- 1Institute of Entomology, Guizhou University, Guiyang, China
- 2Institute of Fungus Resources, Guizhou University, Guiyang, China
- 3College of Tea Sciences, Guizhou University, Guiyang, China
Background: The genus Lecanicillium W.Gams & Zare is a recognized insect pathogen but members of the genus have been found parasitizing various hosts including arthropods, nematodes, plants, and fungi. The new classification system for fungi proposed to reject Lecanicillium and transfer some of the species to the genus Akanthomyces. However, the attribution problem of most species in the original genus Lecanicillium remains unsolved. The current study aimed to improve understanding of the pivotal internal phylogeny in Lecanicillium by estimating the divergence times of Lecanicillium to provide additional insights into the status of this genus within the family Cordycipitaceae.
Results: Dating analyses support the supposition that the ancestor of Lecanicillium was in the Cretaceous period (84.36 Mya, 95% HPD: 72.12–94.74 Mya). After originating from a common ancestor, eight clades of Lecanicillium were derived and evolved independently in parallel with other genera of Cordycipitaceae. Based on the clear divergence age estimates, Lecanicillium clade 8 originated earlier as an independent group in the Cretaceous period (75.61 Mya, 95% HPD: 63.31–87.54 Mya), while Lecanicillium clades 1–7 originated later as an independent group in the boundary of the Cretaceous and Paleogene periods (64.66 Mya, 95% HPD: 52.75–76.74 Mya). Lecanicillium huhutii formed an independent branch in a polytomy together with a clade containing Lecanicillium tenuipes (BI posterior probabilities 1, ML bootstrap 100%).
Conclusion: The pivotal internal phylogeny, origin, and evolutionary history of Lecanicillium in the family Cordycipitaceae were investigated. Phylogenetic and morphological analyses indicated that there are eight representative clades (four representative branches of evolutionary history), including clade 1 (members have a relatively uniform sporulation structure comprising globose heads with a higher number of conidia), clade 8 (including all members of Gamszarea), clades 2–5 (the differences of the divergence time estimations were smaller compared with other clades), and clade 6–7 (members are close to Gibellula, Hevansia, and Ascopolyporus). Based on the above findings, a novel spider-pathogenic fungus, Lecanicillium huhutii, is described. All other species in Lecanicillium clade 1 (Lecanicillium araneogemum, L. nodulosum, L. pissodis, and L. uredinophilum) should be transferred to the genus Akanthomyces. Furthermore, the monotypic genus Parengyodontium should be merged with the genus Gamszarea. More novel species need to be discovered to thoroughly resolve the attribution problem of Lecanicillium. Finally, no major lineages of Lecanicillium were correlated with the nearby Cretaceous-Tertiary extinction event, indicating that the diversity of Lecanicillium is more likely to be caused by long-term environmental adaptation and coevolution with insects rather than by dramatic extinction events.
Introduction
Although members of the genus have been found parasitizing various hosts including arthropods, nematodes, plants, and fungi, the genus Lecanicillium W. Gams and Zare is recognized and largely known as an insect pathogen (Goettel et al., 2008; Sukarno et al., 2009; Su et al., 2019). Members of the genus Lecanicillium have the potential for development as effective biological control agents against numerous plant diseases, insect pests, and plant-parasitic nematodes (Goettel et al., 2008). To date, approximately 15 commercial preparations based on Lecanicillium spp. have been, or are currently being developed (Faria and Wraight, 2007). Species of the genus Lecanicillium are characterized by the formation of slender phialides, mostly from procumbent or prostrate aerial hyphae, singly or in terminal and intercalary whorls, and erect conidiophores with one or several whorls of phialides may also occur (Zare and Gams, 2001). This genus was established to accommodate entomogenous and fungicolous verticillium-like anamorphs of the family Clavicipitaceae (order Hypocreales) previously classified in Verticillium section Prostrata (Gams and Zare, 2001). The genus is typified by Lecanicillium lecanii with Torrubiella confragosa as the sexual morph. All insect pathogens formerly included in Verticillium were reclassified in this genus (Zare and Gams, 2001; Wijayawardene et al., 2017). Contemporaneously, the genus Simplicillium W. Gams and Zare was established to accommodate some species that were similar to those of the genus Lecanicillium but formed a distinct monophyletic group based on molecular data (Zare and Gams, 2001).
Sung et al. (2007) demonstrated the phylogenetic classification of Cordyceps and the Clavicipitaceous fungi and divided the Clavicipitaceae into Clavicipitaceae, Cordycipitaceae, and Ophiocordycipitaceae. Therefore, Lecanicillium was transferred into the family Cordycipitaceae along with other asexual genera like Beauveria Vuill., Akanthomyces Lebert., and Simplicillium. This work supported the findings of Zare and Gams (2001) that Lecanicillium is paraphyletic and morphological characters of these genera, especially the phialides and conidia, are difficult to distinguish a similar taxonomic group (Zare and Gams, 2008; Sukarno et al., 2009).
Kepler et al. (2017) revisited the taxonomic affinities of the family Cordycipitaceae (order Hypocreales) and resolved competing names based on the principles of priority, recognition of monophyletic groups, and the practical use of affected taxa. The type species of Lecanicillium was placed within the genus Akanthomyces, creating a conflict between the usage of the two generic names. In accordance with the reframing of Article 59 of the International Code of Nomenclature for algae, fungi, and plants (ICN) (McNeill et al., 2012). Kepler et al. (2017) proposed to maintain nine generic names, including Akanthomyces Lebert, Ascopolyporus Möller, Beauveria Vuill., Cordyceps Fr., Engyodontium de Hoog, Gibellula Cavara, Hyperdermium J.F. White, R.F. Sullivan, Bills and Hywel-Jones, Parengyodontium C.C. Tsang, J.F.W. Chan, W.M. Pong, J.H.K. Chen, A.H.Y. Ngan, Cheung, C.K.C. Lai, D.N.C. Tsang, S.K.P. Lau, and P. C.Y. Woo, and Simplicillium Gams and Zare, and two new genera including Blackwellomyces Spatafora and Luangsa-ard and Hevansia Luangsaard, Hywel-Jones and Spatafora (Kepler et al., 2017). Eight generic names were rejected, including Evlachovaea B.A. Borisov and Tarasov, Granulomanus de Hoog and Samson, Isaria Pers., Lecanicillium W. Gams and Zare, Microhilum H.Y. Yip and A.C. Rath, Phytocordyceps C.H. Su and H.H. Wang, Synsterigmatocystis Constantin, and Torrubiella Boud.
