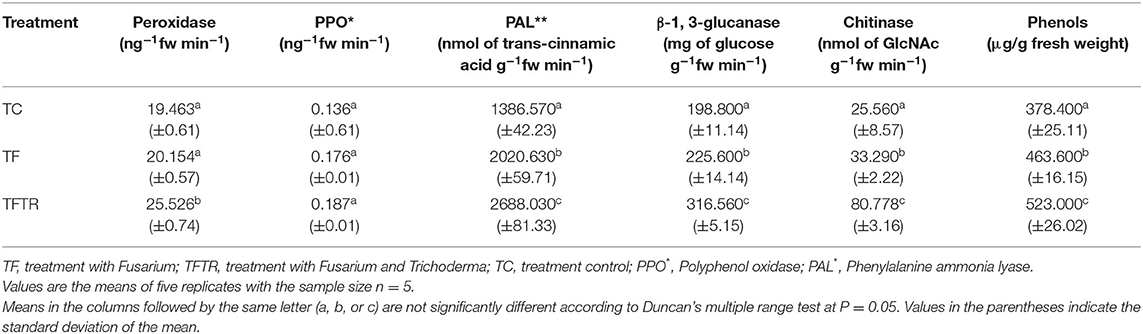
Table 4. Profiling changes in activities of defense enzymes in Trichoderma reesei (CSR-T-3)-inoculated banana plantlets challenged with pathogenic isolate CSR-F-1 of Foc TR-4.
Development of Bioformulation and Field Evaluation of CSR-T-3 (T. reesei) Bioformulation
Bioformulation developed using the T. reesei CSR-T-3 isolate that showed promising results in pot culture studies under greenhouse conditions (Figure 2) in terms of disease suppression was taken to the field level evaluation in the hotspot region of the wilt disease. Efficacy of CSR-T-3 formulation was tested on sick plots in the hot spot area of Sohawal (26° 46′ 3.3852″ N latitude; 81° 59′ 42.7992″ E longitude) where there were severe disease incidences during 2017 and 2018 (Damodaran et al., 2019c). The CSR-T-3 T. reesei isolate was formulated in the form of bioformulation using a loopful of a 2 mm disc in the CSR patent protected bio-media and incubated for 72 h. After 72 h, the plants under the treatment were drenched with 500 ml/plant with 3% of the formulation at 2, 4, 6, and 8 months after planting.
Typical wilting symptoms of yellowing in the older leaves started in the untreated control plants from the 4th month (November) after planting followed by pseudo stem splitting at the 6th month (January) after planting with the disease spreading rapidly over 12–14 months after planting (July-August) corresponding with the flowering and fruiting stages. High disease incidence percentage (46.50%) (Table 5) was recorded in the plants that were not treated with CSR-T-3 T. reesei. However, there was a significant reduction in the disease incidence percentage (10.58%) of the plants treated with Trichoderma isolate CSR-T-3. At harvest, the disease severity index of the Trichoderma-treated plants were significantly lower (1.14) compared with those that were not treated (3.46). The plant height (230.73 cm), girth (43.9 cm), and leaf area (73.13 cm2) of the treated plant group were significantly higher than the other untreated control group. The mean yield (30.57 kg/plant) of the treated plants were significantly higher compared to the untreated plants (7.71 kg). However, there was no significant difference in the number of leaves and number of hands between the treated and untreated plants. There was a significant increase in bunch size and finger size that was attributed to enhanced yield over the diseased plant. The yield under treatment was found to be on par with the average yield of the locality that ranged from 25 to 30 kg/plant. Thus, the drenching of T. reesei isolate CSR-T-3 not only had an antifungal effect in the control of the disease but also had a growth promoting effect in increasing certain growth parameters, such as plant vigor, finger, and bunch size besides yield (bunch weight) which increased 2-fold over untreated control plants.
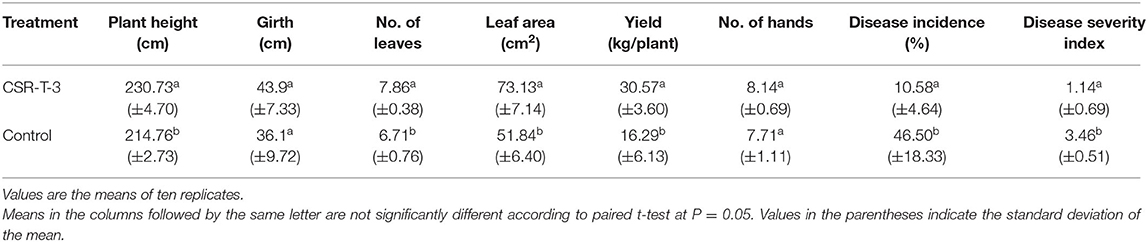
Table 5. Effect of Trichoderma reesei (CSR-T-3) on Fusarium wilt and the growth of banana plantlets under field experiment in sick fields.
Discussion
The global banana production system has been exposed to serious threats in the mid-twentieth century by the devastating wilt disease caused by F. oxysporum f. sp. cubense (Stover, 1962). Popularly known as Panama disease it wiped out the Gros Micheal cultivation in Central America (Ploetz, 1992). The banana industry was sustained with the identification of the Cavendish (AAA) cultivar as a resistant source for the Foc race 1 pathogen. However, the choice varieties of the Indian sub-continent comprising of Pisang Awak (ABB), Rasthali/Amritpani (AAB), and Malbhog (AAB) still remained highly susceptible to the Foc race 1 and are threatened with extinction (Thangavelu et al., 2001). In the early 1990s, a new race was identified in the tropics, in a much more virulent form, devastating the Cavendish cultivars and designated as tropical race 4 (TR4) that causes 70–100% loss (Ploetz and Churchill, 2011) and also economic loss amounting to US$ 388.1 million (FAO, 2017). The perennial nature of the pathosystem complicated the development of sustainable control measures (Ploetz, 2007). The chance of obtaining a tolerant variety was very difficult due to the existence of extremely poor fertility in the Cavendish cultivars (Ortiz, 2013). However, the utilization of antagonistic microorganisms from suppressive soils, such as Pseudomonas, Bacillus, and Trichoderma species can aid in the curtailment of the growth and development of the pathogen (Deng et al., 2013). Even among the antagonistic microbes, Trichoderma has been widely exploited as the potential biocontrol agent owing to their distinct properties in the control of soil borne disease and non-eco-toxic nature of the genus (Benitez et al., 2004). The fast growing and mycoparasitism ability of the Trichoderma isolates place them as potential antagonistic microbes (Monfil and Casas-Flores, 2014; Sellamani et al., 2016). Prominent inhibitory effects of different Trichoderma isolates against Fusarium diseases by mycotoxin production were established and indicated the further need of their ability through field validation (Tian et al., 2018).
