- 1Key Laboratory for Forest Resources Conservation and Utilization in the Southwest Mountains of China, Ministry of Education, Southwest Forestry University, Kunming, China
- 2College of Biodiversity Conservation, Southwest Forestry University, Kunming, China
Wood-inhabiting fungi play a significant role in wood degradation and the cycle of matter in the ecological system. In the present study, three new wood-inhabiting fungal species, Trechispora bambusicola, Trechispora fimbriata, and Trechispora fissurata spp. nov., are nested in Trechispora, which are proposed based on a combination of morphological features and molecular evidence. Sequences of internal transcribed spacer (ITS) and large subunit (nLSU) regions of the studied samples were generated, and the phylogenetic analyses were performed with maximum likelihood, maximum parsimony, and Bayesian inference methods. The phylogenetic analyses inferred from ITS showed that T. bambusicola was sister to Trechispora stevensonii, T. fimbriata grouped with Trechispora nivea, and T. fissurata grouped with Trechispora echinospora. The phylogenetic tree based on ITS + nLSU sequences demonstrated that T. bambusicola formed a single lineage and then grouped with Trechispora rigida and T. stevensonii. T. fimbriata was sister to T. nivea. T. fissurata grouped with Trechispora thelephora.
Introduction
Trechispora P. Karst. (Hydnodontaceae, Trechisporales) was typified with Trechispora onusta P. Karst. (Karsten, 1890). It is characterized by the resupinate to effused basidiomata with smooth to hydnoid to poroid hymenophore, a monomitic or dimitic hyphal structure with clamped generative hyphae having typical ampullaceous septa, and short cylindric basidia and smooth to verrucose or aculeate basidiospores (Karsten, 1890; Bernicchia and Gorjón, 2010). About 49 species are currently known in Trechispora worldwide (Liberta, 1966, 1973; Larsson, 1994, 1995, 1996; Ryvarden, 2002; Trichiès and Schultheis, 2002; Miettinen and Larsson, 2006; Ordynets et al., 2015; Xu et al., 2019) and Index Fungorum1 and MycoBank2.
Larsson (2007) addressed the classification of corticioid fungi and revealed that Trechispora farinacea (Pers.) Liberta grouped with Trechispora hymenocystis (Berk. and Broome) K.H. Larss., in which both species nested within the family Hydnodontaceae Jülich. Based on the large subunit nuclear ribosomal RNA gene (nLSU) datasets, Albee-Scott and Kropp (2010) supported to transfer Hydnodon thelephorus (Lév.) Banker to Trechispora as Trechispora thelephora (Lév.) Ryvarden. The order Trechisporales was studied employing the internal transcribed spacer (ITS) and nLSU regions, in which it suggested that Porpomyces Jülich, Sistotremastrum J. Erikss., Subulicystidium Parmasto, and Trechispora belonged to a highly supported clade and Trechispora belongs to Hydnodontaceae and was closely related to Brevicellicium K.H. Larss. and Hjortstam (Telleria et al., 2013). A phylogenetic study of Trechispora was addressed and demonstrated that Trechispora cyatheae Ordynets, Langer and K.H. Larss. and Trechispora echinocristallina Ordynets, Langer and K.H. Larss. clustered into Trechispora as new members, inferred from the combined data of the ITS and LSU datasets (Ordynets et al., 2015). The phylogeny of Trechisporales was inferred from a combined dataset of ITS-nLSU sequences and showed that Porpomyces, Scytinopogon Singer, and Trechispora grouped together and nested within family Hydnodontaceae (Liu et al., 2019). Phylogram generated from analysis of ITS sequence dataset of Trechispora showed that Trechispora echinospora Telleria was sister to the clade formed by Trechispora araneosa (Hohn. and Litsch.) K.H. Larss., T. farinacea, T. hymenocystis, and Trechispora mollusca (Pers.) Liberta with a low support (Phookamsak et al., 2019). The ITS + nLSU dataset comprised 22 species and revealed that Trechispora yunnanensis C.L. Zhao formed a monophyletic lineage within Trechispora and was closely related to Trechispora byssinella (Bourdot) Liberta and Trechispora laevis K.H. Larss. (Xu et al., 2019).
During the studies on wood-inhabiting fungi in southern China, three species of Trechispora could not be assigned to any described species. Obtaining sequences from the new taxa, the authors examine taxonomy and phylogeny of three new species within the genus Trechispora, based on the ITS and nLSU sequences.
Materials and Methods
Morphology
The studied specimens are deposited at the herbarium of Southwest Forestry University (SWFC), Kunming, Yunnan Province, China. Macromorphological descriptions were based on field notes. Color terms follow Petersen (1996). Micromorphological data were obtained from the dried specimens and observed under a light microscope following Dai (2012). The following abbreviations were used: KOH = 5% potassium hydroxide, CB = Cotton Blue, CB− = acyanophilous, IKI = Melzer’s reagent, IKI− = both inamyloid and indextrinoid, L = mean spore length (arithmetic average for all spores), W = mean spore width (arithmetic average for all spores), Q = variation in the L/W ratios between the studied specimens, n (a/b) = number of spores (a) measured from given number (b) of specimens, spore measurements do not include ornamentation.
Molecular Phylogeny
Cetyltrimethylammonium bromide (CTAB) rapid plant genome extraction kit-DN14 (Aidlab Biotechnologies Co., Ltd., Beijing, China) was used to obtain genomic deoxyribonucleic acid (DNA) from dried specimens, according to the manufacturer’s instructions following Zhao and Wu (2017). ITS region was amplified with primer pair ITS5 and ITS4 (White et al., 1990). Nuclear LSU region was amplified with primer pair LR0R and LR73. The polymerase chain reaction (PCR) procedures for ITS and nLSU following Zhao and Wu (2017). The PCR products were purified and directly sequenced at Kunming Tsingke Biological Technology Limited Company, Kunming, Yunnan Province, China. All newly generated sequences were deposited at GenBank (Table 1).
Sequencher 4.6 (GeneCodes, Ann Arbor, United States) was used to edit the DNA sequence. Sequences were aligned in MAFFT 74 using the “G-INS-I” strategy and manually adjusted in BioEdit (Hall, 1999). The sequence alignment was deposited in TreeBase (submission ID 25879). Sequences of Fibrodontia alba Yurchenko and Sheng H. Wu and Fibrodontia gossypina Parmasto retrieved from GenBank were used as an outgroup in the ITS + nLSU analyses by following Ordynets et al. (2015).