Currently, 35 species of the genus Lecanicillium have been formally described and are listed in the Index Fungorum.1 However, only five of these species were transferred to Akanthomyces by Kepler et al. (2017), and the attribution of most species in the original genus Lecanicillium remains an unsolved problem. Moreover, some novel species described after Kepler’s proposal were still placed in the genus Lecanicillium, such as L. cauligalbarum (Zhou et al., 2018), L. coprophilum (Su et al., 2019), L. gracile (Ponizovskaya et al., 2020), and L. praecognitum (Crous et al., 2020). Consequently, there is a question as to whether all members of the genus Lecanicillium should be transferred into the genus Akanthomyces or whether more new genera should be established, or whether there is an alternative solution to accommodate other species of Lecanicillium? Some scholars have made attempts to address this question. For example, Zhang et al. (2020) transferred five species of the genus Lecanicillium (L. coprophilum, L. kalimantanense, L. restrictum, L. testudineum, and L. wallacei) to a new genus, Gamszarea (for contributions of Walter Gams and Rasoul Zare to the taxonomic study of Lecanicillium W. Gams and Zare) based on ITS and multilocus (TEF, RPB1, RPB2, LSU, and SSU) sequence data. In addition, Wang et al. (2020) transferred three species of Lecanicillium (L. acerosum, L. primulinum, and L. subprimulinum) to a new genus, Flavocillium, based on ITS sequence data. Further work is needed to attempt to fully resolve the attribution problem of Lecanicillium.
New generic names for these species in the family Cordycipitaceae need introducing, supported by more detailed morphological and phylogenetic evidence combined with a larger sampling of taxa (Wang et al., 2020). Phylogenetic analysis revealed that LSU and SSU could not be used for identifying the strains alone because of the small interspecific differences (Zhou et al., 2020). The current study was performed to obtain a better understanding of the pivotal internal phylogeny in Lecanicillium, to execute accurate phylogenetic research, to estimate evolutionary timescales using the molecular clock, and to improve understanding of evolutionary processes across all taxonomic levels. A phylogenetic analysis was conducted with four loci (ITS, TEF, RPB1, and RPB2) from 97 species, including almost all species of the genus Lecanicillium. The calibration point in the molecular clock was based on the fossil record of Paleoophiocordyceps coccophagus G. H. Sung, Poinar, and Spatafora, a fungal animal parasite that had morphological features similar to the asexual states of Hirsutella and Hymenostilbe and belonged to the family Ophiocordycipitaceae (Sung et al., 2008). In addition, a detailed characterization of a novel species of the genus Lecanicillium was performed.
Materials and Methods
Specimen Collection and Fungus Isolation
The novel asexual morph species of Lecanicillium was isolated from spider cadavers hidden in a hole of a tree in Huaxi park of Guizhou province, China, on 5 October 2013. The isolated strains GZUIFRhuhu and GZUIFRhuhu1 were deposited at the Institute of Fungal Resources of Guizhou University (GZAC).
Strain Culture and Observations
Isolated strains were inoculated on potato dextrose agar (PDA) at 25°C for 20 days under 12-h light/12-h dark conditions. For optical microscopy observations and imaging, fresh hyphae were stained with lactophenol cotton blue solution or normal saline, and the samples were observed with an optical microscope (BK5000, OPTEC, United States).
DNA Extraction, PCR Amplification, and Sequencing
Genomic DNA was extracted using a previously described method (Chiriví-Salomón et al., 2015; Zou et al., 2016). PCR amplification of the ITS region, TEF, RPB1, and RPB2 used primers and PCR conditions described in previous research (Zhou et al., 2018). For phylogenetic analysis of Lecanicillium, sequences of selected taxa based on recent phylogenetic studies of Lecanicillium (Crous et al., 2020; Ponizovskaya et al., 2020; Wang et al., 2020; Zhang et al., 2020), Akanthomyces, and other genera of Cordycipitaceae (Sung et al., 2007; Kepler et al., 2017; Mongkolsamrit et al., 2018) were downloaded from the National Center for Biotechnology Information (NCBI) databases.2 A total of 135 accessions were selected for this study (Supplementary Table 1).
Sequence Alignment and Phylogenetic Analyses
The DNA sequences used in this study were edited using LASERGENE software (version 6.0; DNASTAR, Madison002C WI, United States). Multiple sequence alignments for TEF, RPB1, and RPB2 were performed in MAFFT (Katoh et al., 2009) with the default settings. Multiple sequence alignments for ITS were conducted using the MUSCLE algorithm (Edgar, 2004) from MEGA 6 (Tamura et al., 2013). A multiple alignment of the combined partial ITS + TEF + RPB1 + RPB2 sequences was assembled with MEGA 6 (Tamura et al., 2013) and SEQUENCEMATRIX 1.7.8 (Vaidya et al., 2011). The command “hompart” in PAUP* 4.0b10 was used for the assessment of concordance amongst the genes and the ITS region (Swofford, 2001). Bayesian inference (BI) was performed using MRBAYES 3.2 (Ronquist et al., 2012) and maximum likelihood (ML) analysis was performed using RAxML (Muvea et al., 2014) to analyze the combined data, which were divided into twelve separate partitions (Kepler et al., 2017; Mongkolsamrit et al., 2018). Nucleotide substitution models were determined by MrModeltest 2.3 (Nylander et al., 2004). For BI, 10,000,000 generations were performed with one tree selected every 500th generation and the GTR + I + G evolutionary model was used. For ML, the model GTRGAMMA was used and a bootstrap analysis with 500 replicates was performed to assess statistical support for the tree topology. Phylogenetic trees were viewed with TREEGRAPH.
Divergence Time Estimation of Lecanicillium in Cordycipitaceae
The Bayes MCMC algorithm was performed to estimate divergence time with the BEAST v. 2.4.5 software package (Bouckaert et al., 2014). An evolutionary event was used in the analysis as a soft constraint following a uniform limitation of one internal calibration point corresponding to the fossil record P. coccophagus, which was a fungal parasite of a scale insect from the Cretaceous period (99–105 Mya) (Sung et al., 2008; Berbee and Taylor, 2010). The tree topology was estimated by RAxML in the final step of the process. The data by gene was partitioned using the general time reversible (GTR) substitution model for each partition with jModelTest v. 2.1.7. A relaxed clock log normal model was used for BEAST. BEAST analyses were run for 50,000,000 generations, logging parameters, and trees every 1,000 generations. Convergence, mixing, and the effective sample sizes (ESS) of parameters were checked in Tracer v.1.6.5 (Zhang et al., 2018). Three repeat analyses were performed for accuracy and LogCombiner (Bouckaert et al., 2014) was used to combine the runs. The first 20% of trees representing the starting and unreliable results were removed from the analysis and a maximum clade tree was created with TreeAnnotator v. 2.4.5 (Bouckaert et al., 2014).