Success of biological control in the field depends on the correct identification of potential strains and application in the field (Chaves et al., 2011). Therefore, isolation and screening of the highly efficient microorganisms form the base of biocontrol. An attempt was made to assess the efficacy of three Trichoderma isolates that were isolated from the rhizosphere of biotic (banana wilt suppressive) and abiotic (banana salt stressed) stress eco-system (Damodaran and Mishra, 2016; Damodaran et al., 2019c). These antagonists exert differential inhibition potential to the pathogen growth that was evident from the in vitro assay and the dominating role of disease suppression with isolate CSR-T-3 was successfully demonstrated in the in vivo pot culture study. PGPR strains, such as Pseudomonas fluorescens PF-1 and Trichoderma species TRC-54 were earlier reported to have controlled the mycelia growth of Foc race 1 of banana under a dual culture study through the direct action of enzymes and metabolites produced by PGPR (O'Sullivan et al., 1992; Nagarajkumar et al., 2004). CSR-T-3 was the first strain of Trichoderma reesei isolated from the salt affected rhizosphere to show disease suppression against the banana Foc TR4. The antifungal property was further demonstrated in this study using a pot and field experiment. Application of the isolate successfully reduced the disease severity index significantly when compared to Foc TR4-infected plants and also promoted the growth of the banana crop successfully. Earlier, there were successful reports on the biological control of Fusarium wilt disease in bananas using bacterial strains like bacterial isolate ITBB B5 1 (Tan et al., 2015), Streptomyces strain CB75 (Chen et al., 2018), and Bacillus flexus strain Typr1 (Thangavelu and Gopi, 2015) under pot experiments. But none of these reports have indicated field level application and validation of these strains for commercial use. We report here the first successful report of a fungal strain CSR-T-3 controlling Foc TR4 causing wilt disease in bananas at a field level besides pot culture studies.
Trichoderma spp. are known to produce a wide range of bioactive secondary metabolites that are known to have antifungal, antibacterial, and toxic properties to control a wide range of phytopathogens, such as Alternaria alternata, Botrytis cinerea, Fusarium spp., Pythium spp., Rhizoctonia solani, Sclerotinia sclerotiorum, and Ustilago maydis (Hyder et al., 2017; Sood et al., 2020). Trichoderma spp. is reported as an inexhaustible source of antibiotic derivatives (from the acetaldehydes to alpha-pyrones) terpenes, polyketides, isocyanide derivatives, piperacines, and complex families of peptaibols (Keszler et al., 2000; Sood et al., 2020). Therefore, in order to characterize and profile the metabolites, LCMS analysis was performed which aided in the identification of microbial metabolites who were produced by both the pathogen (CSR-F-1) and the biocontrol agent (CSR-T-3). The treatment involving Foc TR4 challenge inoculation and T. reesei isolate CSRT-3 showed the production of a wide range of bioactive antifungal and antioxidant metabolites with high intensity m/z peaks. They include peptaibols, fenigycin, xyloglucan, soyasapanogenol rhamnosyl glucuronide, iturin C, ß-caryophyllene, and antioxidants like catechin-O-galate. The presence of Fusarium related metabolites like fusaristatin, fusarin C, chlamydosporal, enniatin C, etc. were also observed but the intensity of the m/z peaks were comparatively lower than the antifungal and antioxidant metabolites in this treatment. However, significant production of Fusarium-related metabolites like Fusarin C with a high intensity m/z peak was observed in the TF treatment where only the Foc TR4 isolate was inoculated. This implies that there is a suppression of toxins produced by Foc TR4 in the presence of the antifungal metabolites produced by T. reesei isolate CSR-T-3 in this present study. Peptaibols are small peptides of α-aminoisobutyric acid (Aib) produced by the genus Trichoderma that are economically important for their anti-microbial and anti-cancer properties as well as their ability to induce systemic resistance in plants against microbial invasion (Mukherjee et al., 2013). The involvement of these compounds in mycoparasitism established with their role in induced systemic resistance in plants by both apoptosis and autophagy suggest their capability in preventing the invasion of root-invading fungi (Schuster and Schmoll, 2010). The intensity of mass peaks assigned for iturin C and fengycin in the TFTR treatment in our study explicitly show the high antifungal potency of the T. reseei CSR-T-3 isolate. The cyclic heptapeptide iturin with ß-amino fatty acid was known to have high hemolytic and antifungal properties while fengycin, a cyclic depsipeptide with 10 amino acids and a ß-hydroxy fatty acid tail, is known for its excellent antifungal potential against filamentous fungi (Winkelmann et al., 1983; Vanittanakom et al., 1986). Pathak and Keharia (2014), reported iturin C and fengycin as an antifungal compound produced by Bacillus subtilis. Phenolic compounds have crucial roles in inducing the plant defense mechanism through free radical termination by providing hydrogen to reduce free radicals. The presence of phenolic compounds, such as gallocatechin-o-gallate in our LCMS fractions in TFTR and TF treatment signifies the activities of plant defense systems when exposed to biotic stress. The bacterization of tomato plants with beneficial strains showed antifungal activity through the deposition of phenolic compounds and restricting the growth of F. oxysporum f. sp. lycopersici (M'Piga et al., 1997).
Our study has demonstrated the possibility of the increase in the plant defense system through an increase in the activities of phenols and enzymes like peroxidase (POD) and phenylalanine ammonia lyase (PAL). Peroxidase is a defense enzyme implicated in the last enzymatic step of lignin biosynthesis that oxidizes the hydroxyl cinnamyl alcohols into free radicals that are later coupled with lignin polymer (Gross, 1980). POD is known to inhibit the spore germination and mycelia growth of some pathogenic fungi like Pseudocercospora abelmoschi and Pseudocercospora cruenta (Joseph et al., 1998). Enhanced POD and PAL activities were reported by Kavino et al. (2007) in bacterial strain-treated banana plantlets that showed tolerance to Banana bunchy top virus. PAL is an enzyme that plays an important role in the biosynthesis of defense chemicals in phenyl propanoid metabolism (Daayf et al., 1997). Another prominent group of enzymes and compounds that are commonly correlated with the tolerance mechanism exhibited by the Trichoderma spp. are ß-glucanase and chitinase (Chet, 1987). ß-glucans are polysaccharides that stimulate the host immune response on interaction (Garfoot et al., 2014). The higher activity of ß-glucanase and chitinase in the T. reesei-treated plants was observed in biochemical assays of our study which could be another additional mechanism attributing to increasing the host immune response to the pathogen. Radjacommare et al. (2004) assessed the chitinoyltic enzyme against Rhizoctonia solani in rhizobacteria-treated rice which induced a defense response in the host by bacterial co-inoculants of Pseudomonas sp. Higher activity of ß-glucanase and chitinase was present in F. oxysporum f. sp. cubense race 1 tolerant cultivars and hybrids of bananas (Kavino et al., 2009). Previous reports by Migheli et al. (1996) on the biocontrol of P. debaryanum by Arthrobacter spp. elucidated the role of glucanase and protease in the lysis of mycelium.
