- 1Center for Yunnan Plateau Biological Resources Protection and Utilization, Yunnan Engineering Research Center of Fruit Wine, College of Biological Resource and Food Engineering, Qujing Normal University, Qujing, China
- 2Section of Genetics, Institute for Research and Development in Health and Social Care, Battaramulla, Sri Lanka
- 3Centre for Mountain Futures, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 4Department of Economic Plants and Biotechnology, Yunnan Key Laboratory for Wild Plant Resources, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 5Research Centre of Microbial Diversity and Sustainable Utilization, Chiang Mai University, Chiang Mai, Thailand
As a result of an ongoing survey of microfungi associated with garden and ornamental plants in Qijing, Yunnan, China, several saprobic fungal taxa were isolated from Magnolia grandiflora. Both morphological and combined SSU, LSU, ITS, tef1, and rpb2 locus phylogenetic analyses (maximum-likelihood and Bayesian analyses) were carried out to identify the fungal taxa. Three new species are introduced in Pleosporales, viz., Lonicericola qujingensis (Parabambusicolaceae), Phragmocamarosporium magnoliae, and Periacma qujingensis (Lentitheciaceae). Botryosphaeria dothidea, Diplodia mutila, and Diplodia seriata (in Botryosphaeriaceae) are reported from Magnolia grandiflora for the first time in China. Angustimassarina populi (Amorosiaceae) is reported for the first time on M. grandiflora from China, and this is the first report of a member of this genus outside Europe. Shearia formosa is also reported for the first time on M. grandiflora from China.
Introduction
Discovering missing taxa in the fungal tree (or in Kingdom Fungi) is one of the popular topics among taxonomists. Recent species estimation studies have predicted that tropical regions harbor higher fungal diversity than previously expected (Hawksworth and Lücking, 2017; Hyde et al., 2020). Several studies (which were based on DNA sequence analyses) described numerous fungal species during the last decade from tropical countries such as, Thailand and India (Chaiwan et al., 2020; Calabon et al., 2021; Rajeshkumar et al., 2021). A large number of fungal species have also been described from subtropical China, especially in Yunnan and Guizhou provinces (Luo et al., 2018; Lu et al., 2021; Ren et al., 2021; Wang et al., 2021; Wijayawardene et al., 2021). These studies mentioned that a large number of fungal species are waiting to be discovered in Southwestern China, including the Guizhou and Yunnan Provinces.
Magnolia grandiflora (Southern magnolia) is an evergreen tree that is widely used as an ornamental plant in landscaping (Liu et al., 2019) and a fungal-rich host plant genus and is reported with over 1,000 records of taxa in Farr and Rossman (2022). Seventy-two (72) records have been mainly listed from different substrates of Magnolia alba, M. delavayi, M. denudate, and M. grandiflora from China (Farr and Rossman, 2022). Recently, Wanasinghe et al. (2020) studied fungi associated with Magnolia species in the Kunming Botanical Garden and predicted rich fungal diversity.
In this study, we collected ascomycetous fungi (both sexual and asexual morphs) that occur on different substrates of M. grandiflora from Qujing Normal University garden, Qujing, Yunnan province, China. Based on morpho-molecular analyses and previous literature, two new species of Phragmocamarosporium Wijayaw et al. (in the family Lentitheciaceae) and one new species of Lonicericola Phookamsak et al. (in the family Parabambusicolaceae) have been introduced. Besides, five new host/geographical records (Botryosphaeria dothidea, Diplodia mutila, and D. seriata in Botryosphaeriaceae, Botryosphaeriales, and Angustimassarina populi and Shearia formosa in Amorosiaceae and Longiostiolaceae respectively in the order Pleosporales) are herein reported. All the taxa are provided with illustrations and morphological descriptions. Furthermore, possibilities of revealing novel taxa of the respective genera and their distribution are also discussed.
Materials and methods
Sample collection, isolation, and identification
Samples were collected from aerial and ground litter (i.e., leaves, branches, and stems) of Magnolia grandiflora from September 2019 to June 2021 from Qujing Normal University garden in Yunnan, China. The specimens were stored in paper bags and transferred to the laboratory. The samples were examined with a stereomicroscope, and microscopic images of the samples were taken using a Canon EOS700D digital camera (Canon Inc., Ota, Tokyo, Japan) with a Nikon ECLIPSE Ni (Nikon Instruments Inc., Melville, NY, United States) compound microscope. Microcharacters were observed using a digital camera fitted onto a Nikon ECLIPSE 80i compound microscope. Measurements were per-formed with Tarosoft (R) Image Frame Work (v.0.9.7). More than 20 asci and ascospores (in sexual fungi) and more than 30 conidia and conidiogenous cells (in asexual fungi) were measured. Plates were prepared using the Adobe Photoshop CS6 (Adobe Systems, San Jose, CA, United States) software.
A single-spore isolation was carried out to isolate the taxa as described in Chomnunti et al. (2014), and we used water agar as the medium. A spore suspension was prepared using conidiomata or ascomata, and then the suspension was transferred with a sterile pipette onto the surface of a Petri dish with water agar. Germinated spores (approximately 12 h later) were transferred to a new potato dextrose agar (PDA) medium for purification. Dried specimens and living cultures were deposited at the herbarium and culture collection of Guizhou Medical University, Guizhou Province, China.
DNA extraction, polymerase chain reaction amplification, and sequence analysis
The total genomic DNA of microfungi was extracted from fresh mycelia grown on PDA at 25–27°C using the Biospin Fungus Genomic DNA Extraction Kit (BioFlux®, Hangzhou, People's Republic of China) according to the manufacturer's instructions (Dai et al., 2019).
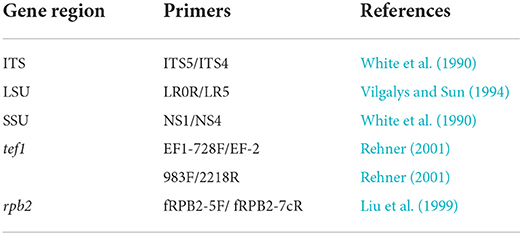
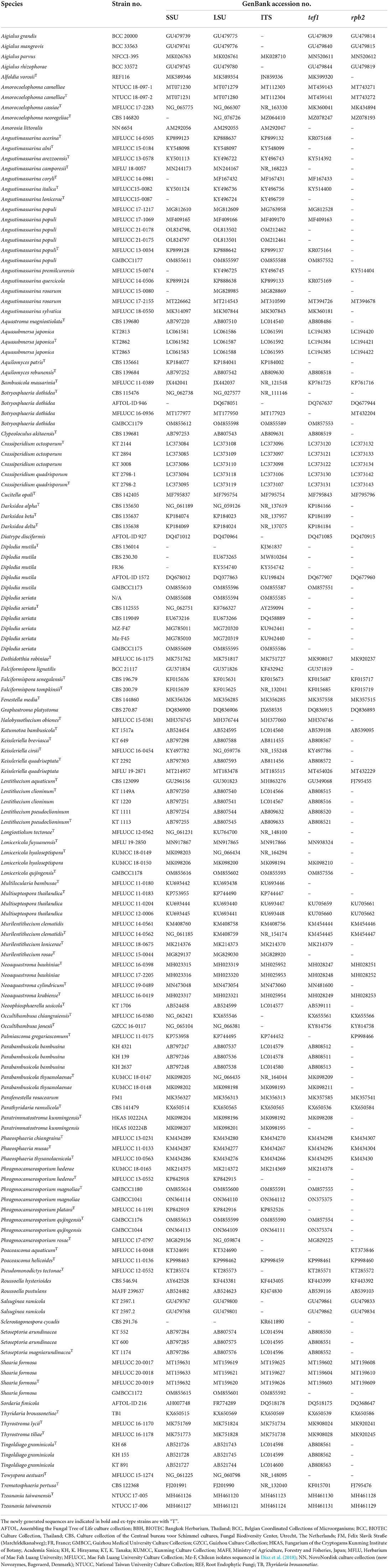
The primers used for amplification are listed in Table 1. PCR amplification conditions are those as followed by Dai et al. (2017). PCR products were sent for sequencing at Shanghai Sangon Biological Engineering Technology & Services Co. (Shanghai, People's Republic of China). All newly generated sequences are deposited in GenBank, and accession numbers are obtained (Table 2).