Results
Phylogenetic Analyses
The sequence data set consisted of 1783 characters, including inserted gaps (ITS: 511 bp; TEF: 404 bp; RPB1: 499 bp; RPB2: 369 bp). There was no noticeable difference in topology between the BI and ML phylogenies and their topologies reflected the same evolutionary relationships. The trees were reconstructed with almost all species of the genus Lecanicillium (only Lecanicillium evansii could not be found in the NCBI database). All major clades in Cordycipitaceae were strongly supported with ML bootstrap proportion and Bayesian posterior probabilities (Figure 1) and the overall topology of the best tree generated in ML analysis was essentially congruent with previous phylogenetic studies of Cordycipitaceae. Lecanicillium huhutii formed an independent branch in a polytomy together with a clade containing L. tenuipes (BI posterior probabilities 1, ML bootstrap 100%). The L. huhutii lineage received maximum statistical support (BI posterior probabilities 1, ML bootstrap 100%).
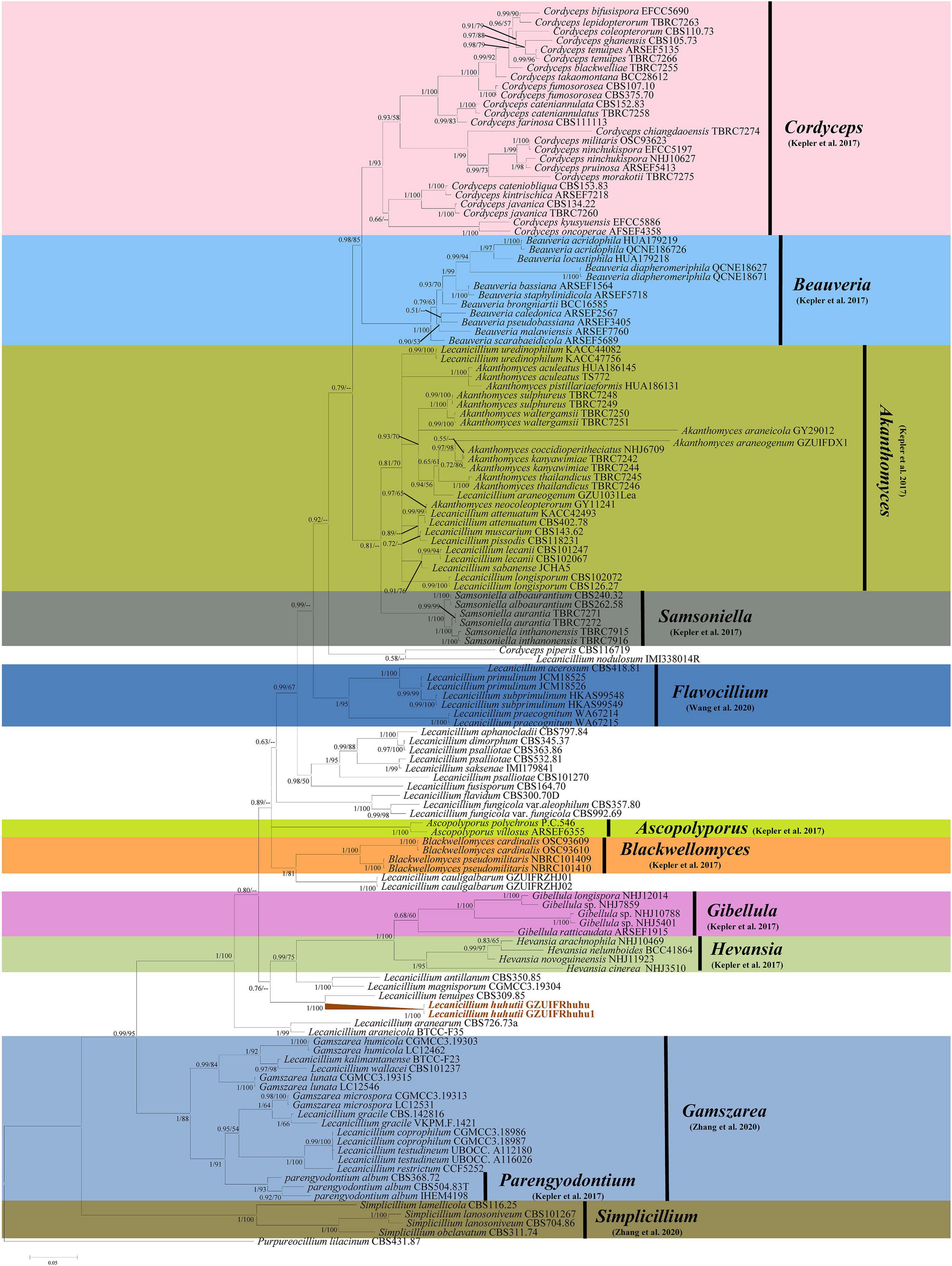
Figure 1. Phylogenetic relationships of the form genera Lecanicillium, Cordyceps, Beauveria, Akanthomyces, Samsoniella, Flavocillium, Ascopolyporus, Blackwellomyces, Gibellula, Hevansia, Gamszarea, Parengyodontium, and Simplicillium in the family Cordycipitaceae. Numbers near the nodes (bootstrap percentage values) correspond to the Bayesian posterior probabilities (BPP) and the ML methods. Bootstrap percentage values are based on 1,000,000 replicates and only the bootstrap proportions ≥ 0.50 are provided. ML values < 50% are shown as “–.” The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree.
Dating and Evolution of Lecanicillium in the Family Cordycipitaceae
To explore the evolution of internal phylogeny, origin, and evolutionary history of Lecanicillium, all species of Lecanicillium and other members of the Cordycipitaceae based on previous phylogenetic studies were selected. The clustering of taxa of genera in the family Cordycipitaceae (Figure 2) almost matched the topology of the resulting consensus phylogenetic tree (Figure 1). Dating analyses supported the supposition that the ancestor of the genus Lecanicillium was in the Cretaceous period (84.36 Mya, 95% HPD: 72.12–94.74 Mya). After originating from a common ancestor, eight clades of Lecanicillium were derived and evolved independently in parallel with other genera of Cordycipitaceae (Figure 2 and Table 1). Based on the clear divergence age estimates, Lecanicillium clade 8 originated earlier as an independent group in the Cretaceous period (75.61 Mya, 95% HPD: 63.31–87.54 Mya), while Lecanicillium clades 1–7 originated later as an independent group in the boundary of the Cretaceous and Paleogene periods (64.66 Mya, 95% HPD: 52.75–76.74 Mya). These analyses also provided a clear direction for the attribution of Lecanicillium (Figure 2 and Table 1).