The genome of T. reesei has been reported to possess genes encoding for 2 cellobiohydrolases, 5 endo-ß-1,4-glucanases, and different isoforms of ß-glucosidases, hemicellulases, and accessory enzymes (Martinez et al., 2008; Hakkinen et al., 2012; Dita et al., 2018). CBH encodes for exo-cellobiohydrolases or exoglucanases which are involved in the hydrolysis of 1,4-beta-D-glucosidic bonds in the external structure of cellulose and aid in cell wall degradation (Antonieto et al., 2014; Nogueira et al., 2015). CBH1 is reported to be an inducible gene in the presence of inducers, such as cellulose, sephorose, lactose, and cellobiose (Dos Santos et al., 2014; Nogueira et al., 2015); which was confirmed in the present study as pathogen interaction with CSR-T-3 induced the expression. On the contrary, endo-β-1,6-glucanase is one of the important genes for mycoparastic activity of Trichoderma sp which catalyzes cell wall degradation by breaking β-1,6 linkages internally (De la Cruz et al., 1995; Gruber and Seidl-Seiboth, 2011). In the present study, the upregulation of CBH1/CBH2, EGL1, and XYN1 genes in TFTR have ascertained the mycoparasitism activity of CSR-T-3. Other signal transduction pathway genes were also found to be upregulated in the treatments involving CSR-T-3. The TGA1 gene encodes for a G protein α subunit, which is involved in mycoparasitic coiling, conidiation (Rocha-Ramirez et al., 2002), formation of chitinase, and the production of antifungal metabolites (Reithner et al., 2005). VEL1 controls the regulation of the pheromone system, mating partner sensing, and sexual and asexual development (Bazafkan et al., 2015). The VEL1 gene encodes for velvet protein 1, a part of the VELVET complex composed of LAE1, VEL1/VELA, and VEL2/VELB proteins. This VEL1 protein is also responsible for morphogenetic traits, is a key regulator of biocontrol in T. virens, and is highly conserved in different species of Trichoderma (Supplementary Figure 2) (Mukherjee and Kenerley, 2010). It is also reported to have a role in the induction of cellulase gene family expression which includes cellobiohydrolases, glucanases, chitinases, and xylanases (Karimi Aghcheh et al., 2016). The TMK1 gene encodes for a mitogen activated protein kinase (MAPK) which plays an important role in mycoparasitism by the regulation of mycoparasitic host attack, formation of aerial hyphae, and conidiation (Reithner et al., 2007; Schmoll, 2008; Swain and Mukherjee, 2020). Generally, MAPK are involved in signal transduction pathways that converge into the accumulation of secondary metabolites. Earlier, Reithner et al. (2007) reported that tmk1 deletion caused reduced mycoparasitic activity against Rhizoctonia solani and Botrytis cinerea and were also associated with the regulation of production 6-pentyl-a-pyrone and peptaibol antibiotics. The upregulation of the TGA1, TMK1, and VEL1 genes indicate the possibility of enhanced chitinase activity. This information also supports the result of high chitinase and phenol activities in TFTR and TTR treatments in the study.
Biocontrol mechanisms of CSR-T-3 (T. reesei) against Fusarium wilt pathogen Foc TR4 (CSR-F-1) are considered to be mainly because of mycoparasitism, the production of antifungal metabolites, such as iturin C, ß-carophyllene etc., and a reduction in the fungal toxins like fusaristatin A, fusarin C, beauveric acid, and chlamydosporal production (Figure 6). This well-established concept of biocontrol reported to occur in Trichoderma species was confirmed by gene expression profiling of the fusaric acid biosynthesis and cellulolytic enzymes of Fusarium species and T. reesei, respectively. Trichoderma species colonize the epidermal root surface and also induce the plant to produce secondary metabolites that include compounds with low molecular weight inducing the expression of genes involved in the defense response against the pathogen (Hermosa et al., 2012; Malmierca et al., 2014; Sood et al., 2020). Earlier whole genome transcriptome analysis indicated that defense genes associated with CEBiP, BAK1, NB-LRR proteins, PR proteins, transcription factor, and cell wall lignification were expressed strongly in resistant variety Yueyoukang 1 indicating that these genes play important roles against Foc TR4 infection in bananas (Bai et al., 2013).
Developing a bioformulation using the CSR-T-3 T. reesei isolate based on the antagonistic effect on the pathogen and the epidemiology of the disease in the hotspot region, we standardized the application schedule and mode of application. A field evaluation of CSR-T-3 bioformulation in the sick plots of a hot spot area has clearly demonstrated the successful control of the pathogen which is evident from the reduced disease incidence at the same time contributed to the successful management of the disease. Earlier, Damodaran et al. (2019b) demonstrated successful control of Foc TR4 using ICAR-FUSICONT. To date, no product is available in India for biological control of Fusarium wilt disease Foc TR4 and this study is the first of its kind to find evidence for successful management.
Conclusion
The current gaps in devising management schedules for Fusarium wilt disease include the scarcity of resistant varieties, no effective chemical control measure, the lack of a virulent biological control agent that could work at field level, and also their clear mode of action. Our study has been a solution to the latter category of biological control of Fusarium wilt disease of bananas caused by Foc TR4. We report here an effective virulent CSR-T-3 isolate of T. reesei that was successfully proven in a pot culture experiment and field evaluation in sick fields of hot spot areas of plants affected by Foc TR4. This biocontrol agent has been proven to have a good effect on the fruit yield and weight besides the suppression of the pathogen. The mechanism of mycoparasitism and reduction in toxin accumulation was also elucidated using gene expression and LC-MS analysis which has established the role of cell wall degrading enzymes. Signal transduction pathways inducing secondary metabolites with antifungal properties in Trichoderma has resulted in the suppression of toxins produced by Foc TR4 and thus conferred induced tolerance to banana plants. Field evaluation of the CSR-T-3 showed the potential of this bioagent in the management of the banana Fusarium wilt disease on a large scale. This study is a holistic approach to the technological advancement of T. reesei from lab to field in the management of Fusarium disease in bananas.
Data Availability Statement
All raw data analyzed supporting this article will be made available by the authors. Public domain genome sequence data sets were used for primer designing and the accession numbers are provided in the article.
Author Contributions
TD conceived, planned, and designed the experiments. SR designed the statistical methodology and analyzed the data. MM designed the gene expression studies, designed primers for expression analysis, and contributed equally to the manuscript preparation with TD. RG and KY performed the experiments of dual culture, pot, and field experiment. SK performed the molecular wet lab studies. IA performed the molecular identification of isolates. NK assisted in the disease screening experiments. SKJ and VKM performed the soil analysis of the pot soil used in the screening trial. All authors contributed to the article and approved the submitted version.
Funding
This work has been initially supported from Application of Microorganisms in Agriculture and Allied Sectors (AMAAS) project operated at ICAR-Central Institute of Soil Salinity Research, Regional Research Station, Lucknow and Flagship project of Indian Council of Agricultural Research operated at ICAR Central Institute for Subtropical Horticulture, Lucknow and then further supported by financial aid from Extra mural fund (EMF).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The Director General, ICAR was acknowledged for critically evaluating the entire program and guiding us toward perfection. The authors thank Dr. A. K. Singh, Deputy Director General, Indian Council of Agricultural Research (ICAR) for providing financial support under the ICAR Extramural Project for taking up the research work and also for motivating the execution of the work. Thanks are due to the Director, ICAR-Central Soil Salinity Research Institute, Karnal, India for providing all logistic support in conducting the experiment. The Director, ICAR-National Institute of Agriculturally Important Micro-organism, Mau, Uttar Pradesh, India, was acknowledged for partial funding under the AMAAS network scheme for the execution of the initial microbiological studies. We thank Dr. N. K. Krishnakumar, Bioversity International, Dr. B. K. Pandey, ADG in-charge (Horticultural Sciences-II), ICAR, and Dr. S. Uma, Director, ICAR-National Research Centre of Banana for their guidance and motivation in the execution of the program. Also, we thank Dr. P. Chakrabarthy, ADG (Plant Protection) for providing critical input into the characterization of Trichoderma reesei. We dedicate the work in memory of the late Dr. R. B. Rai who was the inspiring force for this work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.595845/full#supplementary-material
References
Aasen, J. A. B., Hardstaff, W., Aune, T., and Quilliam, M. A. (2006). Discovery of fatty acid ester metabolites of spirolide toxins in mussels from Norway using liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 20, 1531–1537. doi: 10.1002/rcm.2501
Aguilar Moran, J. F. (2013). Improvement of Cavendish Banana Cultivars through conventional breeding. Acta Hortic. 986, 205–208. doi: 10.17660/ActaHortic.2013.986.21
Amorim, E. P., dos Santos-Serejo, J. A., Amorim, V. B. O., Ferreira, C. F., and Silva, S. O. (2013). Banana breeding at Embrapa cassava and fruits. Acta Hortic. 986, 171–176. doi: 10.17660/ActaHortic.2013.986.18
Antonieto, A. C. C., dos Santos Castro, L., Silva-Rocha, R., Persinoti, G. F., and Silva, R. N. (2014). Defining the genome-wide role of CRE1 during carbon catabolite repression in Trichoderma reesei using RNA-Seq analysis. Fungal Genet. Biol. 73, 93–103. doi: 10.1016/j.fgb.2014.10.009
Bai, T. T., Xie, W. B., Zhou, P. P., Wu, Z. L., and Xiao, W. C. (2013). Transcriptome and expression profile analysis of highly resistant and susceptible banana roots challenged with Fusarium oxysporum f. sp. cubense tropical race 4. PLoS ONE 8:e73945. doi: 10.1371/journal.pone.0073945
Bazafkan, H., Dattenböck, C., Böhmdorfer, S., Tisch, D., Stappler, E., and Schmoll, M. (2015). Mating type-dependent partner sensing as mediated by VEL1 in Trichoderma reesei. Mol. Microbiol. 96, 1103–1118. doi: 10.1111/mmi.12993
Benitez, T., Rincon, A. M., Limon, M. C., and Codon, A. C. (2004). Biocontrol mechanism of Trichoderma strains. Int. Microbiol. 7, 249–260.