Maximum parsimony (MP) analyses were applied to the ITS + nLSU dataset sequences. Approaches to phylogenetic analysis followed Zhao and Wu (2017), and the tree construction procedure was performed in PAUP∗ version 4.0b10 (Swofford, 2002). All characters were equally weighted and gaps were treated as missing data. Trees were inferred using the heuristic search option with tree-bisection reconnection (TBR) branch swapping and 1000 random sequence additions. Max-trees were set to 5000, branches of zero length were collapsed, and all parsimonious trees were saved. Clade robustness was assessed using a bootstrap (BT) analysis with 1000 replicates (Felsenstein, 1985). Descriptive tree statistics tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated for each Maximum Parsimonious Tree generated. Datamatrix was also analyzed using maximum likelihood (ML) approach with RAxML-HPC2 through the Cipres Science Gateway with GTR + I + G molecular evolution model5 (Miller et al., 2009). Branch support (BS) for ML analysis was determined by 1000 BT replicates.
MrModeltest 2.3 (Nylander, 2004) was used to determine the best-fit evolution model (GTR + I + G) for each data set for Bayesian inference (BI) of the phylogeny. BI was calculated with MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003). Four Markov chains were run for two runs from random starting trees for 1 million generations and trees were sampled every 100 generations; the first one-fourth of generations were discarded as burn-in. A majority rule consensus tree of all remaining trees was calculated. Branches were considered as significantly supported if they received ML BT values > 75%, MP BT values > 75%, or Bayesian posterior probabilities (PP) > 0.95.
Results
Molecular Phylogeny
In the ITS dataset, the sequences from 43 fungal specimens representing 25 species were included. The dataset had an aligned length of 1034 characters, of which 521 characters are constant, 86 are variable and parsimony-uninformative, and 427 are parsimony-informative. MP analysis yielded 26 equally parsimonious trees (TL = 2048, CI = 0.4561, HI = 0.5439, RI = 0.6174, RC = 0.2816). Best model for the ITS dataset estimated and applied in the Bayesian analysis: GTR + I + G, lset nst = 6, rates = invgamma; prset statefreqpr = dirichlet (1,1,1,1). Bayesian analysis and ML analysis resulted in a similar topology to MP analysis, with an average standard deviation of split frequencies = 0.009985 (BI).
The phylogeny (Figure 1) inferred from ITS sequences showed that Trechispora bambusicola was sister to Trechispora stevensonii (Berk. and Broome) K.H. Larss, and Trechispora fimbriata grouped with Trechispora nivea. T. fissurata grouped with T. echinospora Telleria, M. Dueñas, I. Melo, and M.P. Martín.

Figure 1. Maximum Parsimony strict consensus tree illustrating the phylogeny of three new species and related species in Trechispora based on ITS sequences. Branches are labeled with maximum likelihood bootstrap values > 70%, parsimony bootstrap proportion values > 50%, and Bayesian posterior probabilities > 0.95, respectively.
In the ITS + nLSU dataset, it included sequences from 49 fungal specimens representing 27 species. The dataset had an aligned length of 2256 characters, of which 1387 characters are constant, 188 are variable and parsimony-uninformative, and 681 are parsimony-informative. MP analysis yielded 100 equally parsimonious trees (TL = 2811, CI = 0.4963, HI = 0.5037, RI = 0.6409, RC = 0.3180). Best model for the ITS dataset estimated and applied in the Bayesian analysis: GTR + I + G, lset nst = 6, rates = invgamma; prset statefreqpr = dirichlet (1,1,1,1). Bayesian analysis and ML analysis resulted in a similar topology to MP analysis, with an average standard deviation of split frequencies = 0.009991 (BI).
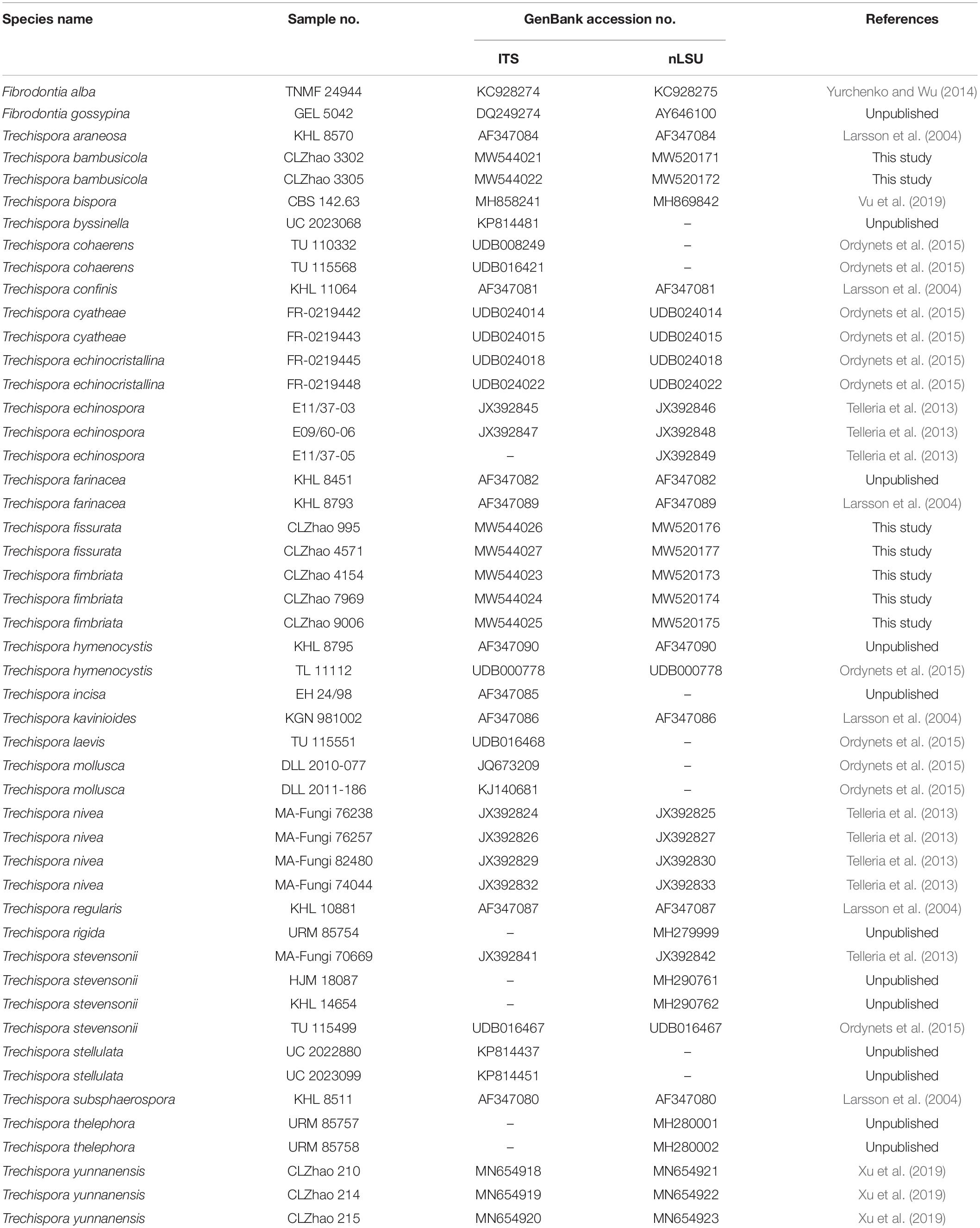
The phylogenetic tree (Figure 2) inferred from ITS + nLSU sequences demonstrated 27 species of Trechispora and revealed that T. bambusicola formed a single lineage and then grouped with Trechispora rigida (Berk.) K.H. Larss. and T. stevensonii. T. fimbriata was sister to T. nivea. T. fissurata grouped with T. thelephora (Lév.) Ryvarden.