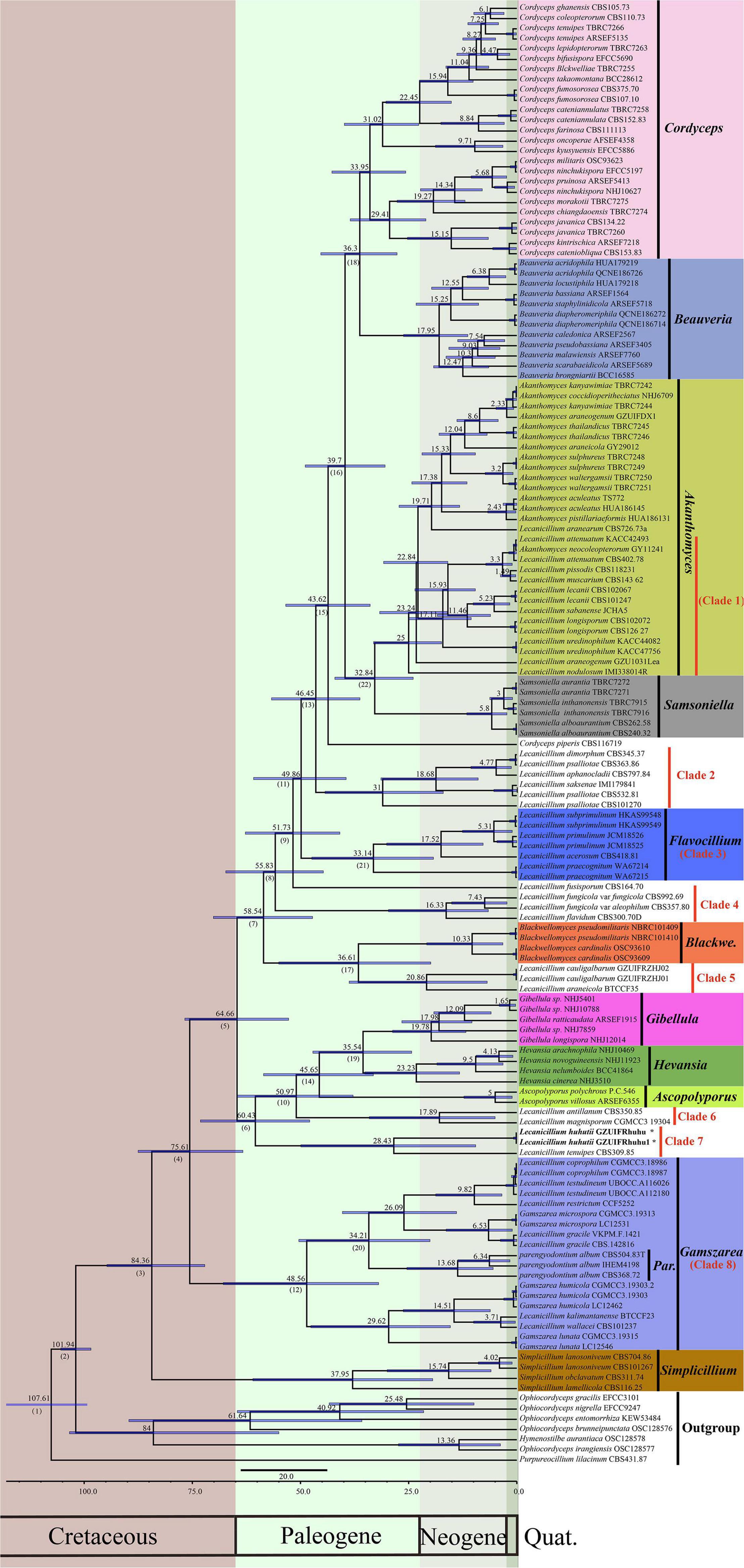
Figure 2. Maximum clade credibility chronogram of Lecanicillium in Cordycipitaceae evolution. The chronogram results from the BEAST analysis are based on the topology of the ML tree. Each node represents the mean divergence time estimate (displayed above the nodes), while bars show the 95% HPD. Numbers corresponding to dated groups shown in Table 1 are indicated in square brackets above the nodes. The proposed novel species of the genus Lecanicillium is indicated in bold font and marked with “*”.
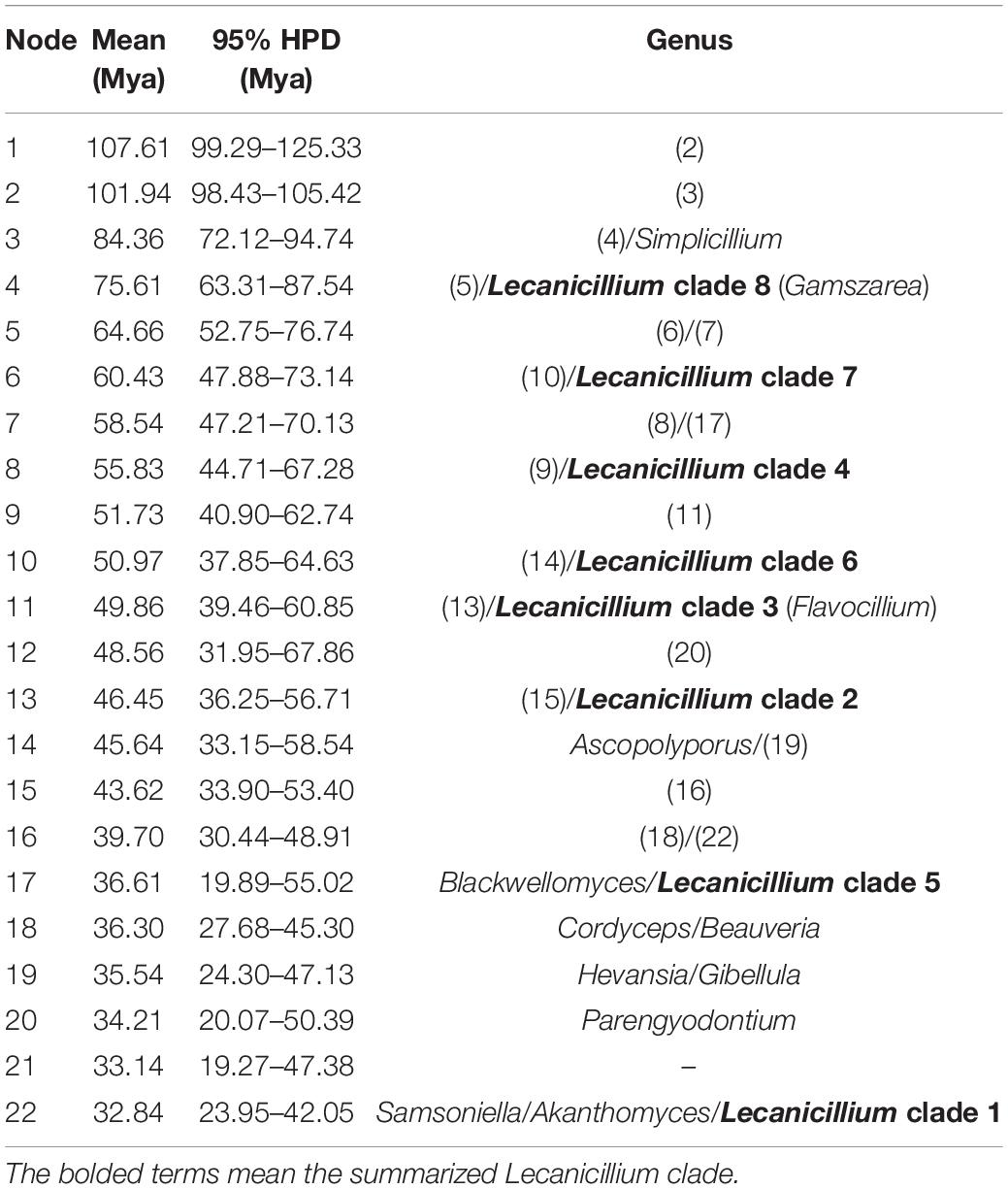
Table 1. Mean and range (95% HPD) divergence time estimations (Mya) of the genus Lecanicillium in the family Cordycipitaceae.