Boller, T., and Mauch, F. (1988). Colorimetric assay for chitinase. Methods Enzymol. 161, 430–435. doi: 10.1016/0076-6879(88)61052-4
Buddenhagen, I. (2009). Understanding strain diversity in Fusarium oxysporum f. sp. cubense and history of introduction of ‘tropical race 4’ to better manage banana production. Acta Hortic. 828, 193–204. doi: 10.17660/ActaHortic.2009.828.19
Carvalhais, L. C., Henderson, J., Rincon-Florez, V. A., O'Dwyer, C., Czislowski, E., Aitken, E. A. B., et al. (2019). Diagnostics of banana Fusarium wilt targeting secreted-in-xylem genes. Front. Plant Sci. 10:547. doi: 10.3389/fpls.2019.00547
Chaves, J. Q., Pires, E. S., and Vivoni, A. M. (2011). Genetic diversity, antimicrobial resistance and toxigenic profiles of Bacillus cereus isolated from food in Brazil over three decades. Int. Food Microbiol. 147, 12–16. doi: 10.1016/j.ijfoodmicro.2011.02.029
Chen, Y., Zhou, D., Qi, D., Gao, Z., Xie, J., and Luo, Y. (2018). Growth promotion and disease suppression ability of a Streptomyces sp. CB-75 from banana rhizosphere soil. Front. Microbiol. 8:2704. doi: 10.3389/fmicb.2017.02704
Chet, I. (1987). “Trichoderma-application, mode of action, and potential as biocontrol agent of soil borne plant pathogenic fungi.” in Innovative Approaches to Plant Disease Control, ed I. Chet (New York, NY: John Wiley & Sons), 137–160.
Daayf, F., Schmitt, A., and Belenger, R. R. (1997). Evidence of phytoalexins in cucumber leaves infected with powdery mildew following treatment with leaf extracts of Reynoutria sachalinesis. Plant Physiol. 113:719727. doi: 10.1104/pp.113.3.719
Damodaran, T. (2018). “Banana production in subtropics–strategies, challenges and research initiatives.” in National Conference on Strategies and Challenges in Doubling Farmer's Income Through Horticultural Technologies in Subtropics (ICAR-Central Institute for Sub-tropical Horticulture) (Lucknow), 99–101.
Damodaran, T., Gopal, R., Yadav, K., Mishra, V. K., and Jha, S. K. (2019c). “CSR-T-3 (Trichoderma reesei)–a potential cellulolytic and proteolytic isolate used for inducing tolerance to biotic and abiotic stress in commercial agri-horticulture crops,” in Souvenir on A Golden Jubilee International Salinity Conference: Resilient Agriculture in Saline Environment Under Changing Climate: Challenges and Opportunities (Karnal), 86–87.
Damodaran, T., Kumar, N., and Kavino, M. (2009). Breeding and evaluation of Musa hybrids resistant to Fusarium oxysporum f. sp. cubense race 1. Fruits 64, 1–16. doi: 10.1051/fruits/2008044
Damodaran, T., and Mishra, V. K. (2016). “Rhizosphere engineering: an innovative approach for sustainable crop production in sodic soils,” in Innovative Saline Agriculture, eds J. Dagar, P. Sharma, D. Sharma, and A. Singh (New Delhi: Springer Publications), 105–117. doi: 10.1007/978-81-322-2770-0_5
Damodaran, T., Rajan, S., Gopal, R., Yadav, A., Yadav, K., Shukla, P. K., et al. (2019b). Successful community based management of banana wilt caused by Fusarium oxysporum f. sp. cubense Tropical race-4 through ICAR- FUSICONT. J. Appl. Hortic. 21, 37–41. doi: 10.37855/jah.2019.v21i01.06
Damodaran, T., Rajan, S., Mishra, V. K., Jha, S. K., Gopal, R., and Ahmad, I. (2018). First report of Fusarium wilt in banana caused by Fusarium oxysporum f. sp. cubense tropical race 4 in India. Plant Dis. 103:1022. doi: 10.1094/PDIS-07-18-1263-PDN
Damodaran, T., Rajan, S., Mishra, V. K., Jha, S. K., Gopal, R., and Sharma, P. C. (2019a). “ICAR-FUSICONT–an innovative technology for the control of Fusarium wilt (TR4) of banana var. Grand Naine in normal and reclaimed sodic soils,” in Souvenir on A Golden Jubilee International Salinity Conference: Resilient Agriculture in Saline Environment Under Changing Climate: Challenges and Opportunities (Karnal), 153.