Sequencing and sequence alignment
Sequences generated from different primers of non-translated loci and protein-coding regions are analyzed with other sequences retrieved from GenBank (Table 2). Sequences with high similarity indices were determined by a BLAST search to find closest matches with taxa in Dothideomycetes and from recently published data (e.g., Thambugala et al., 2015; Tibpromma et al., 2017; Wanasinghe et al., 2018, 2020; Hyde et al., 2020). The multiple alignments of all consensus sequences, as well as the reference sequences, were automatically generated with MAFFT v. 7 (Katoh et al., 2017), and were improved manually when necessary using BioEdit v. 7.0.5.2 (Hall, 1999).
Phylogenetic analyses
Analysis 1 (SSU, LSU, ITS, tef1, and rpb2 multi-sequence analyses of Amorosiaceae, Botryosphaeriaceae, Lentitheciaceae, Longiostiolaceae, and Parabambusicolaceae)
Single-locus data sets were examined for topological incongruence among loci for members of the relevant families. Conflict-free alignments were combined in to the final multi-gene dataset for analyses using BioEdit and concatenated into a multi-locus alignment that was subjected to maximum-likelihood (ML) and Bayesian (BI) phylogenetic analyses. The CIPRES Science Gateway platform (Miller et al., 2010) was used to perform RAxML and Bayesian analyses. ML analyses were performed with RAxML-HPC2 on XSEDE v. 8.2.10 (Stamatakis, 2014) using a GTR + I + G model with 1,000 bootstrap repetitions. Evolutionary models for Bayesian analysis were selected independently for each locus using MrModeltest v. 2.3 (Nylander et al., 2008) under the Akaike Information Criterion (AIC) implemented in both PAUP v. 4.0b10, and GTR + I + G was selected as the best-fit model for all three analyses. MrBayes analyses were performed setting GTR + I + G, 2 M generations, sampling every 100th generation and ending the run automatically when the standard deviation of split frequencies dropped below 0.01 with a burn-in fraction of 0.25.
Analysis 2 (ITS and tef1 sequence analyses of Botryosphaeria sensu stricto)
The ITS and tef1 data sets were examined for topological incongruence among loci for selected members of Botryosphaeria. Conflict-free alignments were concatenated into a multi-locus alignment that was subjected to maximum-likelihood (ML) phylogenetic analysis. The CIPRES Science Gateway platform (Miller et al., 2010) was used to perform RAxML. ML analyses were performed with RAxML-HPC2 on XSEDE v. 8.2.10 (Stamatakis, 2014) using the GTR + I + G model with 1,000 bootstrap repetitions. A Bayesian analysis was performed using SYM + I + G for ITS and GTR + I for tef1 in the final command with 1 M generations. Sampling was conducted on every 100th generation, ending the run automatically when the standard deviation of split frequencies dropped below 0.01 with a burn-in fraction of 0.25.
Analysis 3 (ITS and tef1 sequence analyses of Diplodia sensu stricto)
The ITS and tef1 data sets were examined for topological incongruence among loci for selected members of Diplodia. Conflict-free alignments were concatenated into a multilocus alignment that was subjected to maximum-likelihood (ML) phylogenetic analysis. The CIPRES Science Gateway platform (Miller et al., 2010) was used to perform RAxML. ML analyses were performed with RAxML-HPC2 on XSEDE v. 8.2.10 (Stamatakis, 2014) using the GTR + I + G model with 1,000 bootstrap repetitions. A Bayesian analysis was performed using GTR + I + G for ITS and HKY + G for tef1 in the final command with 1 M generations. Sampling was conducted on every 100th generation, ending the run automatically when the standard deviation of split frequencies dropped below 0.01 with a burn-in fraction of 0.25.
Analysis 4 (ITS, LSU, SSU, and tef1 sequence analyses of Amorosiaceae)
The ITS, LSU, SSU, and tef1 data sets were examined for topological incongruence among loci for selected members of Amorosiaceae. Conflict-free alignments were concatenated into a multi-locus alignment that was subjected to maximum-likelihood (ML) phylogenetic analysis. The CIPRES Science Gateway platform (Miller et al., 2010) was used to perform RAxML. ML analyses were performed with RAxML-HPC2 on XSEDE v. 8.2.10 (Stamatakis, 2014) using the GTR + I + G model with 1,000 bootstrap repetitions. MrBayes analyses were performed setting GTR + I + G, 1 M generations, sampling every 100th generation, ending the run automatically when the standard deviation of split frequencies dropped below 0.01 with a burn-in fraction of 0.25.
Analysis 5 (SSU, LSU, tef1, and ITS sequence analyses of Lentitheciaceae)
Raw sequences were combined using SeqMan and subjected to BLAST in GenBank. The SSU, LSU, tef1, and ITS sequence data closely related to our taxa were retrieved from the NCBI GenBank and are listed in Table 1. Single gene sequence alignment was generated with the MAFFT v. 7 online program (http://mafft.cbrc.jp/alignment/server/) (Katoh et al., 2017). FASTA alignment formats were changed to PHYLIP and NEXUS formats with Aliview 2.11. The single-gene datasets were examined for topological incongruence among loci and the conflict-free alignments were concatenated into a multi-locus alignment that was subjected to ML and BI phylogenetic analyses. The CIPRES Science Gateway platform (Miller et al., 2010) was used to perform RAxML. ML analyses were conducted with RAxML-HPC2 on XSEDE v. 8.2.10 (Stamatakis, 2014) using GTR + I + G model with 1,000 bootstrap repetitions. MrBayes analyses were performed setting GTR + I + G, two parallel runs were conducted, using the default settings, six simultaneous Markov chains were run for 1 M generations, and trees were sampled every 100th generation. The run ended automatically when the standard deviation of split frequencies dropped below 0.01 with a burn-in fraction of 0.2. Phylograms were visualized with the FigTree v1.4.0 program (Rambaut, 2012) and reorganized in Microsoft PowerPoint (2019) and Adobe Illustrator® CS5 (Version 15.0.0, Adobe®, San Jose, CA).