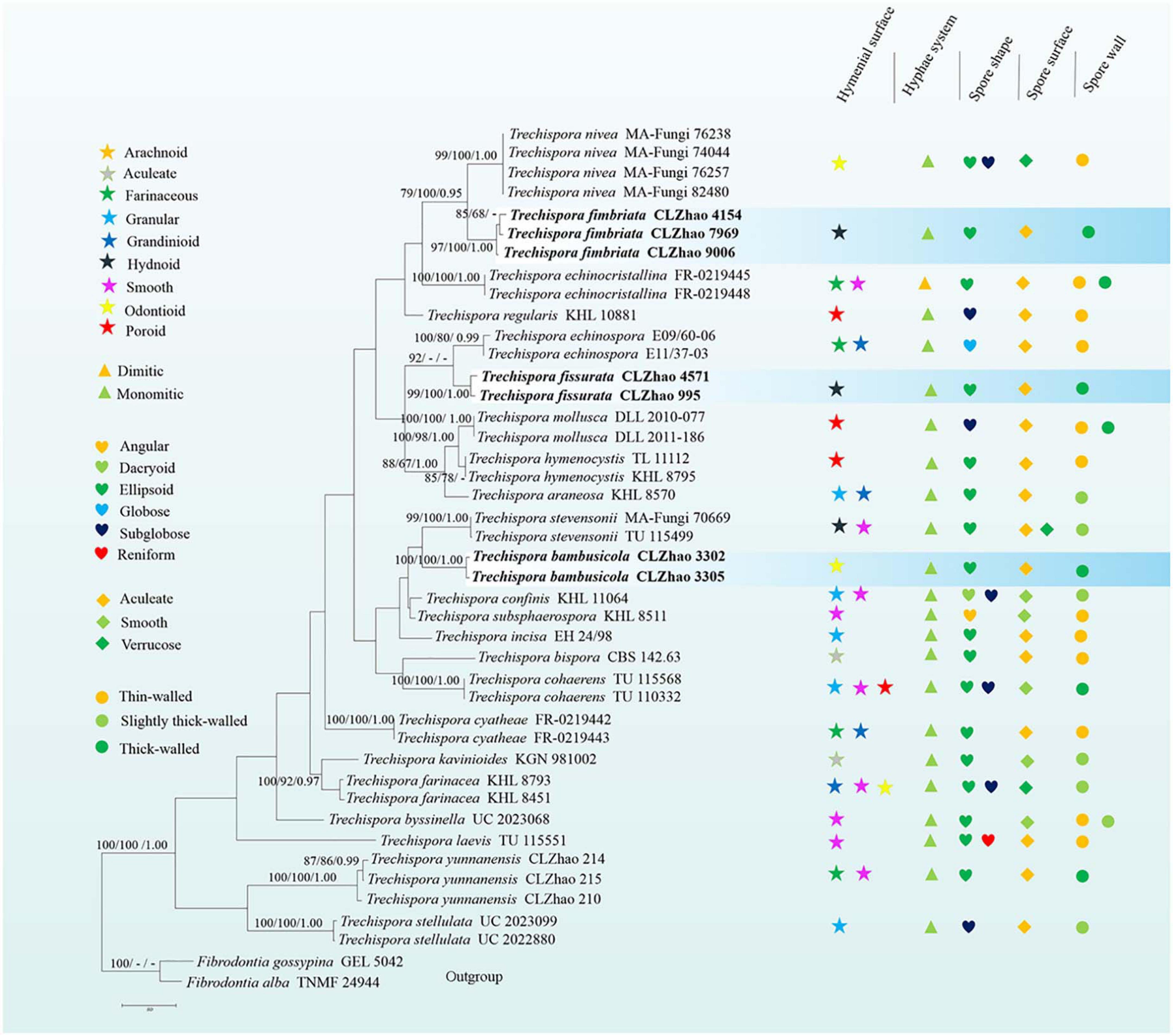
Figure 2. Maximum Parsimony strict consensus tree illustrating the phylogeny of three new species and related species in Trechispora based on ITS + nLSU sequences. Branches are labeled with maximum likelihood bootstrap values > 70%, parsimony bootstrap proportion values > 50%, and Bayesian posterior probabilities > 0.95, respectively.
Taxonomy
Trechispora bambusicola C.L. Zhao, sp. nov.
MycoBank no.: MB 838612 (Figures 3, 4).