Taxonomy
Lecanicillium huhutii X. Zou and Y.M. Zhou, sp.nov-MycoBank: MB835961
Etymology: The epithet “huhutii” refers to the infant name of the discoverer (a 4-year-old boy).
Type: China. Guizhou Province: Guiyang City, Huaxi park, 5 Oct 2013, Sequences from isolated strain GZUIFRhuhu have been deposited in GenBank (ITS: MN944445; TEF: MT006068; RPB1: MT006058; RPB2: MT006063). The information of each sequence of strain GZUIFRhuhu1 was the same as that of strain GZUIFRhuhu.
Description: Colonies on PDA are 72 mm in diameter after 20 days at 25°C, and circular, white, partly yellowish, and reverse yellowish-white. Mycelia are 0.9–1.8 μm, wide, hyaline, and smooth. There are two kinds of conidiogenous cells: (1) arising from aerial hyphae, solitary or 2–3 whorls, 3.5–12 × 0.5–1 μm; (2) very short, arising from conidiogenous cells or aerial hyphae, 0.7–1.2 × 0.3–0.5 μm. Conidia are oval, aseptate, and 1.5–2.4 × 1–1.5 μm. In culture, both phialides and conidia are of similar general shape and size to those found on the host stemborer (Figure 3).
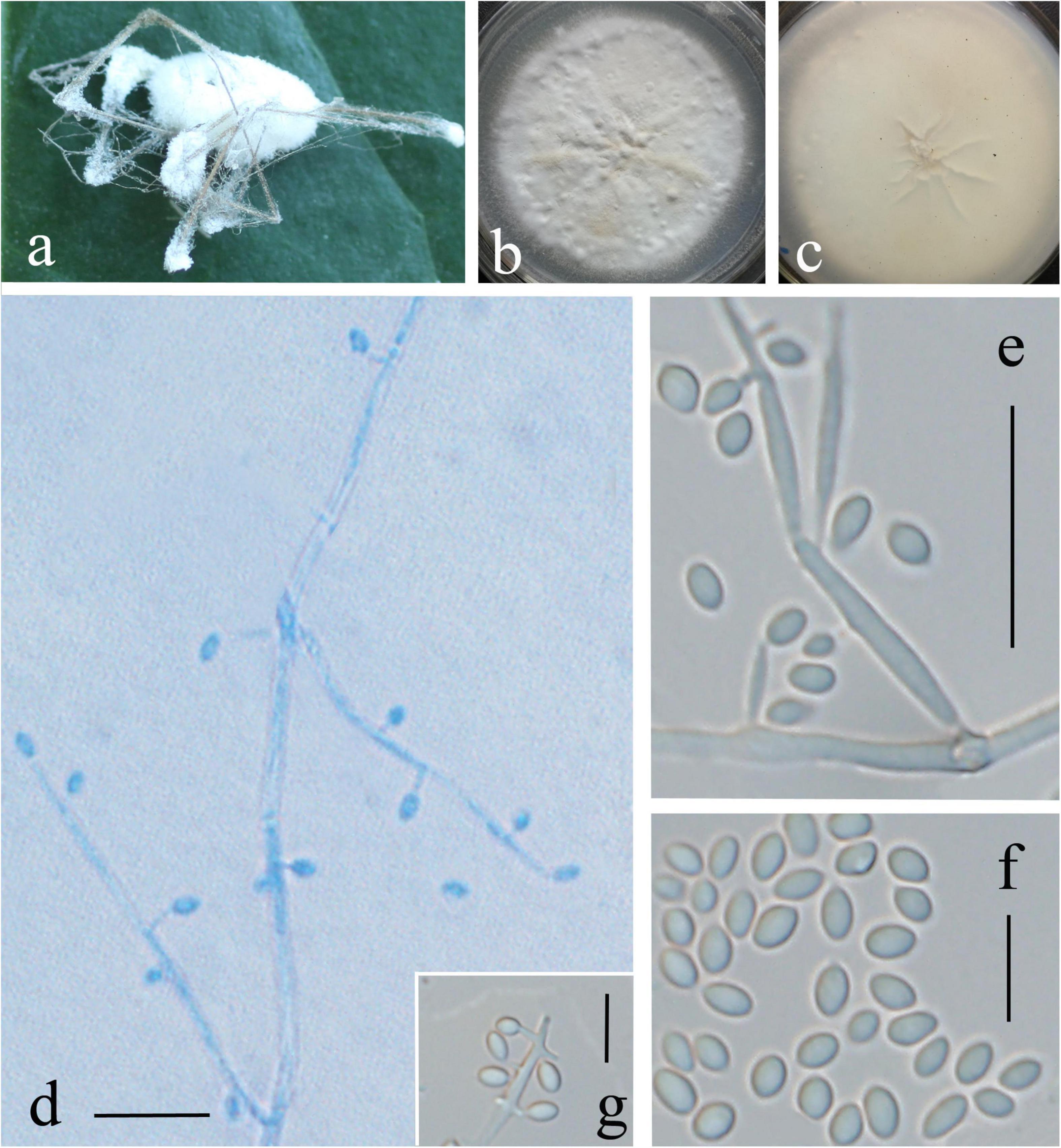
Figure 3. Lecanicillium huhutii. (a) Synnemata emerged from the corpse of a spider. (b,c) Culture plate, cultured on PDA medium. (d–g) Conidiophores, conidiogenous cells, and conidia. Bars: (d,e) = 10 mm; (f,g) = 5 μm.