Results
Phylogenetic analyses
Analysis 1 (SSU, LSU, ITS, tef1, and rpb2 multi-sequence analyses of Amorosiaceae, Botryosphaeriaceae, Lentitheciaceae, Longiostiolaceae, and Parabambusicolaceae): The concatenated dataset (SSU, LSU, ITS, tef1, and rpb2 loci) contained 140 isolates, and the tree was rooted to Diatrype disciformis (AFTOL-ID 927), Graphostroma platystoma (CBS 270.87), and Sordaria fimicola (AFTOL-ID 216). The final alignment contained 4,399 characters used for the phylogenetic analyses, including alignment gaps, which were treated as missing data. The RAxML analysis of the combined datasets yielded a best-scoring tree with a final ML optimization likelihood value of −62,337.358653. The matrix had 2,549 distinct alignment patterns, with 26.87% undetermined characters or gaps. Parameters for the GTR + I + G model of the combined amplicons were as follows: estimated base frequencies; A = 0.241265, C = 0.251518, G = 0.269739, and T = 0.237478; substitution rates AC = 1.482469, AG = 3.530918, AT = 1.543663, CG = 1.180062, CT = 7.309441, and GT = 1; proportion of invariable sites I = 0.428895; gamma distribution shape parameter α = 0.610427. Based on the results of MrModel Test, dirichlet base frequencies and the GTR + I + G model were used for the Bayesian analysis. The Bayesian analyses generated 9,001 trees (saved every 100th generation), from which 6,751 were sampled after 25% of the trees were discarded as burn-ins. The alignment contained a total of 2,551 unique site patterns. In the combined multigene phylogenetic analysis, Phragmocamarosporium species (P. hederae, P. platani, and P. rosae) clustered in one clade (87% ML/1 PP, Figure 1), sister to Murilentithecium (100 ML/1 PP, Figure 1). The strain “Sclerostagonospora cycadis” (CBS 291.76) was also nested with Phragmocamarosporium species. The two strains of Phragmocamarosporium hederae (MFLUCC 13-0552 and KUMCC 18-0165) were not monophyletic. The four strains of Phragmocamarosporium (GMBCC1041, GMBCC1044, GMBCC1176, and GMBCC1180) isolated in this study formed a basal terminal clade in Phragmocamarosporium with <70% ML and <0.95 BYPP both single locus and concatenated datasets. In this clade, GMBCC1044 and GMBCC1176 constituted a monophyletic clade with 100 ML/1 PP support values (Figure 1). GMBCC1041 and GMBCC1180 also displayed a strongly supported monophyletic lineage (100% ML/1 PP, Figure 1). These two new lineages are presented here as new species, viz., Phragmocamarosporium magnoliae sp. nov. (GMBCC1041 and GMBCC1180) and P. qujingensis sp. nov. (GMBCC1044 and GMBCC1176).
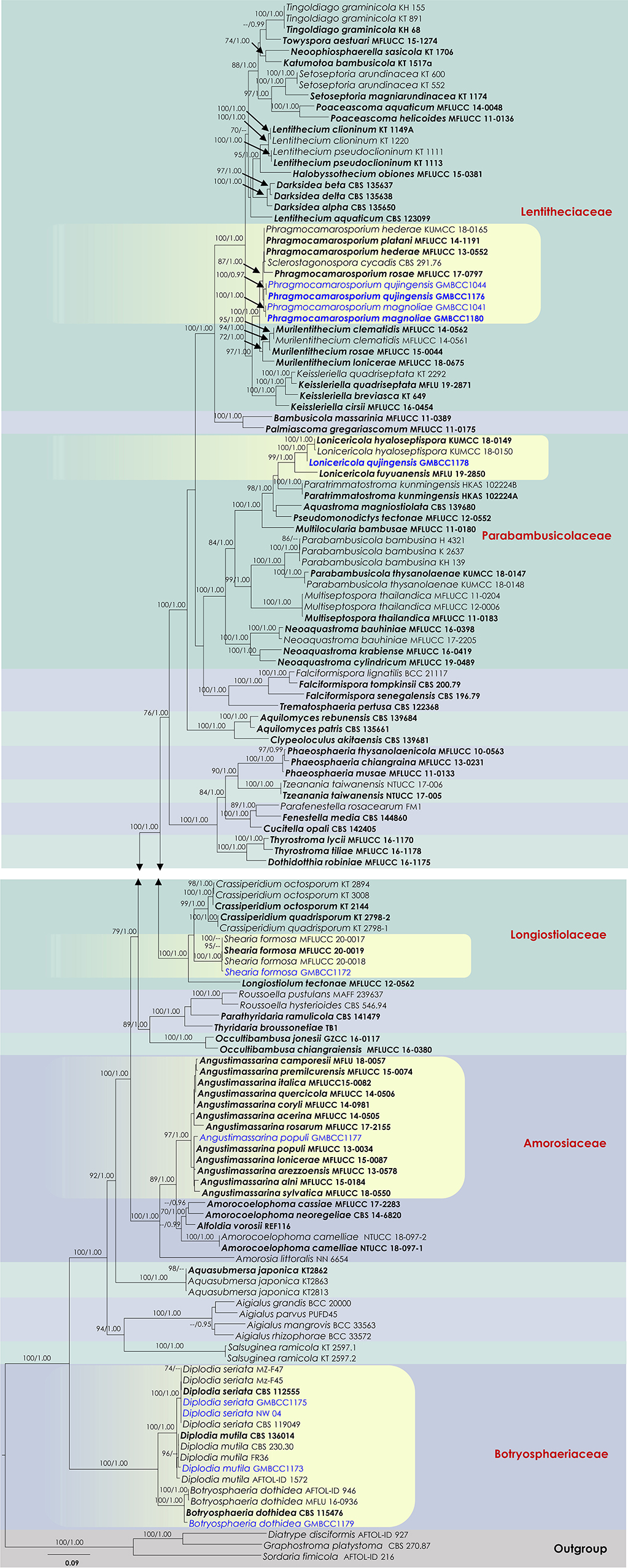
Figure 1. RAxML tree based on a combined dataset of partial SSU, LSU, ITS, tef1, and rpb2 DNA sequence analyses. Bootstrap support values for ML equal to or >70%, Bayesian posterior probabilities (BYPP) equal to or >0.95, are shown as ML/BI above the nodes. The type of strains is in bold, and new isolates are in blue. The scale bar represents the expected number of nucleotide substitutions per site.
The family Parabambusicolaceae was resolved into eight distinct clades representing species in Aquastroma, Lonicericola, Multilocularia, Multiseptospora, Neoaquastroma, Parabambusicola, Paratrimmatostroma, and Pseudomonodictys. Our new strain, GMBCC1178, constituted a strong monophyletic relationship with Lonicericola fuyuanensis (MFLU 19-2850) and L. hyaloseptispora (KUMCC 18-0150 and KUMCC 18-0149). This new lineage (GMBCC1178) is presented here as the new species Lonicericola qujingensis sp. nov.
The Shearia formosa (GMBCC1172) isolated in this study nested in a well-supported clade (100% ML/1 BYPP) with other isolates of S. formosa (MFLUCC 20-0017, MFLUCC 20-0018, and MFLUCC 20-0019), which were used by Wanasinghe et al. (2020) to describe the species, therefore confirming the identification of the studied species. The family Amorosiaceae is composed of four clades, which correspond to known genera Alfoldia, Amorocoelophoma, Amorosia, and Angustimassarina. Our new strain, GMBCC1177, grouped with another 12 Angustimassarina strains with 97% ML and 1 BYPP statistical support values. However, the interspecific relationships of these Angustimassarina species have not received a clear phylogenetic resolution. Our new isolate has a close phylogenetic affinity to Angustimassarina populi.