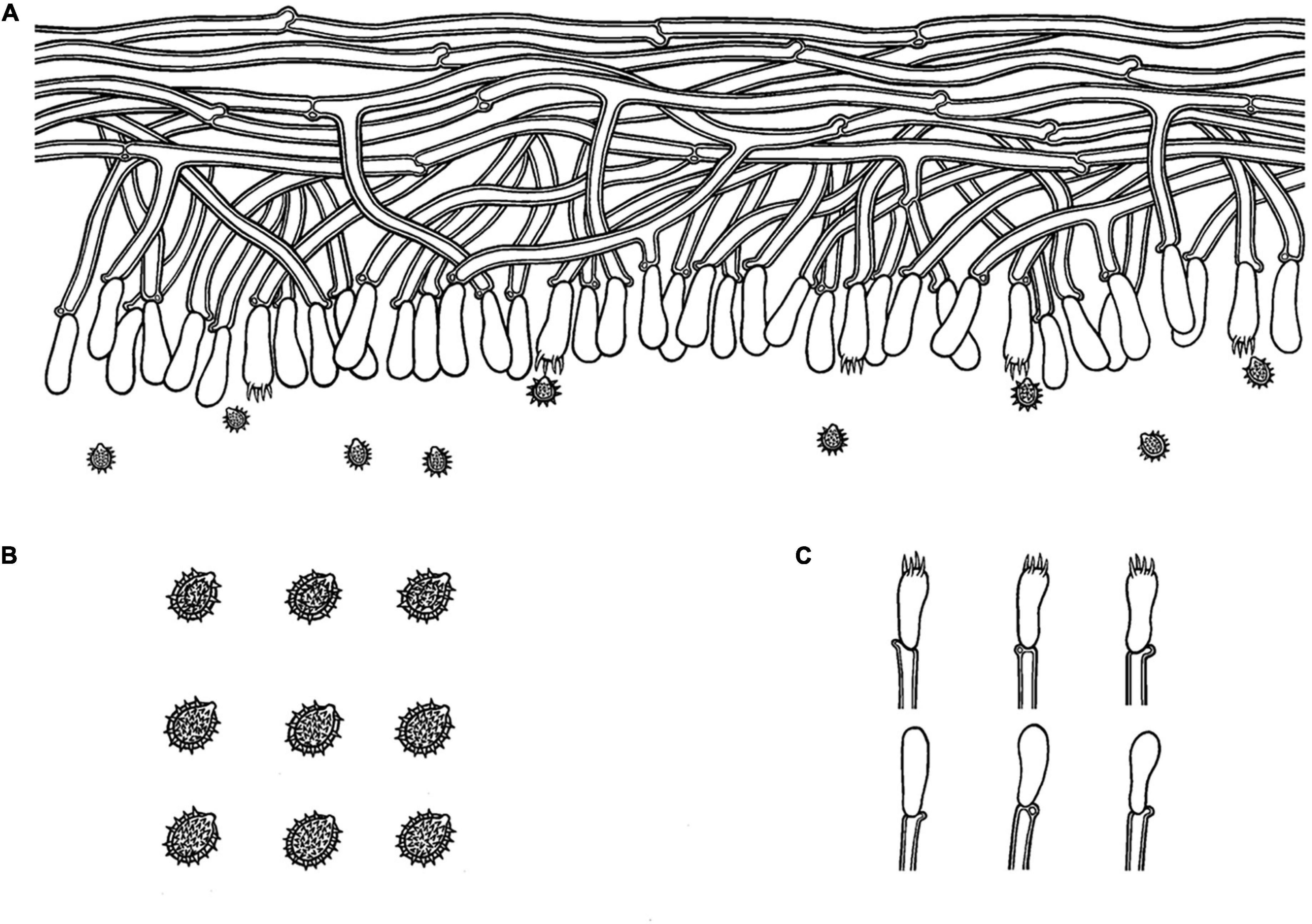
Figure 4. Microscopic structures of Trechispora bambusicola (drawn from the holotype): (A) Section of hymenium. (B) Basidiospores. (C) Basidia and basidioles. Bars: (A,C) 10 μm; (B) 5 μm.
Holotype—China, Yunnan Province, Pu’er, Laiyanghe National Forest Park, on dead bamboo, 30 September 2017, CLZhao 3305 (SWFC).
Etymology—Bambusicola (Lat.): referring to occurrence on bamboo stump.
Basidiomata
Annual, adnate, soft, and fragile, without odor or taste when fresh, becoming granulose upon drying, up to 15 cm long and 5 cm wide, 50–300 μm thick. Hymenial surface odontioid, aculei cylindrical to conical, blunt, 0.3–0.5 mm long, white to cream when fresh, turn to cream to buff upon drying. Margin white to cream.
Hyphal structure
Monomitic, generative hyphae with clamp connections, hyaline, thick-walled, up to 0.7 μm, richly branched, 2–3 μm in diameter, IKI−, CB−; hyphae unchanged in KOH.
Hymenium
Cystidia and cystidioles absent; basidia shortly cylindrical to subclavate with median constriction, with 4-sterigmata and a basal clamp connection, 9–13 × 2.5–5 μm, basidioles dominant, in shape similar to basidia, but slightly smaller.
Basidiospores
Ellipsoid, hyaline, thick-walled, ornamented, sparse aculei, sharp, IKI−, CB−, (2.6−)2.9–3.5(−3.9) × 2–2.7 μm, L = 3.18 μm, W = 2.41 μm, Q = 1.26–1.38 (n = 60/2).
Type of rot
White rot.
Additional specimen examined
CHINA, Yunnan Province, Pu’er, Laiyanghe National Forestry Park, on dead bamboo, 30 September 2017, CLZhao 3302 (SWFC).
Trechispora fimbriata C.L. Zhao, sp. nov.
MycoBank no.: MB 838613 (Figures 5,6).
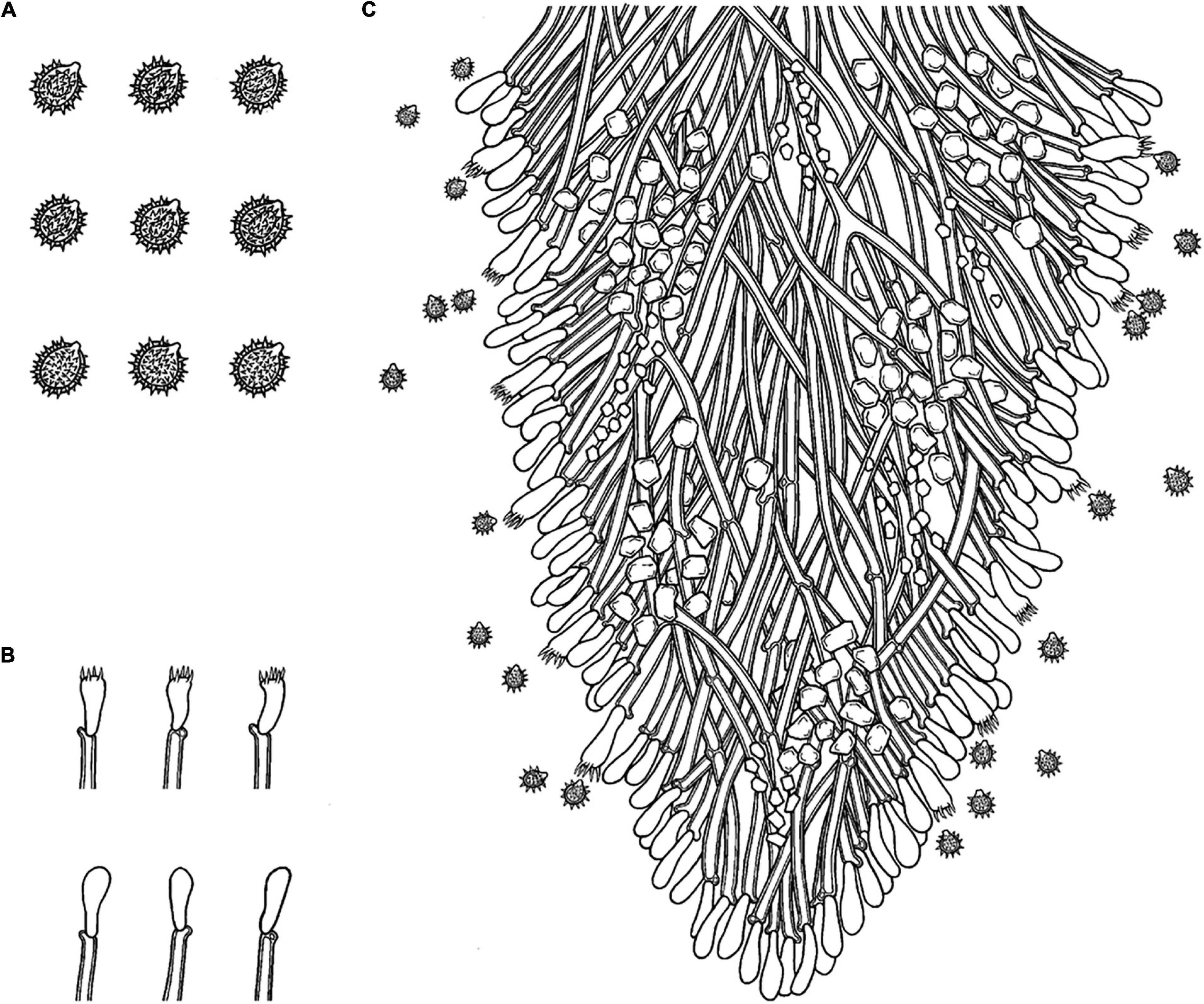
Figure 6. Microscopic structures of Trechispora fimbriata (drawn from the holotype). (A) Basidiospores. (B) Basidia and basidioles. (C) Section of hymenium. Bars: (A) 5 μm; (B,C) 10 μm.