Host: Spider
Habitat and distribution: Hidden in a hole of a tree in Huaxi park of Guizhou province, China.
Teleomorph: Not known.
Remarks: Regarding phylogenetic relationships, L. huhutii is closely related to L. tenuipes. The two strains (GZUIFRhuhu and GZUIFRhuhu1) formed a distinct lineage. All species of Lecanicillium were included in the phylogenetic analysis except for L. evansii for which sequence data could not be located in public databases, although Zare and Gams (2001) published ITS sequences. The morphological features of L. evansii include brownish-cream to brown reverse colonies and two types of conidia, slightly falcate with pointed end macroconidia, 4.5–7.5 × 0.8–1.2 μm, and slightly curved microconidia, 2.0–3.0 × 0.8–1.2 μm (Zare and Gams, 2001). L. evansii is distinct from L. huhutii, which has one type of conidia (1.5–2.4 × 1–1.5 μm) and two types of phialides.
Regarding the morphology of Lecanicillium, only L. dimorphum is reported to have two kinds of conidiogenous cells. However, L. huhutii can be clearly distinguished by the size of the phialides (A, 14–30 × 1.0–1.5 μm; B, 5–12 × 0.7–1.5 μm) and conidia (A, 6–11 × 1.5–2.5 μm; B, 2.5–4.5 × 1.0–1.5 μm) (Zare and Gams, 2001).
Discussion
The genus Lecanicillium was previously shown to be a polyphyletic group (Zare and Gams, 2001; Kepler et al., 2017), which is supported by the phylogeny in the current study. Eight clades of Lecanicillium were derived and evolved independently in parallel with other genera of the family Cordycipitaceae. The Lecanicillium species were transferred to Akanthomyces by Kepler et al. (2017) and were all included in Lecanicillium clade 1 based on the phylogeny in the current study. All other species in this group (Lecanicillium pissodis, L. araneogemum, L. uredinophilum, and L. nodulosum) should be transferred to Akanthomyces, as mentioned in our previous work (Zhou et al., 2018). Significantly, all Lecanicillium clade 1 members are distinct from other members of the genus Akanthomyces, with the Lecanicillium clade 1 members possess a relatively uniform sporulation structure of globose heads with a higher number of conidia.
As more members of the genus are discovered in the future, it will be interesting to ascertain whether Lecanicillium clade 1 should be re-established as a separate genus. Alternatively, it may be worth revisiting this clade in detail, as in the case of Isaria, which was rejected by the same recommendation of (Kepler et al., 2017; Mongkolsamrit et al., 2018). Zhang et al. (2020) transferred five species of Lecanicillium to a novel genus with the name Gamszarea, which is also supported by the phylogeny of Lecanicillium clade 8 in the current study. Significantly, the genus Parengyodontium is included in the Gamszarea. Considering Parengyodontium album is an important human pathogen (Tsang et al., 2016), it will be interesting to determine whether other species of the genus Gamszarea can also cause human disease, warranting further investigation in the future. In addition, the phylogeny in the current study indicates that L. gracile (Ponizovskaya et al., 2020) should be transferred to the genus Gamszarea. Wang et al. (2020) transferred three species of Lecanicillium to a new genus as Flavocillium based on ITS sequence data. This is also supported by the phylogeny in the current study, with Lecanicillium clade 3 including L. pracecognitum (Crous et al., 2020). Significantly, the differences in the divergence time estimations of Lecanicillium clades 2-5 were smaller compared with those of other genera of the Cordycipitaceae. Considering the morphological and structural similarities of Lecanicillium clade 2-5, members of these clades are likely to change as more new species are discovered in the future. Therefore, more species are needed to resolve the attribution problem of Lecanicillium spp.
In the current study, a direct relationship between Lecanicillium and the Cretaceous-Tertiary extinction event was not detected (Schulte et al., 2010). This suggests that the diversity of Lecanicillium is more likely to be caused by long-term environmental adaptation and coevolution with insects, rather than by dramatic extinction events. In the Cretaceous period, the radiation of diverse monocot and eudicot lineages began to change the organisms that interacted with each other, including diversification of the pollinating insects, phytophagous insects, and entomogenous fungi (Labandeira and Sepkoski, 1993; Soltis and Soltis, 2004; Sung et al., 2008).
Spider-pathogenic fungi are widely distributed globally. The first records of spider-pathogenic fungi appear in 1856 (Gray, 1858). Evans and Samson (1987) note that all confirmed and unconfirmed parasitic spider fungi belong to the order Hypocreales. Interestingly, spider-pathogenic fungi are almost restricted to the families of Cordycipitaceae and Ophiocordycipitaceae within this order, with no reports of any spider-pathogenic fungi belonging to Clavicipitaceae within Hypocreales (Shrestha et al., 2019). Statistically, spider-pathogenic fungi are predominantly concentrated in the family Cordycipitaceae of Hypocreales with 8 genera and 75 species out of 13 genera and 86 species (Shrestha et al., 2019). Among them, there are 5 spider-pathogenic species of Lecanicillium, including L. lecanii, L. tenuipes, L. aranearum (Zare and Gams, 2001), L. araneicola (Sukarno et al., 2009), and L. araneogenum (Chen et al., 2017). With the readjustment of the Cordycipitaceae, spider pathogens with Cordyceps-, Isaria-, Lecanicillium-, or Torrubiella-like morphs have been described in the genus Akanthomyces (Kepler et al., 2017). However, since the taxonomic status of the genus Lecanicillium has not been completely solved, only L. lecanii and L. araneogenum have been placed into Akanthomyces at present. In addition, Kepler’s suggestion to re-use the name Engyodontium is another problem to consider. According to previous descriptions, L. aranearum (Petch) Zare and W. Gams (≡ Cephalosporium aranearum Petch, = Engyodontium arachnophilum), associated with Torrubiella alba Petch, and L. tenuipes (Petch) Zare and W. Gams (≡ Acremonium tenuipes Petch, = Engyodontium aranearum) (Shrestha et al., 2019). Given that there is only one recognized species of Engyodontium, which is E. parvisporum (Kepler et al., 2017), whether these two kinds of appellation (L. tenuipes and L. aranearum) should be restired requires further discussion. Although the novel species described in the current study is close to L. tenuipes in the phylogenetic tree, we think this species is clearly distinguished from Engyodontium by morphology. We also treat the novel species as Lecanicillium, considering the small sample and the unknown teleomorph. Thus, based on the present molecular phylogeny, derived from nuclear and ribosomal DNA sequence data, together with morphological evidence, a distinct novel species of the genus Lecanicillium is proposed, L. huhutii, a new spider-pathogenic fungus.