Analysis 2 (ITS and tef1 sequence analyses of Botryosphaeria sensu stricto): The concatenated dataset (ITS and tef1 loci) contained 33 isolates, and the tree was rooted to Macrophomina phaseolina (CBS 227.33). The final alignment contained 761 characters used for phylogenetic analyses, including alignment gaps. The RAxML analysis of the combined dataset yielded a best-scoring tree with a final ML optimization likelihood value of −1,861.242225. The matrix had 161 distinct alignment patterns, with 6.84 % undetermined characters or gaps. Parameters for the GTR + I + G model of the combined amplicons were as follows: estimated base frequencies; A = 0.210798, C = 0.292947, G = 0.259679, and T = 0.236576; substitution rates AC = 0.435178, AG = 1.502643, AT = 1.277448, CG = 0.440383, CT = 4.352625, and GT = 1; proportion of invariable site I = 0.701032; gamma distribution shape parameter α = 0.87371. In the combined sequence data analyses of the ITS and tef1 loci, our new strain, GMBCC1179, clustered with 16 other strains of Botryosphaeria dothidea (Supplementary Figure 1). Two strains of Botryosphaeria auasmontanum (MFLUCC 15-0923 and MFLUCC 17-1071) also grouped in B. dothidea. However, the Botryosphaeria dothidea clade is statistically not well-supported.
Analysis 3 (ITS and tef1 sequence analyses of Diplodia sensu stricto): The concatenated ITS and tef1 loci contained 67 isolates, and the tree was rooted to Lasiodiplodia lignicola (MFLUCC 11-0656). The final alignment contained 882 characters used for the phylogenetic analyses, including alignment gaps. The RAxML analysis of the combined dataset yielded a best-scoring tree with a final ML optimization likelihood value of −3,628.405258. The matrix had 321 distinct alignment patterns, with 12.65 % undetermined characters or gaps. Parameters for the GTR + I + G model of the combined amplicons were as follows: estimated base frequencies; A = 0.206552, C = 0.298475, G = 0.261435, and T = 0.233538; substitution rates AC = 1.148622, AG = 3.343902, AT = 1.039052, CG = 1.718372, CT = 4.817617, and GT = 1; proportion of invariable site I = 0.441349; gamma distribution shape parameter α = 0.65846. In our analysis of selected Diplodia species, the new strain GMBCC1173 clustered with Diplodia mutila (MFLUCC 15-0918, CBS 230.30, CBS 112553, CBS 136014, and MFLUCC 15-0917). Particularly, GMBCC1173 has a close phylogenetic affinity to MFLUCC 15-0917, which was introduced by Dissanayake et al. (2017) from Italy on Acer negundo. The two collections (GMBCC1175 and NW04) isolated in this study formed a basal terminal lineage in the Diplodia seriata clade that includes thirteen strains. This Diplodia seriata clade also did not receive a strong phylogenetic support (Supplementary Figure 2).
Analysis 4 (ITS, LSU, SSU, and tef1 sequence analyses of Amorosiaceae): Twenty-five strains are included in the sequence analysis and comprise 2,106 characters with gaps. A single gene analysis was carried out and compared with each species to compare the topology of the tree and clade stability. Botryosphaeria dothidea (CBS 115476 and AFTOL-ID 946) was used as the outgroup taxon. The tree topology of the ML analysis was similar to the BYPP. The best-scoring RAxML tree with a final likelihood value of −5144.567766 is presented. The matrix had 247 distinct alignment patterns, with 18.63% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.243552, C = 0.248710, G = 0.270808, and T = 0.236930; substitution rates AC = 0.682136, AG = 1.407781, AT = 1.275132, CG = 0.809575, CT = 6.927148, and GT = 1; gamma distribution shape parameter alpha = 0.654596 (Figure 2). The family Amorosiaceae is composed of four clades, which correspond to known genera Alfoldia, Amorocoelophoma, Amorosia, and Angustimassarina. Our new strain, GMBCC1177, grouped with another Angustimassarina strains with low statistical support values (Figure 2). However, the interspecific relationships of these Angustimassarina species have not received a clear phylogenetic resolution. The strain GMBCC1177 showed a close phylogenetic affinity with Angustimassarina populi strains clustered together with A. arezzoensis (MFLUCC 13-0578) and A. sylvatica (MFLUCC 18-0550).

Figure 2. Phylogram generated from maximum likelihood analysis based on combined SSU, LSU, tef1, and ITS partial sequence data. Bootstrap support values for ML equal to or >50% and BYPP values equal to or >0.9 are given above the nodes. The newly generated sequence is in red.
Analysis 5 (SSU, LSU, tef1, and ITS sequence analyses of Lentitheciaceae): Twenty-nine strains are included in the sequence analysis and comprise 3,086 characters with gaps. A single gene analysis was carried out and compared with each species to compare the topology of the tree and clade stability. Massarina cisti (CBS 266.62) and Massarina eburnea (H 3953) are used as outgroup taxa. The tree topology of the ML analysis was similar to the BYPP. The best-scoring RAxML tree with a final likelihood value of −10,433.446109 is presented. The matrix had 492 distinct alignment patterns, with 25.16% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.237977, C = 0.252206, G = 0.270764, and T = 0.239052; substitution rates AC = 0.947399, AG = 1.742628, AT = 0.987565, CG = 1.240193, CT = 6.545005, and GT = 1; gamma distribution shape parameter alpha = 0.608667 (Figure 3). The family Lentitheciaceae comprises eight genera that show distinct phylogenetic lineages (Figure 3). Separation of Phragmocamarosporium species is agreement with morphological evidence (Table 4), but the new collections GMBCC1180 and GMBC1041 clustered with ex-types of P. hederae and P. platani with low bootstrap values (which are indicated in blue in Figure 3). The ex-types of P. hederae and P. platani are lacking ITS loci in the GenBank; thus, we suggest that including more gene regions of these two species will facilitate a better understanding of intraspecific segregation. However, here, we follow the morphological evidence to introduce novel species (i.e., P. magnoliae) (see below under the Section Taxonomy).
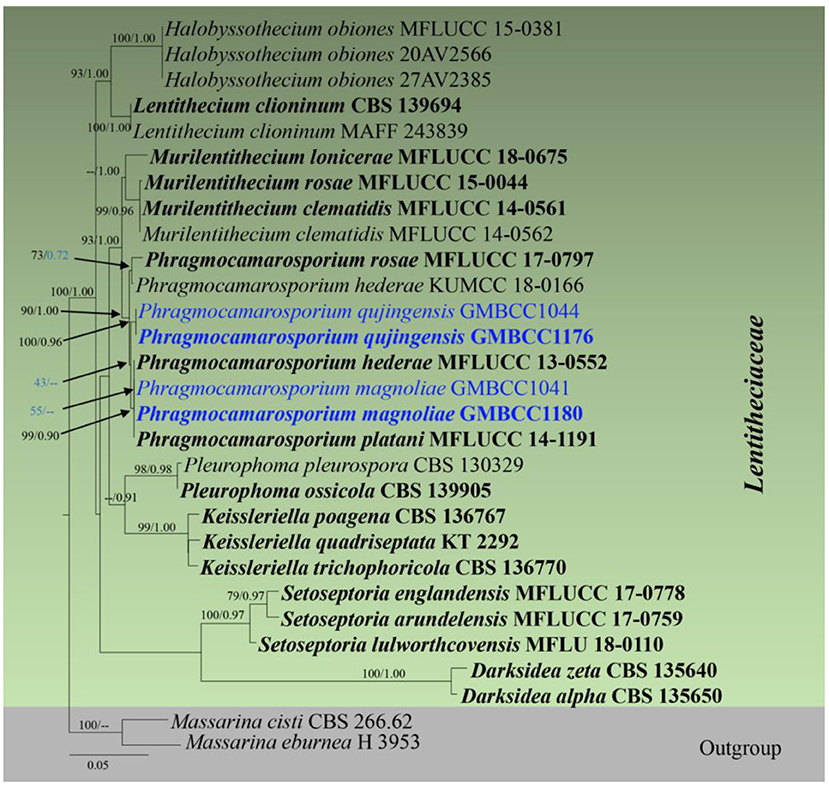
Figure 3. RAxML tree based on a combined dataset of partial SSU, LSU, tef1, and ITS DNA sequence analyses. Bootstrap support values for ML equal to or >50% and Bayesian posterior probabilities (BYPP) equal to or >0.9 are shown as ML/BI above/below the nodes (except for those values in blue in Phragmocamarosporium clade). The type of strains is in bold, and new isolates are in blue. The scale bar represents the expected number of nucleotide substitutions per site.