Holotype—China, Yunnan Province, Puer, Jingdong County, Wuliangshan National Nature Reserve, on the angiosperm trunk, October 5, 2017, CLZhao 4154 (SWFC).
Etymology—Fimbriata (Lat.): refers to the fimbriate margin of the basidiomata.
Basidiomata
Annual, adnate, without odor or taste when fresh, becoming fragile upon drying, up to 10 cm long and 3 cm wide, 100–200 μm thick. Hymenial surface hydnoid, with aculei, cylindrical, blunt, 0.4–0.7 mm long, white to pink when fresh, turn to pink to buff upon drying. Margin white to cream, thinning out, fimbriate.
Hyphal system
Monomitic, generative hyphae with clamp connections, hyaline, thick-walled, up to 0.6 μm, branched, 2–4 μm in diameter, IKI−, CB−; hyphae unchanged in KOH.
Hymenium
Cystidia and cystidioles absent; basidia shortly cylindrical with median constriction, with 4–6 sterigmata and a basal clamp connection, 7–11.5 × 3.5–5 μm, basidioles dominant, in shape similar to basidia, but slightly smaller.
Basidiospores
Ellipsoid, hyaline, thick-walled, ornamented, sparse aculei, sharp, IKI−, CB−, (2.5−)3–3.6(−3.8) × 2.4–3.2 μm, L = 3.25 μm, W = 2.63 μm, Q = 1.17–1.38 (n = 90/3).
Type of rot
White rot.
Additional specimens examined
China, Yunnan Province, Yuxi, Xinping County, Mopanshan National Forestry Park, on living tree of angiosperm, August 9, 2018, CLZhao 7969 (SWFC); on angiosperm trunk, October 15, 2018, CLZhao 9006 (SWFC).
Trechispora fissurata C.L. Zhao, sp. nov.
MycoBank no.: MB 838614 (Figures 7, 8).
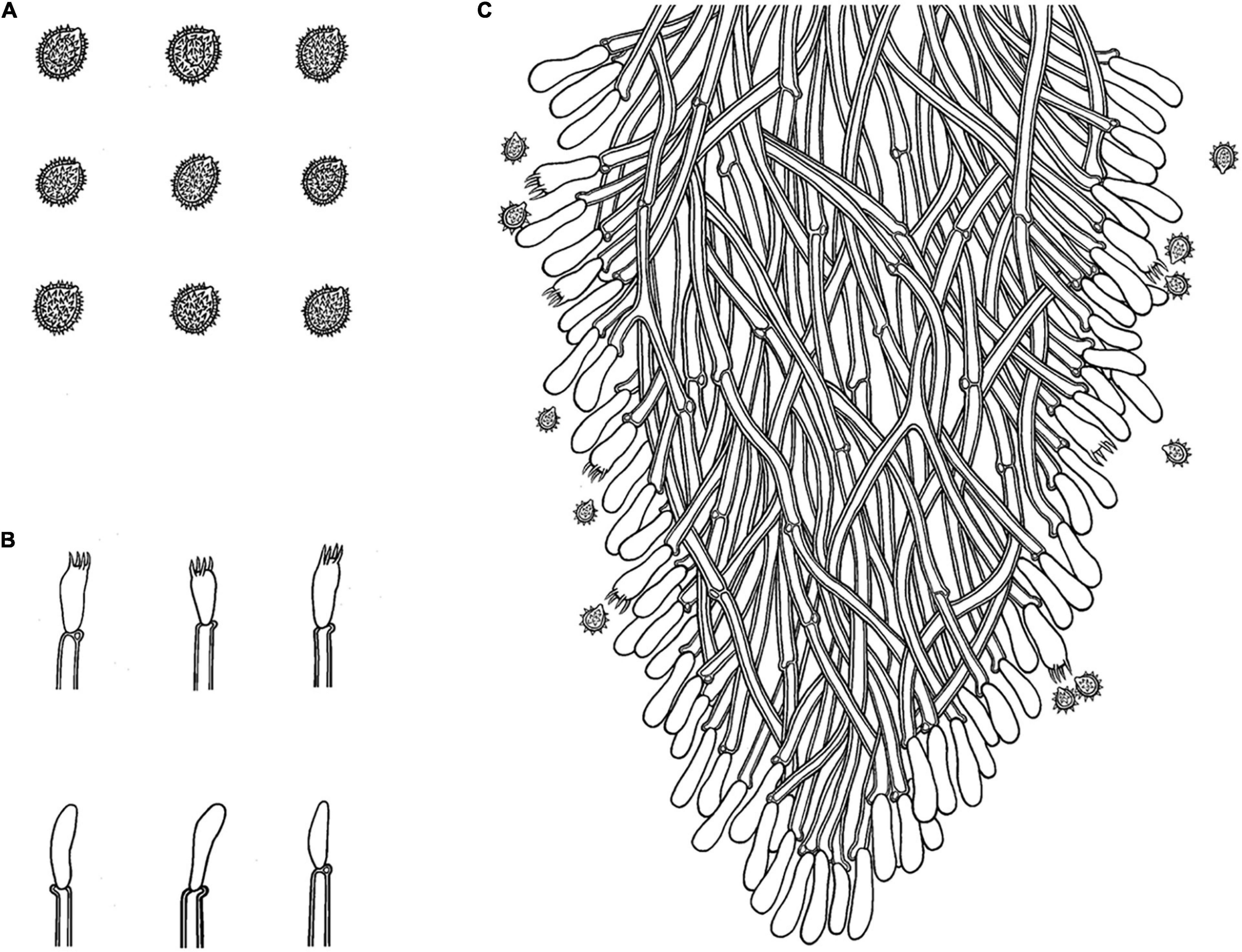
Figure 8. Microscopic structures of Trechispora fissurata (drawn from the holotype). (A) Basidiospores. (B) Basidia and basidioles. (C) Section of hymenium. Bars: (A) 5 μm; (B,C) 10 μm.