De la Cruz, J., Pintor-Toro, J. A., Benítez, T., and Llobell, A. (1995). Purification and characterization of an endo-beta-1, 6-glucanase from Trichoderma harzianum that is related to its mycoparasitism. J. Bacteriol. 177, 1864–1871. doi: 10.1128/JB.177.7.1864-1871.1995
de Matos, A. P., Cordeiro, Z. J. M., de Oliveira e Silva, S., Amorim, E. P., and Ferreira, D. M. V. (2011). Reaction of diploid (AA) and tetraploid (AAAB) banana hybrids to Fusarium wilt under field conditions. Acta Hortic. 897, 387–390. doi: 10.17660/ActaHortic.2011.897.53
Deng, X., Li, Q. F., Hou, X. W., and Wu, C. Y. (2013). Soil microbial functional diversity from different infection grades of banana Fusarium wilt (Fusarium oxysporum f. sp. cubense). Appl. Mech. Mater. 2301, 2274–2280. doi: 10.4028/www.scientific.net/AMM.295-298.2274
Dita, M., Barquero, M., Heck, D., Mizubuti, E. S. G., and Staver, C. P. (2018). Fusarium wilt of Banana: current knowledge on epidemiology and research needs toward sustainable disease management. Front. Plant Sci. 9:1468. doi: 10.3389/fpls.2018.01468
Dong, F., Wang, S., Yu, M., Sun, Y., Xu, J., and Shi, J. (2017). Natural occurrence of deoxynivalenol and deoxynivalenol-3-glucoside in various wheat cultivars grown in Jiangsu Province, China. World Mycotoxin J. 10, 285–293. doi: 10.3920/WMJ2016.2158
Dos Santos, C. L., Pedersoli, W. R., Antonieto, A. C. C., Steindorff, A. S., Silva-Rocha, R., Martinez-Rossi, N. M., Rossi, A., et al. (2014). Comparative metabolism of cellulose, sophorose and glucose in Trichoderma reesei using high-throughput genomic and proteomic analyses. Biotechnol. Biofuels 7:41. doi: 10.1186/1754-6834-7-41
Druzhinina, I. S., Shelest, E., and Kubicek, C. P. (2012). Novel traits of Trichoderma predicted through the analysis of its secretome. FEMS Microbiol. Lett. 337, 1–9. doi: 10.1111/j.1574-6968.2012.02665.x
Garcez, M., Santos Martins, J. A., and Rodrigues, E. J. R. (2016). Evaluation of different banana genotypes for resistance to panama disease. Biosci. J. 32, 431–435. doi: 10.14393/BJ-v32n2a2016-29818
Garfoot, A. L., Dearing, K. L., VanSchoiak, A. D., Wysocki, V. H., and Rappleye, C. A. (2014). Eng1 and Exg8 are the major β-Glucanases secreted by the fungal pathogen Histoplasma capsulatum. J. Biol. Chem. 292, 4801–4810. doi: 10.1074/jbc.M116.762104
Getha, K., and Vikineswary, S. (2002). Antagonistic effects of Streptomyces violaceusniger strain G10 on Fusarium oxysporum f. sp. cubense race 4: indirect evidence for the role of antibiosis in the antagonistic process. J. Ind. Microbiol. Biot. 28, 303–310. doi: 10.1038/sj.jim.7000247
Gonçalves, Z. S., Haddad, F., de Oliveira Amorim, V. B, Ferreira, C., F., de Oliveira, S., and Amorim, E. P. (2019). Agronomic characterization and identification of banana genotypes resistant to Fusarium wilt race 1. Eur. J. Plant Pathol. 155, 1093–1103. doi: 10.1007/s10658-019-01837-5
Gross, G. G. (1980). The biochemistry of lignification. Adv. Bot. Res. 8, 25–63. doi: 10.1016/S0065-2296(08)60032-4
Gruber, S., and Seidl-Seiboth, V. (2011). Self versus non-self: fungal cell wall degradation in Trichoderma. Microbiology 158, 26–34. doi: 10.1099/mic.0.052613-0
Guo, L., Yang, L., Liang, C., Wang, G., Dai, Q., and Huang, J. (2015). Differential colonization patterns of Bananas (Musa spp.) by physiological Race 1 and Race 4 isolates of Fusarium oxysporum f. sp. cubense. J. Phytopathol. 163, 807–817. doi: 10.1111/jph.12378
Hakkinen, M., Arvas, M., Oja, M., Aro, N., Penttila, M., Saloheimo, M., et al. (2012). Re-annotation of the CAZy genes of Trichoderma reesei and transcription in the presence of lignocellulosic substrates. Microb. Cell Fact. 11:134. doi: 10.1186/1475-2859-11-134
Hammerschmidt, R., Nuckles, E. M., and Kuc, J. (1982). Association of enhanced peroxidase activity with induced systemic resistance of cucumber of Colletotrichum lagenarium. Physiol. Plant Pathol. 20, 73–82. doi: 10.1016/0048-4059(82)90025-X
Harman, G. E., and Kubicek, C. P. (1998). Trichoderma and Gliocladium: Enzymes, Biological Control and Commercial Applications. Vol. 2. London: CRC Press.
Hegge, A., Lonborg, R., Nielsen, D. M., and Sorensen, J. L. (2015). Factors influencing production of Fusaristatin A in Fusarium graminearum. Metabolites 5, 184–191. doi: 10.3390/metabo5020184
Hermosa, R., Belén Rubio, M., Cardoza, R. E., Nicolás, C., Monte, E., and Gutiérrez, S. (2013). The contribution of Trichoderma to balancing the costs of plant growth and defense. Int. Microbiol. 16, 69–80. doi: 10.2436/20.1501.01.181
Hermosa, R., Viterbo, A., Chet, I., and Monte, E. (2012). Plant-beneficial effects of Trichoderma and of its genes. Microbiology 158, 17–25. doi: 10.1099/mic.0.052274-0
Hwang, J. W., Natarajan, S. B., Kim, Y. S., Kim, E. K., Lee, J. W., Moon, S. H., et al. (2017). Biosynthesis of oligomeric anthocyanins from grape skin extracts. Molecules 22:497. doi: 10.3390/molecules22030497
Hyder, S., Inam-ul-Haq, M., Bibi, S., Malik, A., Ghuffar, S., and Iqbal, S. (2017). Novel potential of Trichoderma Spp. as biocontrol agent. J. Entomol. Zool. Stud. 5, 214–222.
Jin, M. C., Yang, Y. W., Su, B. G., and Ren, Q. L. (2006). Rapid quantification and characterization of soyasaponins by high-performance liquid chromatography coupled with electrospray mass spectrometry. J. Chromatogr. 1108, 31–37. doi: 10.1016/j.chroma.2005.12.099
Jin, M. C., Yang, Y. W., Su, B. G., and Ren, Q. L. (2007). Determination of soyasaponins Ba and Bb in human serum by high performance liquid chromatography coupled with electrospray ionization tandem mass spectrometry. J. Chromatogr. 846, 169–175. doi: 10.1016/j.jchromb.2006.08.043
Joseph, L. M., Koon, T. K., and Man, W. S. (1998). Antifungal effects of hydrogen peroxide and peroxidase on spore germination and mycelial growth of Pseudocercospora species. Can. J. Bot. 76, 2119–2124. doi: 10.1139/b98-166
Karimi Aghcheh, R., Németh, Z., Atanasova, L., Fekete, E., Paholcsek, M., Sándor, E., et al. (2016). The VELVET an orthologue VEL1 of Trichoderma reesei regulates fungal development and is essential for cellulase gene expression. PLoS ONE 9:e112799. doi: 10.1371/journal.pone.0112799
Kavino, M., Harish, S., Kumar, N., Saravanakumar, D., Damodaran, T., Soorianathasundaram, K., and Samiyappan, R. (2007). Rhizosphere and endophytic bacteria for induction of systemic resistance of banana plantlets against bunchy top virus. Soil Biol. Biochem. 39, 1087–1098. doi: 10.1016/j.soilbio.2006.11.020
Kavino, M., Kumar, N., Damodaran, T., Harish, S., and Saravanakumar, D. (2009). Biochemical markers as a useful tool for the early identification of Fusarium oxysporumf. sp. cubense, race 1 resistance banana clones. Arch. Phytopathol. Pflanzenschutz. 42, 1069–1078. doi: 10.1080/03235400701622089