GMBCC1176 and GMBCC1044, are grouped as the basal clade to the clade that comprises ex-types of P. hederae, P. magnoliae, and P. platani (Figure 3). Both strains were generated from morphologically similar collections and thus introduced as a new species, i.e., Phragmocamarosporium qujingensis.
The strain named Phragmocamarosporium hederae (KUMCC 18-0165) clustered with the ex-type of P. rosae (MFLUCC 17-0797) but was distinct from the ex-type of P. hederae. This strain was named mistakenly as Phragmocamarosporium hederae and thus needs an extensive study to confirm if it warrants a novel species.
Taxonomy
In this section, we introduce three new pleosporalean species from M. grandiflora in Qujing Normal University, Qujing, Yunnan Province, China. Moreover, five (three in Botryosphaeriales and two in Pleosporales) species are reported as a new host and geographical records from M. grandiflora and China, respectively.
Botryosphaeriales C.L. Schoch, Crous, & Shoemaker (2007)
Botryosphaeriaceae Theiss. & Syd. [as “Botryosphaeriacae”], Annls mycol. 16(1/2): 16 (1918)
Notes
Botryosphaeriaceae (in Botryosphaeriales) is an important family that comprises a broad range of life modes such as saprobes, pathogens, and endophytes and shows a worldwide distribution (Phillips et al., 2013). Wijayawardene et al. (2022) accepted 22 genera in Botryosphaeriaceae. During our collecting programs of fungi inhabiting M. grandiflora, we collected three collections of Botryosphaeriaceae taxa. According to our knowledge, these are the first records of the above mentioned taxa reported from M. grandiflora.
Botryosphaeria Ces. & De Not., Comm. Soc. crittog. Ital. 1(fasc. 4): 211 (1863)
Index Fungorum Registration Identifier IF 635
Notes
The genus Botryosphaeria was introduced by Cesati and De Notaris (1863) who did not designate the type. Barr (1972) proposed B. dothidea (Moug.:Fr.) Ces. & De Not. as the lectotype. Slippers et al. (2004) designated the neotype and epitype of B. dothidea. Phillips et al. (2013) comprehensively revisited the genus and accepted six species including B. dothidea while providing illustration and description for asexual morph.
Botryosphaeria dothidea (Moug.:Fr.) Ces. & De Not. 1863, (Figure 4)
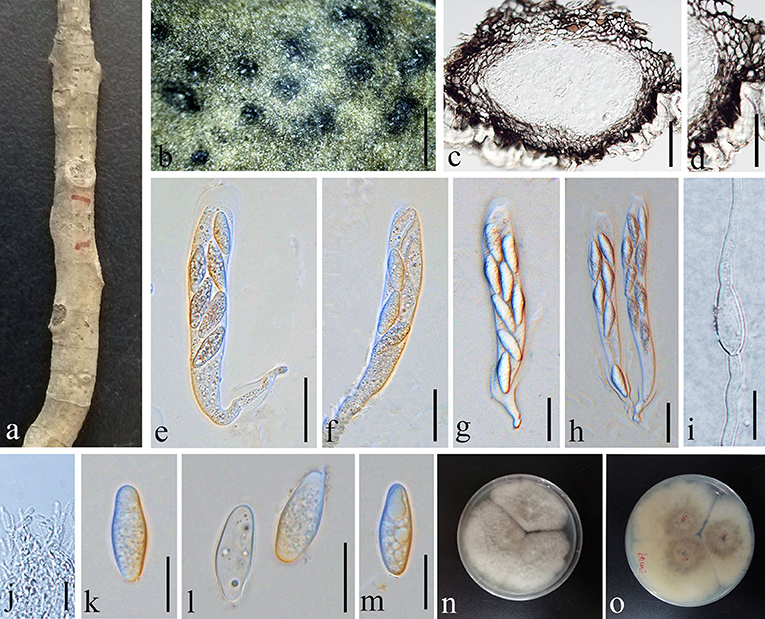
Figure 4. Botryosphaeria dothidea (GMB1387, new host record from China). (a) Host material. (b) Appearance of ascomata on the host. (c) Vertical section of ascoma. (d) Peridium. (e–h) Asci. (i) Germinating ascospore. (j) Pseudoparaphyses. (k–m) Ascospores. (n) Culture on PDA from above. (o) Culture on PDA from the bottom. Scale bars: (b) = 1,000, (c) = 50, (d) = 25, (e–h) =30, (i,k–m) = 15, and (j) =10 μm.
Index Fungorum Registration Identifier IF 183247
Saprobic on dead (hanging) branches of M. grandiflora. Sexual morph: Ascomata 155–460 × 165–315 μm ( = 281.3 × 236.2 μm, n = 10), eustromatic, gregarious, black, uniloculate, with a thick pseudoparenchymatic wall composed of textura angularis or textura globose with the outer layers blackened and their cells more thickened, and erumpent at maturity. Pseudoparaphyses 2.5–3.5 μm ( = 2.9 μm, n = 20) wide, thin-walled, hyaline, aseptate, and constricted at the septa. Asci 65–110 × 15–20 μm ( = 84.4 × 17.7 μm, n = 20), clavate or cylindric-clavate, stipitate, bitunicate, ectotunica thin, endotunica rather thick, 3-layered, with a prominent apical chamber, 8-spored, and developing on a broad basal hymenial layer. Ascospores 20–25 × 7–9 μm ( = 23.8 × 7.6 μm, n = 20), irregularly biseriate in the ascus, hyaline, sometimes becoming pale brown with age, thin-walled, ovoid, fusoid, fusoid-ellipsoid, usually widest in the middle, straight or inequilateral, smooth, one-celled sometimes becoming 1–2 septate with age, contents smooth or granular, and may be guttulate. Asexual morph: undetermined.
Culture characteristics
Ascospores germinating on PDA within 24 h and germ tubes produced from both sides. Colonies growing fast on PDA, reaching 6 cm in 1 week at 28°C, effuse, velvety to hairy, circular, white in the first week, and brown to dark brown after 1 week from above and below.
Materials examined
China, Yunnan Province, Qujing Normal University, 25°52′36.75″N, 103°74′46.73″E, 1,853.8 m, on branch of Magnolia grandiflora L., 18 August 2021, Dong-Qin Dai and Mei-ling Zhu, Ling 47, GMB1387 (new host record), living culture, GMBCC1179.
Notes
Our new collection of B. dothidea from M. grandiflora morphologically resembles the type collection described in Phillips et al. (2013). In phylogenetic analyses, our collection (GMBCC1179) groups with B. dothidea s. str. (Figure 1). According to Deng (1963), Tai (1979), and Farr and Rossman (2022), B. dothidea has not been previously reported from Magnolia species in China. Zlatkovic et al. (2018) reported B. dothidea from M. grandiflora as a pathogenic species (a causal agent of stem and shoot dieback) from Serbia. However, we did not notice any disease symptoms in the host plant that we collected. Nevertheless, it is essential to collect more samples to confirm the impact of B. dothidea on Magnolia species, since it is an important ornamental plant in China. Here, we report B. dothidea from M. grandiflora in China for the first time.