Conclusion
The pivotal internal phylogeny, origin, and evolutionary history of Lecanicillium in the family Cordycipitaceae were studied. Phylogenetic and morphological analyses indicated there were eight representative clades (four representative branches of evolutionary history), including clade 1 (members have a relatively uniform sporulation structure comprising globose heads with a higher number of conidia), clade 8 (including all members of Gamszarea), clades 2–5 (the differences in divergence time estimations were smaller for these clades compared with other clades), and clades 6–7 (members are close to Gibellula, Hevansia and Ascopolyporus). L. pissodis, L. araneogemum, L. uredinophilum, and L. nodulosum in clade 1 should be transferred to the genus Akanthomyces. The monotypic genus Parengyodontium should be merged with the genus Gamszarea. More new species need to be discovered to thoroughly resolve the attribution problem of Lecanicillium. Finally, no major lineages of Lecanicillium were correlated with the nearby Cretaceous-Tertiary extinction event, indicating that the diversity of Lecanicillium is more likely to be caused by long-term environmental adaptation and coevolution with insects rather than by dramatic extinction events.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI GenBank, accession numbers: ITS: MN944445; TEF: MT006068; RPB1: MT006058; RPB2: MT006063.
Author Contributions
Y-MZ performed the experiments, analyzed the data, and wrote the article. J-RZ revised the manuscript. J-JQ analyzed the data. XZ isolated the fungus and revised the manuscript. All authors reviewed the manuscript.
Funding
This study was supported by the National Science Foundation of China (Project Nos. 31860507 and 31860037), Guizhou International Science and Technology Cooperation Base (Project No. G[2016]5802), and the Science and Technology Plan Projects of Guizhou Province (Project No. Qian Ke He Foundation [2020]1Z009).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.859886/full#supplementary-material
Abbreviations
GZAC, Institute of Fungal Resources of Guizhou University; ITS, ITS1-5.8S rDNA-ITS2 region the first and the internal transcribed spacers; LSU, Large Subunits of the rDNA; NCBI, National Center for Biotechnology Information; PDA, potato dextrose agar; RPB1, DNA-directed RNA polymerase II subunit rpb1; RPB2, DNA-directed RNA polymerase II subunit rpb2; SSU, small subunits of the rDNA; TEF, transcription elongation factor-1 α .
Footnotes
References
Berbee, M. L., and Taylor, J. W. (2010). Dating the molecular clock in fungi-how close are we? Fungal Biol. Rev. 24, 1–16. doi: 10.1016/j.fbr.2010.03.001
Bouckaert, R., Heled, J., Kuhnert, D., Vaughan, T., Wu, C. H., Xie, D., et al. (2014). BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10:e1003537. doi: 10.1371/journal.pcbi.1003537.g001
Chen, W. H., Han, Y. F., Liang, Z. Q., and Jin, D. C. (2017). Lecanicillium araneogenum sp. nov., a new araneogenous fungus. Phytotaxa 305, 29–34. doi: 10.11646/phytotaxa.305.1.4
Chiriví-Salomón, J. S., Danies, G., Restrepo, S., and Sanjuan, T. (2015). Lecanicillium sabanense sp. nov. (Cordycipitaceae) a new fungal entomopathogen of coccids. Phytotaxa 234, 63–74. doi: 10.11646/phytotaxa.234.1.4
Crous, P. W., Wingfield, M. J., Chooi, Y. H., Gilchrist, C. L. M., Lacey, E., Pitt, J. I., et al. (2020). Fungal Planet description sheets: 1042-1111. Pers. Mol. Phylogeny Evol. Fungi 44, 301–459. doi: 10.3767/persoonia.2020.44.11
Edgar, R. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Evans, H. C., and Samson, R. A. (1987). Fungal pathogens of spiders. Mycologists 1, 152–159. doi: 10.1016/S0269-915X(87)80107-6
Faria, M. R., and Wraight, S. P. (2007). Mycoinsecticides and Mycoacaricides: A comprehensive list with worldwide coverage and international classification of formulation types. Biol. Control 43, 237–256. doi: 10.1016/j.biocontrol.2007.08.001
Gams, W., and Zare, R. (2001). A revision of Verticillium sect. Prostrata. III. Generic classification°). Nova Hedwigia 72, 47–55.
Goettel, M. S., Koike, M., Kim, J. J., Aiuchi, D., Shinya, R., and Brodeur, J. (2008). Potential of Lecanicillium spp. for management of insects, nematodes and plant diseases. J. Invertebr. Pathol. 98, 256–261. doi: 10.1016/j.jip.2008.01.009
Gray, G. R. (1858). Notices of Insects that are Known to Form the Bases of Fungoid Parasites. London: British Museum.
Katoh, K., Asimenos, G., and Toh, H. (2009). Multiple alignment of DNA sequences with MAFFT. Meth. Mol. Biol. 537, 39–64.
Kepler, R. M., Luangsa-ard, J. J., Hywel-Jones, N. L., Quandt, C. A., Sung, G. H., Rehner, S. A., et al. (2017). A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 8, 335–353. doi: 10.5598/imafungus.2017.08.02.08
Labandeira, C. C., and Sepkoski, J. J. (1993). Insect diversity in the fossil record. Science 261, 310–315. doi: 10.1126/science.11536548
McNeill, J., Barrie, F. F., Buck, W. R., Demoulin, V., Greuter, W., Hawkworth, D. L., et al. (2012). International Code of Nomenclature for Algae, Fungi, and Plants (Melbourne Code). [Regnum Vegetabile No. 154.] Königstein: Koeltz Scientific Books.