Keszler, A., Forgacs, E., Kotai, L., Vizcaíno, J. A., Monte, E., and García-Acha, I. (2000). Separation and identification of volatile components in the fermentation broth of Trichoderma atroviride by solid-phase extraction and gas chromatography-mass spectroscopy. J. Chromatogr. Sci. 38, 421–424. doi: 10.1093/chromsci/38.10.421
Khan, N., Hidalgo, P. M., Ice, T. A., Maymon, M., Humm, E. A., Nejat, N., et al. (2018). Antifungal activity of Bacillus species against Fusarium and analysis of the potential mechanisms used in biocontrol. Front. Microbiol. 9:2363. doi: 10.3389/fmicb.2018.02363
Kumar, V., Lee, J. D., Clark, R. J., and Woodruff, T. M. (2018). Development and validation of a LC-MS/MS assay for pharmacokinetic studies of complement C5a receptor antagonists PMX53 and PMX205 in mice. Sci. Rep. 8:8101. doi: 10.1038/s41598-018-26387-4
Li, C., Zuo, C., Deng, G., Kuang, R., Yang, Q., Hu, C., et al. (2013). Contamination of bananas with beauvericin and fusaric acid produced by Fusarium oxysporum f. sp. cubense. PLoS ONE 8:e70226. doi: 10.1371/journal.pone.0070226
López-Díaz, C., Rahjoo, V., Sulyok, M., Ghionna, V., Martín-Vicente, A., Capilla, J., et al. (2017). Fusaric acid contributes to virulence of Fusarium oxysporum on plant and mammalian hosts. Mol. Plant Pathol. 19, 440–453. doi: 10.1111/mpp.12536
Magdama, F., Monserrate-Maggi, L., Serrano, L., Sosa, D., Geiser, D. M., Jiménez-Gasco, M., et al. (2019). Comparative analysis uncovers the limitations of current molecular detection methods for Fusarium oxysporum f. sp. cubense race 4 strains. PLoS ONE 14:e0222727. doi: 10.1371/journal.pone.0222727
Malmierca, M. G., Barua, J., Mccormick, S. P., Izquierdo-Bueno, I., Cardoza, R. E., Alexander, N. J., et al. (2014). Novel aspinolide production by Trichoderma arundinaceum with a potential role in Botrytis cinerea antagonistic activity and plant defence priming. Environ. Microbiol. 17, 1103–1118. doi: 10.1111/1462-2920.12514
Martinez, D., Berka, R. M., Henrissat, B., Saloheimo, M., Arvas, M., Baker, S. E., et al. (2008). Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotech. 26, 553–560. doi: 10.1038/nbt1403
Martínez-Hidalgo, P., García, J. M., and Pozo, M. J. (2015). Induced systemic resistance against Botrytis cinerea by Micromonospora strains isolated from root nodules. Front. Microbiol. 6:922. doi: 10.3389/fmicb.2015.00922
Martínez-Medina, A., Fernandez, I., Sánchez-Guzmán, M. J., Jung, S. C., Pascual, J. A., and Pozo, M. J. (2013). Deciphering the hormonal signaling network behind the systemic resistance induced by Trichoderma harzianum in tomato. Front. Plant Sci. 4:206. doi: 10.3389/fpls.2013.00206
Matarese, F., Sarrocco, S., Gruber, S., Seidl-Seiboth, V., and Vannacci, G. (2012). Biocontrol of Fusarium head blight: interactions between Trichoderma and mycotoxigenic Fusarium. Microbiology 158, 98–106. doi: 10.1099/mic.0.052639-0
Mayer, A. M., Harel, E., and Shaul, R. B. (1965). Assay of catechol oxidase, a critical comparison of methods. Phytochemistry 5, 783–789. doi: 10.1016/S0031-9422(00)83660-2
Migheli, Q., Friard, O., Del Tedesco, D., Musso, M. R., and Gullino, M. L. (1996). Stability of transformed antagonistic Fusarium oxysporum strains in vitro and in soil microcosms. Mol. Ecol. 5, 641–649. doi: 10.1111/j.1365-294X.1996.tb00359.x
Monfil, V. O., and Casas-Flores, S. (2014). “Molecular mechanisms of biocontrol in Trichoderma spp. and their applications in agriculture,” in Biotechnology and Biology of Trichoderma, eds V. K. Gupta, M. Schmoll, A. Herrera-Estrella, R. S. Upadhyay, I. Druzhinina, and M. G. Tuohy (Dordrecht: Elsevier), 429–453. doi: 10.1016/B978-0-444-59576-8.00032-1
M'Piga, P., Bélanger, R. R., Paulitz, T. C., and Benhamou, N. (1997). Increased resistance to Fusarium oxysporum f. sp. radicis-lycopersiciin tomato plants treated with the endophytic bacterium Pseudomonas fluorescens strain 63-28. Physiol. Mol. Plant Pathol. 50, 301–320. doi: 10.1006/pmpp.1997.0088
Mukherjee, P. K., Horwitz, B. A., Herrera-Estrella, A., Schmoll, M., and Kenerley, C. M. (2013). Trichoderma research in the genome era. Annu. Rev. Phytopathol. 51, 105–129. doi: 10.1146/annurev-phyto-082712-102353
Mukherjee, P. K., and Kenerley, C. M. (2010). Regulation of morphogenesis and biocontrol properties in Trichoderma virens by a VELVET protein, Vel1. Appl. Environ. Microbiol. 76, 2345–2352. doi: 10.1128/AEM.02391-09
Nagarajkumar, M., Bhaaskaran, R., and Velazhahan, R. (2004). Involvement of secondary metabolites and extracellularlytic enzymes produced by Pseudomonas fluorescens in inhibition of Rhizoctonia solani, the rice sheath of blight pathogen. Microbiol. Res. 159, 73–81. doi: 10.1016/j.micres.2004.01.005
Niehaus, E. M., Díaz-Sánchez, V., von Bargen, K. W., Kleigrewe, K., Humpf, H. U., Limón, M. C., et al. (2014). “Fusarins and fusaric acid in fusaria,” in Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites, Fungal Biology, eds Martín J. F., García-Estrada C., and Zeilinger S (New York, NY: Springer), 239–262. doi: 10.1007/978-1-4939-1191-2_11
Nogueira, K. M. V., Costa, M. d. N., de Paula, R. G., Mendonça-Natividade, F. C., Ricci-Azevedo, R., and Silva, R. N. (2015). Evidence of cAMP involvement in cellobiohydrolase expression and secretion by Trichoderma reesei in presence of the inducer sophorose. BMC Microbiol. 15:195. doi: 10.1186/s12866-015-0536-z
Ortiz, R. (2013). Conventional banana and plantain breeding. Acta Hortic. 986, 177–194. doi: 10.17660/ActaHortic.2013.986.19
O'Sullivan, M., Stephens, P. M., and O'Gara, F. (1992). Extracellular protease production by fluorescent Pseudomonas spp. and the colonization of sugar beet roots and soil. Soil Biol. Biochem. 23, 623–627. doi: 10.1016/0038-0717(91)90074-T
Pan, S. Q., Ye, X. S., and Kuc, J. (1991). Association of β-1,3-glucanase activity and isoform pattern with systemic resistance to blue mold in tobacco induced by stem injection with Peronospora tabacina or leaf inoculation with tobacco mosaic virus. Physiol. Mol. Plant Pathol. 39, 25–39. doi: 10.1016/0885-5765(91)90029-H
Papavizas, G. (1985). Trichoderma and Gliocladium: biology, ecology, and potential for biocontrol. Annu. Rev. Phytopathol. 23, 23–54. doi: 10.1146/annurev.py.23.090185.000323
Pathak, K. V., and Keharia, H. (2014). Application of extracellular lipopeptide biosurfactant produced by endophytic Bacillus subtilis K1 isolated from aerial roots of banyan (Ficus benghalensis) in microbially enhanced oil recovery (MEOR). 3 Biotech. 4, 41–48. doi: 10.1007/s13205-013-0119-3
Pérez-Vicente, L., Dita, M. A., and Martínez-De La, P. E. (2014). Technical Manual: Prevention and Diagnostic of Fusarium Wilt (Panama disease) of Banana Caused by Fusarium oxysporum f.sp.Cubense Tropical Race 4 (TR4). Roma: FAO, 75.