Diplodia Fr., In: Mont., Ann. Sci. Nat. Bot., sér. 2, 1: 302. 1834
Index Fungorum Registration Identifier IF 8047
Notes
Montagne (1834) introduced Diplodia with D. mutila (Fr.) Mont. as the type of species. Currently, 28 species are accepted in Wu et al. (2021). Members of Diplodia are distributed worldwide and occur as different life modes such as pathogens, saprobes, and endophytes (Phillips et al., 2013). Approximately, over 60 records of Diplodia species have been reported from China according to Xiao et al. (2021) and Farr and Rossman (2022). Nevertheless, Diplodia species have not been reported from Magnolia species in China. Here, we report Diplodia mutila and D. seriata from M. grandiflora for the first time in China. According to our knowledge, Diplodia species have not been reported from M. grandiflora so far.
Diplodia mutila (Fr.) Mont., Annls Sci. Nat., Bot., sér. 2 1: 302 (1834), (Figure 5)
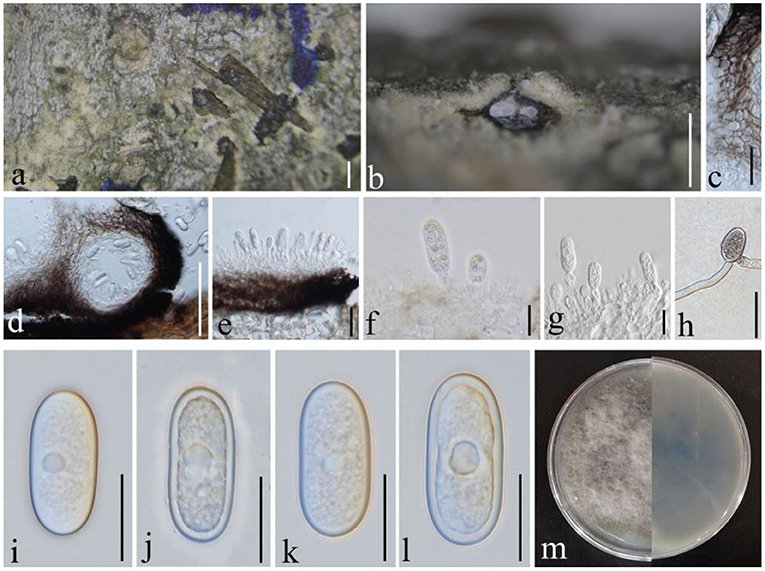
Figure 5. Diplodia mutila (GMB1381, a new host record). (a) Appearance of conidioma on the host. (b,d) Vertical sections of conidiomata. (c) Conidiomata walls. (e–g) Conidia attached to conidiog-enous cells. (h) Germinating conidium. (i–l) Conidia. (m) Cultures on PDA. Scale bars: (a) = 500, (b) = 200, (c,g,h) = 20, (d) = 100, (e) = 25, (f) = 10, and (i–l) = 15 μm.
Index Fungorum Registration Identifier IF 201741
Saprobic on dead branches of M. grandiflora. Conidiomata 120–450 diam. × 160–400 μm high, solitary, immersed, partially erumpent at maturity, black, and globose. Ostiole central, circular, and papillate. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 11–14 × 4–5 μm, holoblastic, discrete, cylindrical, hyaline, and smooth. Conidia 25–30 × 10–15 μm ( = 26.7 × 11.6 μm, n = 20), hyaline and aseptate at immature stage, smooth, thick-walled, oblong to ovoid, straight, both ends broadly rounded, and becoming pale brown at maturity.
Culture characteristics
Conidia germinating on PDA within 24 h and germ tubes produced from one side. Colonies growing fast on PDA, reaching 9 cm in 1 week at 28°C, effuse, velvety to hairy, circular, white in the first week, brown to dark brown from above after 1 week, and dark brown to lividity from below.
Material examined
China, Yunnan Province, Qujing Normal University, 25°52′36.75″N, 103°74′46.73″E, 1,853.8 m, on branch of M. grandiflora L., 7 September 2019, Dong-Qin Dai and Mei-ling Zhu, Ling 31, (GMB1381) (new host record), living culture GMBCC1173.
Notes
In morphology, our new collection closely resembles Diplodia mutila (Phillips et al., 2013) except for conidial width (10–15 vs. 13–14). However, in phylogenetic analyses, a new strain clusters with D. mutila s. str. with high statistical values (Supplementary Figure 2; 83% in ML analysis). Hence, we conclude that our collection is D. mutila, and it is the first report of this species from M. grandiflora.
Diplodia seriata De Not., Mém. R. Accad. Sci. Torino, Ser. 2 7: 26 (1845), (Figure 6)
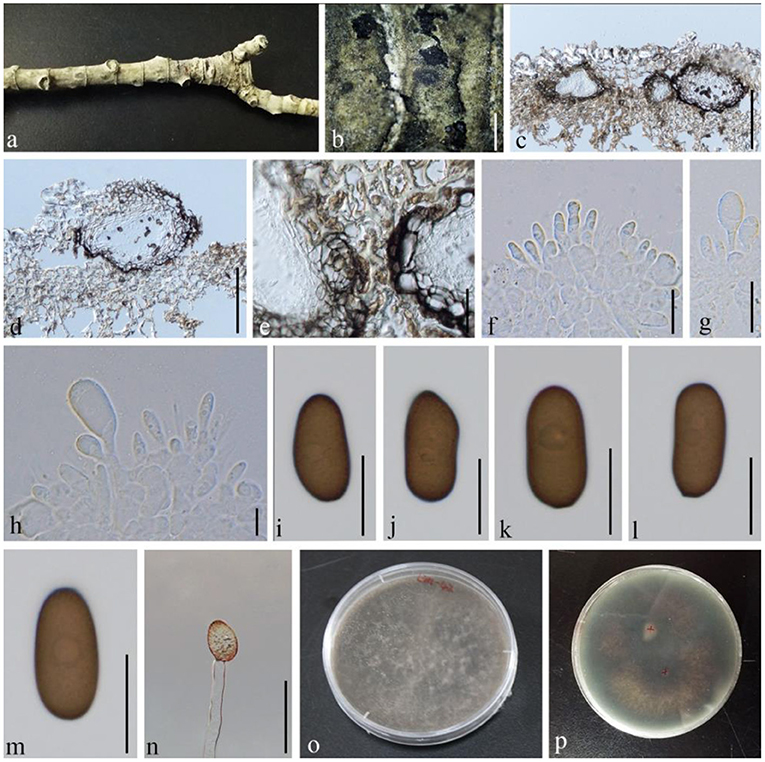
Figure 6. Diplodia seriata (GMB1383, a new host record). (a) M. grandiflora branch. (b) Appearance of conidiomata on the host. (c,d) Vertical sections of conidiomata. (e) Conidiomata walls. (f–h) Conidia attached to conidiogenous cells. (i–m) Mature conidia. (n) Germinating conidium. (o,p) Cultures on PDA. Scale bars: (b) = 1,000, (c,d) = 150, (e) = 30, and (f–n) = 15 μm.
Index Fungorum Registration Identifier IF 201741
Saprobic on dead branches of M. grandiflora. Conidiomata 30–205 diam. × 23–65 μm high, solitary, immersed, partially erumpent at maturity, dark brown, and globose. Ostiole central, circular, and nonpapillate. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 6–10 × 4–6 μm, holoblastic, discrete, cylindrical, hyaline, and smooth. Conidia 20–27 × 10–15 μm ( = 22.7 × 12.6 μm, n = 20), hyaline and aseptate at immature stage, smooth, thick-walled, oblong to ovoid, straight, both ends broadly rounded, and becoming dark brown at maturity.