Holotype—China, Yunnan Province, Puer, Jingdong County, Wuliangshan National Nature Reserve, on angiosperm trunk, October 6, 2017, CLZhao 4571 (SWFC).
Etymology—Fissurata (Lat.): refers to the cracking fissures on hymenial surface.
Basidiomata
Annual, adnate, without odor or taste when fresh, becoming cracking upon drying, up to 8 cm long and 4.5 cm wide, 400–800 μm thick. Hymenial surface hydnoid, with aculei, cylindrical to conical, sharp, 0.5–0.9 mm long, cream to straw yellow when fresh, turn to cream to yellow upon drying. Margin cream to yellow.
Hyphal system
Monomitic, generative hyphae with clamp connections, hyaline, thick-walled, up to 0.8 μm, branched, 2.5–5 μm in diameter, IKI−, CB−; hyphae unchanged in KOH.
Hymenium
Cystidia and cystidioles absent; basidia shortly clavate to tubular, with 4-sterigmata and a basal clamp connection, 8–10.5 × 2.5–4.5 μm, basidioles dominant, in shape similar to basidia, but slightly smaller.
Basidiospores
Ellipsoid, hyaline, thick-walled, ornamented, dense aculei, sharp, IKI−, CB−, (3−)3.3–4(−4.3) × (2.5−)2.8–3.5(−3.9) μm, L = 3.67 μm, W = 3.19 μm, Q = 1.13–1.17 (n = 60/2).
Type of rot
White rot.
Additional specimen examined
CHINA, Yunnan Province, Yuxi, Xinping County, Mopanshan Forestry Park, on the fallen angiosperm branch, January 17, 2017, CLZhao 995 (SWFC).
Discussion
Phylogenetically, Phookamsak et al. (2019) introduced the phylogram generated from BI analysis of ITS sequence dataset of Trechispora sequences and included most taxa in this genus, in which it implied the phylogenetic relationship among species of Trechispora. In the present study, based on the ITS sequences (Figure 1), T. bambusicola was sister to T. stevensonii (Berk. and Broome) K.H. Larss; T. fimbriata grouped with T. nivea; T. fissurata grouped with T. echinospora Telleria, M. Dueñas, I. Melo, and M.P. Martín. Further ITS + nLSU dataset (Figure 2) revealed that T. bambusicola formed a single lineage and then grouped with T. rigida and T. stevensonii; T. fimbriata was sister to T. nivea; T. fissurata grouped with T. thelephora. However, T. rigida differs in its dirty white to buff hymenophore (Larsson, 1996). T. stevensonii is separated from T. bambusicola by the smooth to hydnoid hymenophore and larger basidiospores (4–4.5 × 3–3.5 μm; Larsson, 1995). T. nivea differs from T. fimbriata by the white to light ochraceous hymenial surface (Persoon, 1794). T. echinospora differs from T. fissurata by the farinaceous to grandinioid hymenophore and larger, globose basidiospores (3.3–4 × 2.8–3.5 μm; Phookamsak et al., 2019) and T. thelephora differs in its pileate to stipitate with light yellow brown surface and larger (4–5 × 3.4–4.5 μm; Albee-Scott and Kropp, 2010).
In the present study, three new species, T. bambusicola, T. fimbriata, and T. fissurata spp. nov. are found from rotten wood. Morphologically, T. bambusicola is similar to T. cyatheae Ordynets, Langer and K.H. Larss. by sharing the characteristics of soft and fragile basidiomata. However, T. cyatheae differs from T. bambusicola by having farinaceous to grandinioid hymenophore and thin-walled generative hyphae (Ordynets et al., 2015).
Trechispora fimbriata has similar characteristics of having the fimbriate margin with Trechispora canariensis Ryvarden and Liberta, Trechispora clancularis (Park.-Rhodes) K.H. Larss., Trechispora microspora (P. Karst.) Liberta, Trechispora stellulata (Bourdot and Galzin) Liberta, and Trechispora subhelvetica (Parmasto) Liberta. However, T. canariensis differs in its arachnoid to pelliculose hymenophore and larger basidiospores (5–7 × 3–3.5 μm; Ryvarden and Liberta, 1978); T. clancularis differs in the poroid to irpicoid hymenophore and slightly cyanophilous basidiospores (Larsson, 1994); T. stellulata differs in the arachnoid to byssoid hymenophore with whitish hymenial surface (Liberta, 1966); and T. subhelvetica differs in the narrower basidiospores (3–4 × 2–2.5 μm; Parmasto, 1965).
Trechispora fissurata resembles several species with similar features of having the hydnoid hymenophore and a monomitic hyphal system: T. nivea (Pers.) K.H. Larss., T. stevensonii (Berk. and Broome) K.H. Larss., and Trechispora verruculosa (G. Cunn.) K.H. Larss., but T. nivea by the white to pale ochraceous hymenial surface and thin-walled generative hyphae encrusted with granular crystals (Bernicchia and Gorjón, 2010); T. stevensonii by the white to ochraceous hymenial surface and larger basidiospores (4–4.5 × 3–3.5 μm; Larsson, 1995); T. verruculosa by the slightly cyanophilous and larger basidiospores (4.5–5.5 × 3.5–4.5 μm; Larsson, 1996).
Currently, eight species of Trechispora have been reported from China (Dai, 2011; Xu et al., 2019), Trechispora alnicola, Trechispora cohaerens, T. farinacea, T. microspora, T. nivea, Trechispora polygonospora Ryvarden, Trechispora subsphaerospora (Litsch.) Liberta, and T. yunnanensis, and one species of T. yunnanensis was found in Yunnan Province of China and it differs from three new species by having a smooth to farinaceous hymenial surface and larger basidiospores (7–8.5 × 5–5.5 μm; Xu et al., 2019). Three new taxa do not closely group together in phylogenetic trees, and morphologically, T. bambusicola differs from T. fimbriata and T. fissurata by having granulose basidiomata with cream to buff hymenial surface and growth on dead bamboo. T. fimbriata differs in its fimbriate margin of the basidiomata with pink to buff hymenial surface.