Mongkolsamrit, S., Noisripoom, W., Thanakitpipattana, D., Wutikhun, T., Spatafora, J. W., and Luangsaard, J. (2018). Disentangling cryptic species with isaria-like morphs in cordycipitaceae. Mycologia 110, 230–257. doi: 10.1080/00275514.2018.1446651
Muvea, A. M., Meyhofer, R., Subramanian, S., Poehling, H. M., Ekesi, S., and Maniania, N. K. (2014). Colonization of onions by endophytic fungi and their impacts on the biology of Thrips tabaci. PLoS One 9:e108242. doi: 10.1371/journal.pone.0108242
Nylander, J. A. A., Ronquist, F., Huelsenbeck, J. P., Nieves-Aldrey, J., and Buckley, T. (2004). Bayesian phylogenetic analysis of combined data. Syst. Biol. 53, 47–67. doi: 10.1080/10635150490264699
Ponizovskaya, V. B., Grum-Grzhimaylo, A. A., Georgieva, M. L., Kokaeva, L. Y., and Bilanenko, E. N. (2020). Lecanicillium gracile (Cordycipitaceae), a new species isolated from mineral building materials. Phytotaxa 443, 265–278. doi: 10.11646/phytotaxa.443.3.3
Ronquist, F., Teslenko, M., Van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schulte, P., Alegret, L., Arenillas, I., Arz, J. A., Barton, P. J., Bown, P. R., et al. (2010). The Chicxulub asteroid impact and mass extinction at the Cretaceous-Paleogene boundary. Science 327, 1214–1218. doi: 10.1126/science.1177265
Shrestha, B., Kubátová, A., Tanaka, E., Oh, J., Yoon, D. H., Sung, J. M., et al. (2019). Spider-pathogenic fungi within Hypocreales (Ascomycota): their current nomenclature, diversity, and distribution. Mycol. Prog. 18, 983–1003. doi: 10.1007/s11557-019-01512-3
Soltis, P. S., and Soltis, D. E. (2004). The origin and diversification of angiosperms. Am. J. Bot. 91, 1614–1626. doi: 10.3732/ajb.91.10.1614
Su, L., Zhu, H., Guo, Y., Du, X., Guo, J., Zhang, L., et al. (2019). Lecanicillium coprophilum (Cordycipitaceae, Hypocreales), a new species of fungus from the feces of Marmota monax in China. Phytotaxa 387, 55–62.
Sukarno, N., Kurihara, Y., Park, J. Y., Inaba, S., Ando, K., Harayama, S., et al. (2009). Lecanicillium and Verticillium species from Indonesia and Japan including three new species. Mycoscience 50, 369–379. doi: 10.1007/s10267-009-0493-1
Sung, G. H., Hywel-Jones, N. L., Sung, J. M., Luangsa-ard, J. J., Shrestha, B., and Spatafora, J. W. (2007). Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 57, 5–59. doi: 10.3114/sim.2007.57.01
Sung, G. H., Poinar, G. O., and Spatafora, J. W. (2008). The oldest fossil evidence of animal parasitism by fungi supports a Cretaceous diversification of fungal-arthropod symbioses. Mol. Phylogenet. Evol. 49, 495–502. doi: 10.1016/j.ympev.2008.08.028
Swofford, D. L. (2001). PAUP*: Phylogenetic Analysis Using Parsimony (and other methods) 4.0. b10. Sunderland, MA: Sinauer Associates.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tsang, C. C., Chan, J. F. W., Pong, W. M., Chen, J. H. K., Ngan, A. H. Y., Cheung, M., et al. (2016). Cutaneous hyalohyphomycosis due to Parengyodontium albumgen. et comb. nov. Med. Mycol. 54, 699–713. doi: 10.1093/mmy/myw025
Vaidya, G., Lohman, D. J., and Rudolf, M. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180.
Wang, Y. B., Wang, Y., Fan, Q., Duan, D. E., Zhang, G. D., Dai, R. Q., et al. (2020). Multigene phylogeny of the family Cordycipitaceae (Hypocreales): new taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers. 103, 1–46. doi: 10.1007/s13225-020-00457-3
Wijayawardene, N. N., Hyde, K. D., Rajeshkumar, K. C., Hawksworth, D. L., Madrid, H., Kirk, P. M., et al. (2017). Notes for genera: ascomycota. Fungal Divers. 86, 1–594. doi: 10.1007/s13225-017-0386-0
Zare, R., and Gams, W. (2001). The genera Lecanicillium and Simplicillium gen. nov. Nova Hedwigia 73, 1–50.
Zare, R., and Gams, W. (2008). A revision of the Verticillium fungicola species complex and its affinity with the genus Lecanicillium. Mycol. Res. 112, 811–824. doi: 10.1016/j.mycres.2008.01.019
Zhang, Z. F., Zhao, P., and Cai, L. (2018). Origin of cave fungi. Front. Microbiol. 9:1407. doi: 10.3389/fmicb.2018.01407
Zhang, Z. F., Zhou, S. Y., Eurwilaichitr, L., Ingsriswang, S., Raza, M., Chen, Q., et al. (2020). Culturable mycobiota from Karst caves in China II, with descriptions of 33 new species. Fungal Divers. 106, 29–136. doi: 10.1007/s13225-020-00453-7
Zhou, Y. M., Zhi, J. R., Ye, M., Zhang, Z. Y., Yue, W. B., and Zou, X. (2018). Lecanicillium cauligalbarum sp. nov. (Cordycipitaceae, Hypocreales), a novel fungus isolated from a stemborer in the Yao Ren National Forest Mountain Park, Guizhou. MycoKeys 43, 59–74. doi: 10.3897/mycokeys.43.30203
Zhou, Y. M., Zou, X., Zhi, J. R., Xie, J. Q., and Jiang, T. (2020). Fast recognition of Lecanicillium spp., and its virulence against Frankliniella occidentalis. Front. Microbiol. 11:561381. doi: 10.3389/fmicb.2020.561381
Keywords: entomopathogenic fungi, Lecanicillium spp., new species, origin and evolution, phylogeny
Citation: Zhou YM, Zhi JR, Qu JJ and Zou X (2022) Estimated Divergence Times of Lecanicillium in the Family Cordycipitaceae Provide Insights Into the Attribution of Lecanicillium. Front. Microbiol. 13:859886. doi: 10.3389/fmicb.2022.859886
Received: 21 January 2022; Accepted: 29 March 2022;
Published: 06 May 2022.
Edited by:
George Tsiamis, University of Patras, GreeceReviewed by:
Samantha Chandranath Karunarathna, Qujing Normal University, ChinaNicolaas A. van der Merwe, University of Pretoria, South Africa
Copyright © 2022 Zhou, Zhi, Qu and Zou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao Zou, xzou@gzu.edu.cn
 Ye-Ming Zhou
Ye-Ming Zhou Jun-Rui Zhi1
Jun-Rui Zhi1 Xiao Zou
Xiao Zou