Culture characteristics
Conidia germinating on PDA within 24 h and germ tubes produced from rear side. Colonies growing fast on PDA, reaching 9 cm in 1 week at 28°C, effuse, velvety to hairy, circular, white in the first week, brown to dark brown after 1 week from above, and black in the central and outermost circles with dark brown in the middle from below.
Material examined
China, Yunnan province, Qujing Normal University, 25°52′36.75“N, 103°74′46.73″E, 1,853.8 m, on branch of M. grandiflora L., 18 August 2020, Dong-Qin Dai and Mei-ling Zhu, Ling 42, (GMB1383) (a new host record), living culture GMBCC1175.
Notes
In conidial morphology, our new collection from M. grandiflora is morphologically similar to Diplodia seriata (Phillips et al., 2013). In our phylogenetic analyses, it was accommodated with D. seriata s. str. (Figure 1; Supplementary Figures 1, 2). Hence, we confirmed our new collection as D. seriata. According to Farr and Rossman (2022), D. seriata was not reported as from M. grandiflora. Hence, in here, we report D. seriata from M. grandiflora as a new host record in China.
Pleosporales Luttr. ex M.E. Barr 1987
Amorosiaceae Thambug. & K.D. Hyde, Fungal Diversity 74: 252 (2015)
Index Fungorum Registration Identifier IF 551277
Notes
Thambugala et al. (2015) introduced this family based on Amorosia Mantle & D. Hawksw. (type species: Amorosia littoralis Mantle & D. Hawksw.). At the same time, Thambugala et al. (2015) introduced Angustimassarina with A. populi Thambug. & K.D. Hyde as the type species. Currently, the family comprises five genera (Wijayawardene et al., 2022).
Angustimassarina Thambug., Kaz. Tanaka & K.D. Hyde, Fungal Diversity 74: 253 (2015)
Index Fungorum Registration Identifier IF 551278
Notes
Angustimassarina was introduced by Thambugala et al. (2015) with three species and A. populi as the type species. Twelve records are listed in Index Fungorum (2022) while most of Angustimassarina species have been reported from Germany and Italy (Thambugala et al., 2015; Tibpromma et al., 2017; Hyde et al., 2019). Only one species, A. populi, was reported with both asexual and sexual morphs (Thambugala et al., 2015), while other species have been reported with only a sexual morph.
Angustimassarina populi Thambug. & K.D. Hyde, Fungal Diversity: 10.1007/s13225-015-0348-3, [56] (2015), (Figure 7)
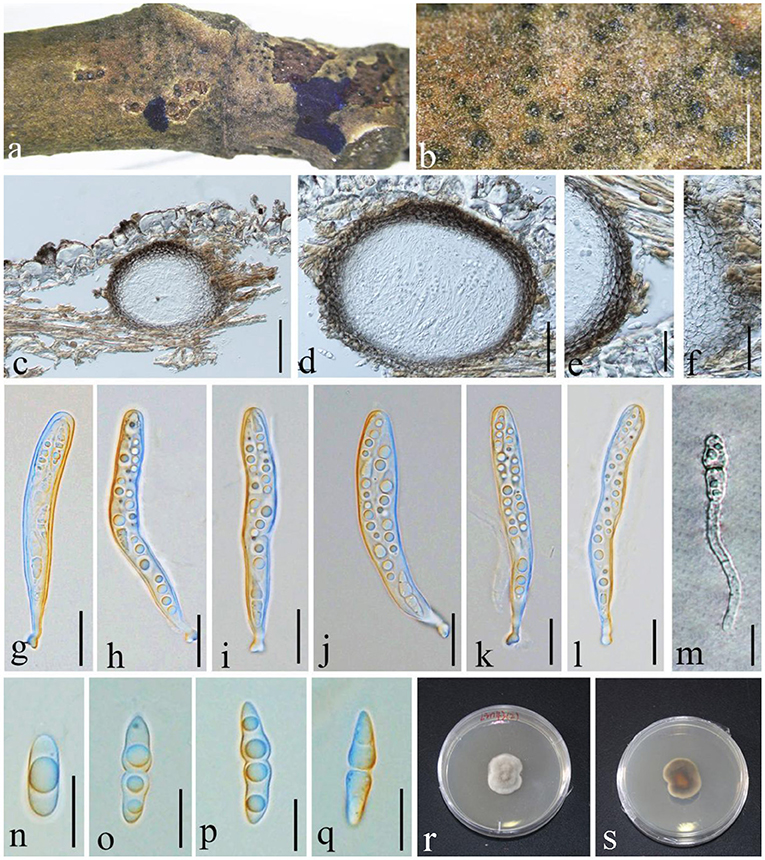
Figure 7. Angustimassarina populi (GMB1385, new host and country record). (a,b) Ascomata on a M. grandiflora branch. (c,d) Vertical sections of ascomata. (e,f) Peridium. (g–l) Asci. (m) Germinating ascospore. (n–q) Ascospores. (r,s) Cultures on PDA [(r) = from above and (s) = from bottom]. Scale bars: (b) = μm, (c) = 100, (d) = 50, (e) = 30, (f) = 25, (g–l) = 20, and (m–q) = 10 μm.
Index Fungorum Registration Identifier IF 551279
Saprobic on branch of M. grandiflora. Sexual morph: Ascomata 100–165 × 100–120 μm ( = 125.2 × 112.7 μm, n = 10), visible black dots, small dome-shaped on host surface, immersed, scattered, globose to sub-globose, uni-loculate, and without ostiole and papilla. Peridium 3–9 × 2–4.5 μm ( = 7.2 × 3.1 μm, n = 20), unequal thickness, thick-walled, composed of several layers of brown to dark brown, and arranged in textura angularis. Hamathecium composed of dense, 1.5–2 μm wide, filamentous, unbranched, septate, pseudoparaphyses, anastomosed between the asci, and embedded in hyaline gelatinous matrix. Asci 80–105 × 9–15 μm ( = 86.2 × 10.2 μm, n = 20), 8-spored, bitunicate, fissitunicate, cylindrical to cylindric-clavate, short with club-shaped pedicellate, and with well-developed ocular chamber. Ascospores 20–23 × 4–6 μm ( = 20.4 × 5.6 μm, n = 20), overlapping 1–2-seriate, fusiform, hyaline, 1 septate at the center, constricted at the septum, the upper cell larger than the lower cell, conical at the ends, guttulate, smooth-walled, and without mucilaginous sheath. Asexual morph: undetermined.
Culture characteristics
Ascospores germinating on PDA within 24 h and germ tubes produced from one side. Colonies grow on PDA at 28°C under normal light, reaching 2.5 cm diam. after 2 weeks, dense, irregular, umbonate, surface smooth, with edge entire, cottony, white to gray from above; white at the margin and dark brown at the center from below, and do not produce pigmentation in PDA.
Material examined
China, Yunnan Province, Qujing Normal University, 25°52′36.75“N, 103°74′46.73″E, 1,853.8 m, on branch of M. grandiflora L., 18 August 2021, Lin 43-2, (GMB1385; a new host and country record), living culture GMBCC1177.