In addition, the ectomycorrhizal fungi (EcM) play an important role in ecosystems based on their mutualistic association with many groups of plants (Heijden et al., 2015). Vanegas-León et al. (2019) discovered the Trechisporales basidiomes and root colonization from T. thelephora basidiome. In the present study, T. fissurata was sister to T. thelephora based on ITS + nLSU phylogenetic analysis (Figure 2), which implied that both species have close evolutionary relationship. However, T. fissurata grows on deeply decayed wood, and T. thelephora is a soil-inhabiting fungus. Therefore, future investigations in both inhabiting types are needed to determine whether the natural selection or other factors pushes the different direction on inhabiting soil/wood among Trechispora.
In the habitat and distribution, Hibbett et al. (2014) revealed that most species of Trechispora is considered as soil-inhabiting. Later, some species were found on deeply decayed wood fungi (Bernicchia and Gorjón, 2010; Dai, 2011). However, some species in Trechispora are a typical feature of ectomycorrhizal fungi as frequently forming basidiomes on soil (Dunham et al., 2007; Vanegas-León et al., 2019). In the neotropical and subtropical region, the ectomycorrhizal basidiomes are found; however, the researches on the new taxa related to wood-decaying fungi of Trechispora from China are poorly reported. Further studies may focus on the relationships between the plants and species from Trechispora and try to better understand the evolutionary directions between soil-inhabiting and decayed wood fungi of Trechispora; many fungal studies on phylogeny and application were from these areas, which will be useful to push future researches for the genus Trechispora (Dai, 2011; Cui et al., 2019; Shen et al., 2019; Zhu et al., 2019; Richter et al., 2019; Angelini et al., 2020; Bao et al., 2020).
Disclosure
All the experiments undertaken in this study comply with the current laws of the People’s Republic of China.
Data Availability Statement
The data presented in the study are deposited in the https://www.ncbi.nlm.nih.gov/GenBank and https://www.mycobank.org/page/Home/MycoBank repository accession number of GenBank (ITS MW544021-MW544027 and nLSU MW520171-MW520177) and MycoBank (MB 838612-MB 838614).
Author Contributions
C-LZ collected the species. WZ performed the molecular phylogenetic analyses. Both authors were responsible for the morphological analysis and description of the collections, planned, organized, and evaluated critically the experimental parts, wrote the manuscript, contributed to the article, and approved the submitted version.
Funding
The research was supported by the Yunnan Fundamental Research Project (Grant No. 202001AS070043), the Key Laboratory of Forest Resources Conservation and Utilization in the Southwest Mountains of China Ministry of Education, Southwest Forestry University (KLESWFU-202003), and the High-level Talents Program of Yunnan Province (YNQR-QNRC-2018-111).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
ITS, internal transcribed spacer; nLSU, large subunit; SWFC, herbarium of Southwest Forestry University, Kunming, China; KOH, 5% potassium hydroxide; CB, Cotton Blue; CB −, acyanophilous; IKI, Melzer’s reagent; IKI −, both inamyloid and indextrinoid; L, mean spore length (arithmetic average for all spores); W, mean spore width (arithmetic average for all spores); Q, variation in the L/W ratios between The studied specimens, n (a/b), number of spores (a) measured from given number (b) of specimens, spore measurements do not include ornamentation; CTAB, cetyltrimethylammonium bromide; DNA, deoxyribonucleic acid; PCR, polymerase chain reaction; MP, maximum parsimony; ML, maximum likelihood; BI, Bayesian inference; TBR, tree-bisection reconnection.
Footnotes
- ^ http://www.indexfungorum.org/Names/Names.asp
- ^ https://www.mycobank.org/Biolomics.aspx?Table=Mycobank&Page=200&ViewMode=Basic
- ^ http://lutzonilab.org/nuclear-ribosomal-dna/
- ^ http://mafft.cbrc.jp/alignment/server/
- ^ www.phylo.org
References
Albee-Scott, S., and Kropp, B. R. (2010). A phylogenetic study of Trechispora thelephora. Mycotaxon 114, 395–399. doi: 10.5248/114.395
Angelini, C., Vizzini, A., Justo, A., Bizzi, A., and Kaya, E. (2020). First report of a neotropical agaric (lepiota spiculata, agaricales, basidiomycota) containing lethal α-amanitin at toxicologically relevant levels. Front. Microbiol. 11:1833. doi: 10.3389/fmicb.2020.01833
Bao, D. F., Mckenzie, E. H. C., Bhat, D. J., Hyde, K. D., and Su, H. Y. (2020). Acrogenospora (acrogenosporaceae, minutisphaerales) appears to be a very diverse genus. Front. Microbiol. 11:1606. doi: 10.3389/fmicb.2020.01606
Bernicchia, A., and Gorjón, S. P. (2010). Fungi Europaei 12: Corticiaceae s.l. Alassio: Edizioni Candusso.
Cui, B. K., Li, H. J., Ji, X., Zhou, J. L., Song, J., Si, J., et al. (2019). Species diversity, taxonomy and phylogeny of Polyporaceae (Basidiomycota) in China. Fungal Divers. 97, 137–392. doi: 10.1007/s13225-019-00427-4
Dai, Y. C. (2011). A revised checklist of corticioid and hydnoid fungi in China for 2010. Mycoscience 52, 69–79. doi: 10.1007/S10267-010-0068-1
Dai, Y. C. (2012). Polypore diversity in China with an annotated checklist of Chinese polypores. Mycoscience 53, 49–80. doi: 10.1007/s10267-011-0134-3
Dunham, S. M., Larsson, K. H., and Spatafora, J. W. (2007). Species richness and community composition of mat-forming ectomycorrhizal fungi in old-and second-growth Douglas-fir forests of the Hj Andrews Experimental Forest, Oregon, USA. Mycorrhiza 17, 633–645. doi: 10.1007/s00572-007-0141-6
Felsenstein, J. (1985). Confidence intervals on phylogenetics: an approach using bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. doi: 10.1021/bk-1999-0734.ch008
Heijden, M. G., Martin, F. M., Selosse, M. A., and Sanders, I. R. (2015). Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol. 205, 1406–1423. doi: 10.1111/nph.13288
Hibbett, D. S., Bauer, R., Binder, M., Giachini, A. J., Hosaka, K., and Justo, A. (2014). “14 Agaricomycetes,” in Systematics and evolution, eds D. J. McLaughlin and J. W. Spatafora, (Berlin: Springer), 373–429.