Perincherry, L., Lalak-Kańczugowska, J., and Stepień, L. (2019). Fusarium-produced mycotoxins in plant-pathogen interactions. Toxins (Basel) 11:664. doi: 10.3390/toxins11110664
Ploetz, R. C. (1992). “Fusarium wilt of banana (Panama disease),” in Plant Diseases of International Importance, Vol. III, eds A. N. Mukhopadhyay, H. S. Chaube, J. Kumar, and U. S. Singh (Upper Saddle River, NJ: Prentice Hall), 270–282.
Ploetz, R. C. (2004). “Biological control of Fusarium wilt: a review and an evaluation,” in Abstract Booklet, International Congress on Musa: Harnessing Research to Improve Livelihoods (Penang: INIBAP Bioversity International), 141.
Ploetz, R. C. (2006a). Fusarium wilt of banana is caused by several pathogens referred to as Fusarium oxysporum f. sp. cubense. Phytopathology 96, 653–656. doi: 10.1094/PHYTO-96-0653
Ploetz, R. C. (2006b). Panama disease: an old nemesis rears its ugly head. Part 2. The Cavendish era and beyond. Plant Health Prog. 7:36. doi: 10.1094/APSnetFeature-2005-1005
Ploetz, R. C. (2007). Diseases of tropical perennial crops: challenging problems in diverse environments. Plant Dis. 91, 644–663. doi: 10.1094/PDIS-91-6-0644
Ploetz, R. C., and Churchill, A. C. L. (2011). Fusarium wilt: the banana disease that refuses to go away. Acta Hortic. 897, 519–526. doi: 10.17660/ActaHortic.2011.897.73
Ploetz, R. C., Haynes, J. L., and Vazquez, A. (1999). Responses of new banana accessions in South Florida to Panama disease. Crop Protec. 18, 445–449. doi: 10.1016/S0261-2194(99)00043-5
Ploetz, R. C., Herbert, J., Sebasigari, K., Hernandez, J. H., Pegg, K. G., Ventura, J. A., et al. (1990). “Importance of Fusarium wilt in different banana-growing regions,” in Fusarium Wilt of Banana, ed R. C. Ploetz (St. Paul, MN: APS Press), 9–26.
Ploetz, R. C., and Pegg, K. G. (2000). “Fusarium wilt.” in Diseases of banana, Abaca and Enset, ed R. Jones (Wallingford: CABI Publishing), 1–34.
Pollier, J., Morreel, K., and Geelen, D. (2011). Metabolite profiling of triterpene saponins in medicago truncatula hairy roots by liquid chromatography fourier transform ion cyclotron resonance mass spectrometry. J. Nat. Products 74, 1462–1476. doi: 10.1021/np200218r
Qi, D., Zou, L., Zhou, D., Feng, R., Gao, Z., and Zhang, X. (2017). Isolation, identification of strain GA1-2 and its antifungal activity against Fusarium oxysporum f. sp. cubense. J. Plant Prot. 44, 809–816.
Qian, Y., Zhong, L., Sun, Y., Sun, N., Zhang, L., Liu, W., et al. (2019). Enhancement of cellulase production in Trichoderma reesei via disruption of multiple protease genes identified by comparative secretomics. Front. Microbiol. 10:2784. doi: 10.3389/fmicb.2019.02784
Qiu, J., Xu, J., and Shi, J. (2019). Fusarium toxins in Chinese wheat since the 1980s. Toxins (Basel) 11:248. doi: 10.3390/toxins11050248
Radjacommare, R., Kandan, A., Nandakumar, R., and Samiyapan, R. (2004). Association of the hydrolytic enzyme chitinase against Rhizoctonia solani in rhizobacteria treated rice plants. J. Phytopathol. 152, 365–370. doi: 10.1111/j.1439-0434.2004.00857.x
Rai, R. B., Damodaran, T., Kannan, R., Rathore, A. P., Sharma, D. K., Mishra, V. K., et al. (2012). Low cost multiplication technology of salt tolerant bio-growth enhancers (Bacillus, Pseudomonads and Trichoderma) for increasing productivity of agri-horti crops in normal and sodic soils. J. Plant Interact. 9, 577–584. doi: 10.1080/17429145.2013.873958
Rangaswamy, R. (1995). A Textbook Book of Agricultural Statistics. New Delhi: New Age International (P) Ltd.
Rebouças, T. A., Haddad, F., Ferreira, C. F., de Oliveira, S. A. S., da Silva Ledo, C. A., and Amorim, E. P. (2018). Identification of banana genotypes resistant to Fusarium wilt race 1 under field and greenhouse conditions. Sci Hortic. 239, 308–313. doi: 10.1016/j.scienta.2018.04.037
Reis, A., Domingues, M. R. M., Amado, F. M. L., Ferrer-Correia, A. J. V., and Domingues, P. (2005). Separation of peroxidation products of diacyl-phosphatidylcholines by reversed-phase liquid chromatography–mass spectrometry. Biomed. Chromatogr. 19, 129–137. doi: 10.1002/bmc.429
Reithner, B., Brunner, K., Schuhmacher, R., Peissl, I., Seidl, V., Krska, R., et al. (2005). The G protein α subunit Tga1 of Trichoderma atroviride is involved in chitinase formation and differential production of antifungal metabolites. Fungal Genet. Biol. 42, 749–760. doi: 10.1016/j.fgb.2005.04.009
Reithner, B., Schuhmacher, R., Stoppacher, N., Pucher, M., Brunner, K., and Zeilinger, S. (2007). Signaling via the Trichoderma atroviride mitogen-activated protein kinase Tmk1 differentially affects mycoparasitism and plant protection. Fungal Genet Biol. 44, 1123–1133. doi: 10.1016/j.fgb.2007.04.001
Ribeiro, L. R., Silva, S. d. O., Oliveira, S. A. S. d., Amorim, E. P., Serejo, J. A. S. S., and Haddad, F. (2018). Sources of resistance to Fusarium oxysporum f. sp. cubense in banana germplasm. Rev. Brasil. Fruticult. 40:e-202. doi: 10.1590/0100-29452018202
Rocha-Ramirez, V., Omero, C., Chet, I., Horwitz, B. A., and Herrera-Estrella, A. (2002). Trichoderma atroviride G-protein alpha-subunit gene tga1 is involved in mycoparasitic coiling and conidiation. Eukaryot. Cell 1, 594–605. doi: 10.1128/EC.1.4.594-605.2002
Ross, W. W., and Sederoff, R. R. (1992). Phenylalanine ammonia-lyase from Loblolly Pine. Plant Physiol. 98, 380–386. doi: 10.1104/pp.98.1.380
Sadasivam, S., and Manickam, A. (1996). Biochemical Methods. 2nd Edn. New Delhi: New Age International (P) Ltd.