Larsson, K. H. (1994). Poroid species in Trechispora and the use of calcium oxalate crystals for species identification. Mycol. Res. 98, 1153–1172. doi: 10.1016/S0953-7562(09)80200-1
Larsson, K. H. (1995). Taxonomy of Trechispora farinacea and proposed synonyms I. Species with a grandinioid or hydnoid hymenophore. Symb. Bot. Ups. 30, 101–118.
Larsson, K. H. (1996). New species and combinations in Trechispora (Corticiaceae, Basidiomycotina). Nord. J. Bot. 16, 83–98. doi: 10.1111/j.1756-1051.1996.tb00218.x
Larsson, K. H. (2007). Re-thinking the classification of corticioid fungi. Mycol. Res. 111, 1040–1063. doi: 10.1016/j.mycres.2007.08.001
Larsson, K. H., Larsson, E., and Koljalg, U. (2004). High phylogenetic diversity among corticioid homobasidiomycetes. Mycol. Res. 108, 983–1002. doi: 10.1017/S0953756204000851
Liberta, A. E. (1973). The genus Trechispora (Basidiomycetes, Corticiaceae). Canad. J. Bot. 51, 1871–1892. doi: 10.1139/b73-240
Liu, S. L., Ma, H. X., He, S. H., and Dai, Y. C. (2019). Four new corticioid species in Trechisporales (Basidi omycota) from East Asia and notes on phylogeny of the order. MycoKeys 48, 97–113. doi: 10.3897/mycokeys.48.31956
Miettinen, O., and Larsson, K. H. (2006). Trechispora elongata species nova from North Europe. Mycotaxon 96, 193–198.
Miller, M. A., Holder, M. T., Vos, R., Midford, P. E., Liebowitz, T., Chan, L., et al. (2009).The CIPRES Portals. – CIPRES. Available online at: http://www.phylo.org/sub_sections/portal (accessed December 7, 2011).
Nylander, J. A. A. (2004). MrModeltest v2. Program distributed by the author. Uppsala: Evolutionary Biology Centre.
Ordynets, A., Larsson, K. H., and Langer, E. (2015). Two new Trechispora species from La Réunion Island. Mycol. Progr. 14:113. doi: 10.1007/s11557-015-1133-0
Parmasto, E. (1965). Corticiaceae U. R. S. S. I. Descriptiones taxorum. Combinationes noval [novarum]. Izv. Akad. Nauk Ėstonsk. SSR, Ser. Biol. 14, 220–223.
Persoon, C. H. (1794). Neuer Versuch einer systematischen Eintheilung der Schwämme. Neues Mag. Bot. 1, 63–80.
Petersen, J. H. (1996). Farvekort. The Danish Mycological Society’s colour-chart. Greve: Foreningen til Svampekundskabens Fremme.
Phookamsak, R., Hyde, K. D., Jeewon, R., Bhat, D. J., Jones, E. B. G., Maharachchikumbura, S., et al. (2019). Fungal diversity notes 929-1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 95, 1–273. doi: 10.1007/s13225-019-00421-w
Richter, C., Yurkov, A. M., Boekhout, T., and Stadler, M. (2019). Diversity of Tilletiopsis-Like Fungi in Exobasidiomycetes (Ustilaginomycotina) and Description of Six Novel Species. Front. Microbiol. 10:2544. doi: 10.3389/fmicb.2019.02544
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: B2017ayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Ryvarden, L. (2002). A note on the genus Hydnodon Banker. Some neotropical wood inhabiting fungi. Syn. Fungorum 15, 31–33.
Ryvarden, L., and Liberta, A. E. (1978). Contribution to the Aphyllophoralles of the Canary Islands 4. Two new species of Trechispora and Xenmastella. Canad. J. Bot. 56, 2617–2619. doi: 10.1139/b78-314
Shen, L. L., Wang, M., Zhou, J. L., Xing, J. H., Cui, B. K., and Dai, Y. C. (2019). Taxonomy and phylogeny of Postia. Multi-gene phylogeny and taxonomy of the brown-rot fungi: Postia and its related genera. Persoonia 42, 101–126. doi: 10.3767/persoonia.2019.42.05
Swofford, D. L. (2002). PAUP∗: Phylogenetic Analysis Using Parsimony (∗and Other Methods). Version 4.0b10. Sunderland, MA: Sinauer Associates.
Telleria, M. T., Melo, I., Dueñas, M., Larsson, K. H., and Paz Martin, M. P. (2013). Molecular analyses confirm Brevicellicium in Trechisporales. IMA Fungus 4, 21–28. doi: 10.5598/imafungus.2013.04.01.03
Trichiès, G., and Schultheis, B. (2002). Trechispora antipus sp. nov., une seconde espèce bisporique du genre Trechispora (Basidiomycota, Stereales). Mycotaxon 82, 453–458.
Vanegas-León, M. L., Sulzbacher, M. A., Rinaldi, A. C., and Mélanie, Roy, and Neves, M. A. (2019). Are trechisporales ectomycorrhizal or non-mycorrhizal root endophytes? Mycol. Progr. 18, 1231–1240. doi: 10.1007/s11557-019-01519-w
Vu, D., Groenewald, M., de Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., et al. (2019). Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 92, 135–154. doi: 10.1016/j.simyco.2018.05.001
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods And Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White, (San Diego, CA: Academic Press), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Xu, T. M., Chen, Y. H., and Zhao, C. L. (2019). Trechispora yunnanensis sp. nov. from China. Phytotaxa 424, 253–261. doi: 10.11646/phytotaxa.424.4.5
Yurchenko, E., and Wu, S. H. (2014). Fibrodontia alba sp. nov. (Basidiomycota) from Taiwan. Mycoscience 55, 336–343. doi: 10.1016/j.myc.2013.12.004
Zhao, C. L., and Wu, Z. Q. (2017). Ceriporiopsis kunmingensis sp. nov. (Polyporales, Basidiomycota) evidenced by morphological characters and phylogenetic analysis. Mycol. Progr. 16, 93–100. doi: 10.1007/s11557-016-1259-8
Keywords: Hydnodontaceae, phylogeny, taxonomy, wood-inhabiting fungi, Yunnan Province
Citation: Zhao W and Zhao C-L (2021) The Phylogenetic Relationship Revealed Three New Wood-Inhabiting Fungal Species From Genus Trechispora. Front. Microbiol. 12:650195. doi: 10.3389/fmicb.2021.650195
Received: 08 January 2021; Accepted: 01 February 2021;
Published: 17 March 2021.
Edited by:
Dhanushka Nadeeshan Wanasinghe, Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, ChinaReviewed by:
Baokai Cui, Beijing Forestry University, ChinaYu-Cheng Dai, Beijing Forestry University, China
Copyright © 2021 Zhao and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang-Lin Zhao, Fungichanglinz@163.com
 Wei Zhao1,2
Wei Zhao1,2 Chang-Lin Zhao
Chang-Lin Zhao