Sagratini, G., Zuo, Y., Caprioli, G., Cristalli, G., Giardina, D., Maggi, F., et al. (2009). Quantification of soyasaponins I and βg in Italian lentil seeds by solid-phase extraction (SPE) and high-performance liquid chromatography-mass spectrometry (HPLC-MS). J. Agric. Food Chem. 57, 11226–11233. doi: 10.1021/jf901707z
Schmoll, M. (2008). The information highways of a biotechnological workhorse-signal transduction in Hypocrea jecorina. BMC Genomics. 9:430. doi: 10.1186/1471-2164-9-430
Schuster, A., and Schmoll, M. (2010). Biology and biotechnology of Trichoderma. Appl. Microbiol. Biot. 87, 787–799. doi: 10.1007/s00253-010-2632-1
Sellamani, M., Kalagatur, N. K., Siddaiah, C., Mudili, V., Krishna, K., Natarajan, G., et al. (2016). Antifungal and zearalenone inhibitory activity of Pediococcus pentosaceus isolated from dairy products on Fusarium graminearum. Front. Microbiol. 7:890. doi: 10.3389/fmicb.2016.00890
Sood, M., Kapoor, D., Kumar, V., Sheteiwy, M. S., Ramakrishnan, M., Landi, M., et al. (2020). Trichoderma: the “Secrets” of a multitalented biocontrol agent. Plants 9:762. doi: 10.3390/plants9060762
Sorensen, J. L., and Giese, H. (2013). Influence of carbohydrates on secondary metabolism in Fusarium avenaceum. Toxins 5, 1655–1663. doi: 10.3390/toxins5091655
Speckbacher, V., and Zeilinger, S. (2018). “Secondary metabolites of mycoparasitic fungi,” in Secondary Metabolites–Sources and Applications, ed R. Vijayakumar, and S. S. S. Raja (London: Intechopen). doi: 10.5772/intechopen.75133
Stover, R. H. (1962). Fusarium Wilt (Panama disease) of Bananas and Other Musa Species. London: Common wealth Mycological Institute.
Swain, H., and Mukherjee, A. K. (2020). “Host–pathogen–trichoderma interaction,” in Trichoderma, Rhizosphere Biology, eds A. Sharma and P. Sharma (Singapore: Springer). doi: 10.1007/978-981-15-3321-1_8
Tala, V. R. S., Silva, V. C., Rodrigues, C. M., Nkengfack, A. E., Santos, L. C., and Vilegas, W. (2013). Characterization of proanthocyanidins from Parkia biglobosa (Jacq.) G. Don. (Fabaceae) by flow injection analysis-electrospray ionization ion trap tandem mass spectrometry and liquid chromatography/electrospray ionization mass spectrometry. Molecules 18, 2803–2820. doi: 10.3390/molecules18032803
Tan, D., Fu, L., Han, B., Sun, X., Zheng, P., and Zhang, J. (2015). Identification of an endophytic antifungal bacterial strain isolated from the rubber tree and its application in the biological control of banana Fusarium wilt. Plos ONE 10:e0131974. doi: 10.1371/journal.pone.0131974
Tava, A., Mella, M., Avato, P., Biazzi, E., Pecetti, L., Bialy, Z., et al. (2009). New triterpenic saponins from the aerial parts of Medicago arabica (L.) huds. J. Agric. Food Chem. 57, 2826–2835. doi: 10.1021/jf8036984
Tava, A., Pecetti, L., Romani, M., Mella, M., and Avato, P. (2011). Triterpenoid glycosides from the leaves of two cultivars of Medicago polymorpha L. J. Agric. Food Chem. 59, 6142–6149. doi: 10.1021/jf2005854
Thangavelu, R., and Gopi, M. (2015). Field suppression of Fusarium wilt disease in banana by the combined application of native endophytic and rhizospheric bacterial isolates possessing multiple functions. Phytopathol. Mediter. 54, 241–252. doi: 10.14601/Phytopathol_Mediterr-15160
Thangavelu, R., Palaniswami, A., Ramakrishnan, G., Doraiswamy, S., Muthukrishnan, S., and Velazhahan, R. (2001). Involvement of fusaric acid detoxification by Pseudomonas fluorescens strain Pf10 in the biological control of Fusarium wilt of banana caused by Fusarium oxysporum f. sp cubense. J. Plant Dis. Prot. 108, 433–445.
Tian, Y., Tan, Y., Liu, N., Yan, Z., Liao, Y., Chen, J., et al. (2016). Detoxification of deoxynivalenol via glycosylation represents novel insights on antagonistic activities of Trichoderma when confronted with Fusarium graminearum. Toxins 8:335. doi: 10.3390/toxins8110335
Tian, Y., Tan, Y., Yan, Z., Liao, Y., Chen, J., Boevre, M. D., Saeger, S. D. and Wu, A. (2018). Antagonistic and detoxification potentials of Trichoderma isolates for control of Zearalenone (ZEN) producing Fusarium graminearum. Front. Microbiol. 8:2710. doi: 10.3389/fmicb.2017.02710
Ting, A. S. Y., Meon, S., Kadir, J., Radu, S., and Singh, G. (2009). Induced host resistance by non-pathogenic Fusarium endophyte as a potential defense mechanism in Fusarium wilt management of banana. Pest Technol. 3, 67–72.
Trikas, E. D., Papi, R. M., Kyriakidis, D. A., and Zachariadi, G. A. (2016). A sensitive LC-MS method for anthocyanins and comparison of by products and equivalent wine content. Separations 3:18. doi: 10.3390/separations3020018
Vanittanakom, N., Loeffler, W., Koch, U., and Jung, G. (1986). Fengycin-a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29-3. J. Antibiot. 39, 888–901. doi: 10.7164/antibiotics.39.888
Vinueza, N. R., Gallardo, V. A., Klimek, J. F., Carpita, N. C., and Kenttamaa, H. I. (2013). Analysis of xyloglucans by ambient chloride attachment ionization tandem mass spectrometry. Carbohydr. Polym. 98, 1203–13. doi: 10.1016/j.carbpol.2013.06.070
Winkelmann, G., Allgaier, H., Lupp, R., and Jung, G. (1983). Iturin AL-a NEW long chain iturin a possessing an unusual high content of C16-β-amino acids. J. Antibiot. 36, 1451–1457. doi: 10.7164/antibiotics.36.1451
Yadav, K., Damodaran, T., Kumari, N., Dutt, K., Gopal, R., and Muthukumar, M. (2020). Characterization of Trichoderma isolates and assessment of antagonistic potential against Fusarium oxysporum f. sp. cumini. J. Appl. Hortic. 22, 38–44. doi: 10.37855/jah.2020.v22i01.08
Keywords: Trichoderma reesei, banana, Fusarium wilt TR4, antifungal, LC-MS, gene expression, field evaluation
Citation: Damodaran T, Rajan S, Muthukumar M, Ram Gopal, Yadav K, Kumar S, Ahmad I, Kumari N, Mishra VK and Jha SK (2020) Biological Management of Banana Fusarium Wilt Caused by Fusarium oxysporum f. sp. cubense Tropical Race 4 Using Antagonistic Fungal Isolate CSR-T-3 (Trichoderma reesei). Front. Microbiol. 11:595845. doi: 10.3389/fmicb.2020.595845
Received: 17 August 2020; Accepted: 26 October 2020;
Published: 16 December 2020.
Edited by:
Lívia Kmetzsch, Federal University of Rio Grande do Sul, BrazilReviewed by:
Chetan Keswani, Banaras Hindu University, IndiaM. Carmen Limon, Sevilla University, Spain
Copyright © 2020 Damodaran, Rajan, Muthukumar, Ram Gopal, Yadav, Kumar, Ahmad, Kumari, Mishra and Jha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muthukumar Manoharan, muthukumarbt@gmail.com; Thukkaram Damodaran, damhort73@gmail.com
 Thukkaram Damodaran
Thukkaram Damodaran Shailendra Rajan2
Shailendra Rajan2 Muthukumar Manoharan
Muthukumar Manoharan Sandeep Kumar
Sandeep Kumar Nidhi Kumari
Nidhi Kumari