Notes
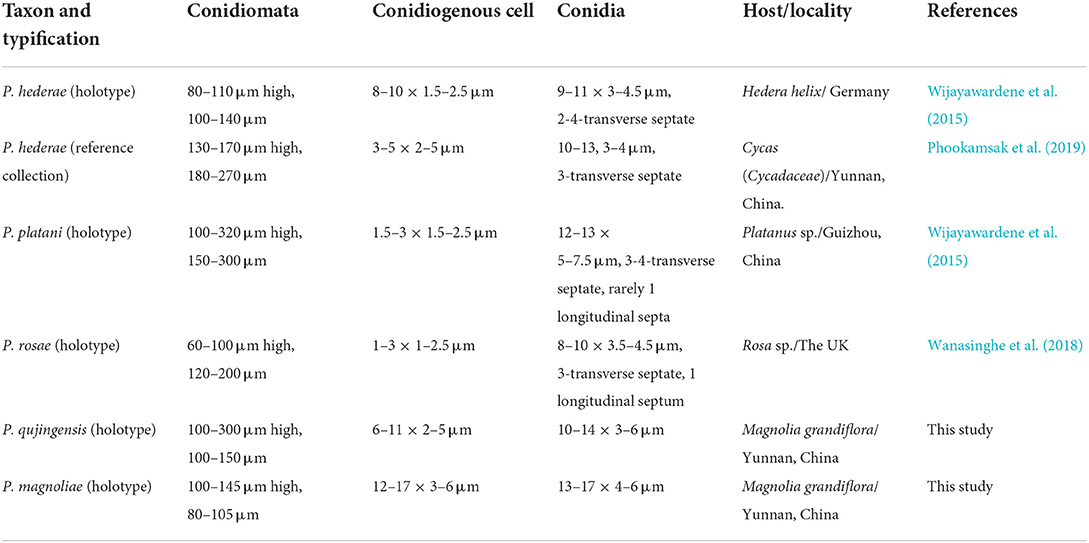
In morphology, our new collection closely resembles Angustimassarina populi (holotype MFLU 14-0588). Angustimassarina populi was introduced by Thambugala et al. (2015), was collected from Italy on dead branches of Populus sp., and was characterized by erumpent, globose to subglobose ascomata with crest-like ostiole, cylindrical asci with fusiform ascospores, 1(−3)-septate, constricted at the central septum, and surrounded by a mucilaginous sheath; asexual morph is hyphomycetous. Based on our phylogenetic analyses of combined SSU, LSU, tef1, and ITS sequence data (Figure 2), our strain (GMBCC1177) clusters with the strains of A. populi at the basal clade of A. arezzoensis and A. sylvatica. Thus, we identify our fresh collection as A. populi. This is the first report of A. populi from China and on M. grandiflora. We also compared the morphology of the sexual morph of our new collection with Angustimassarina species that are morphologically and phylogenetically closely related (Table 3). However, the phylogenetic affinities of Angustimassarina species are not well-resolved (Figure 2). Therefore, to resolve the current status of Angustimassarina, further research is needed together with asexual morph and protein-coding genes.
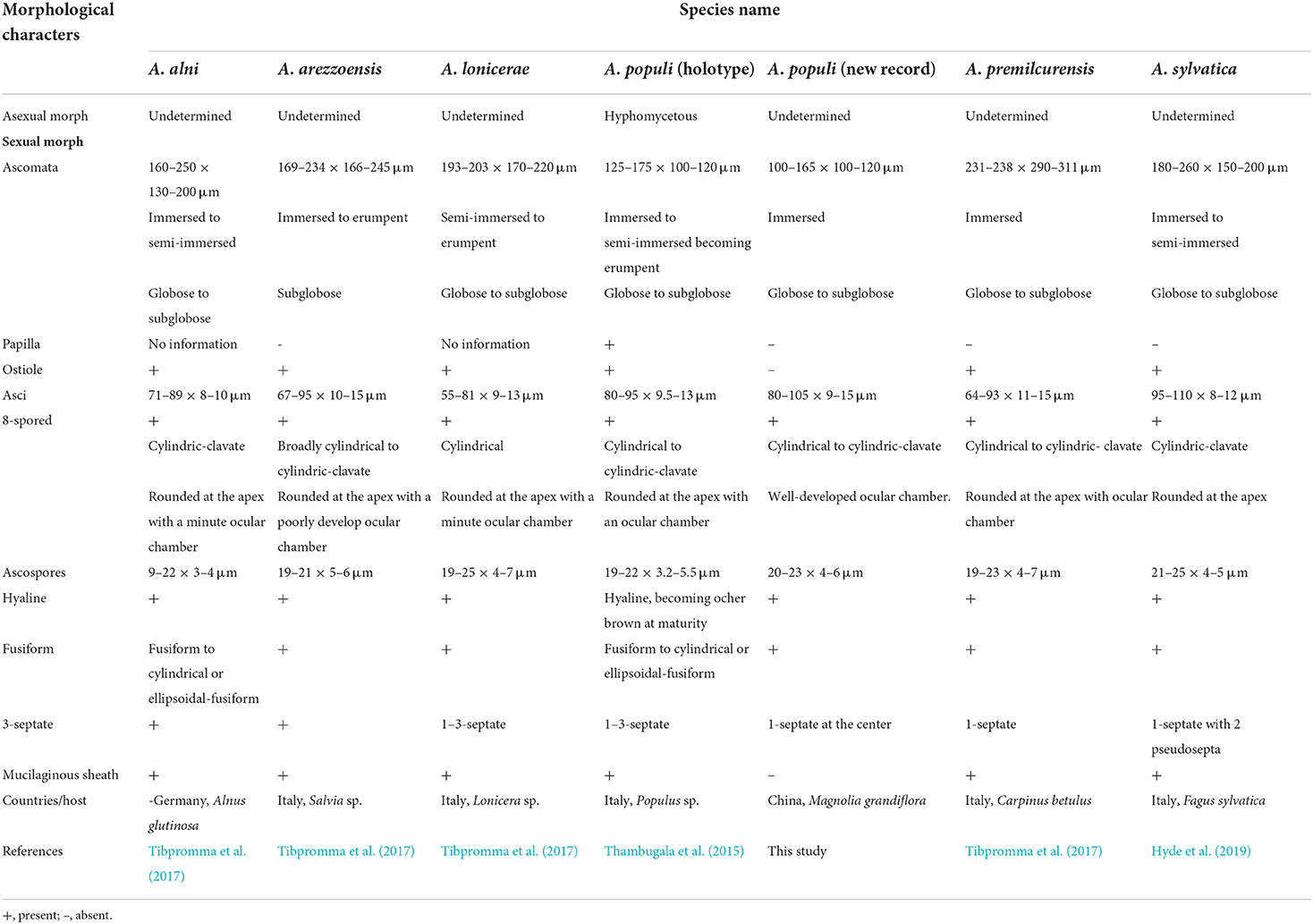
Table 3. Morphological comparisons, location, and hosts of Angustimassarina species phylogenetically related to the new collection.
Parabambusicolaceae Kaz. Tanaka & K. Hiray. Stud. Mycol. 82: 115 (2015)
Index Fungorum Registration Identifier IF 811324
Notes
Tanaka et al. (2015) introduced this family to accommodate two genera, viz., Aquastroma and Parabambusicola (type genus). Members of the family have been mainly reported as saprobes. Currently, the family comprises nine genera (Wijayawardene et al., 2022).
Lonicericola Phookamsak, Jayasiri & K.D. Hyde, Fungal Diversity 95(1): 39 (2019)
Index Fungorum Registration Identifier IF 556139
Notes
Lonicericola was introduced by Phookamsak et al. (2019) with L. hyaloseptispora Phookamsak et al. as the type of species. Later, Yasanthika et al. (2020) introduced the second species, L. fuyuanensis Yasanthika et al. Both have been introduced from Yunnan Province, China as saprobic species. Our new collection is morphologically resembling Lonicericola s. str. Multi-gene phylogenetic analyses and morphological characteristics confirmed that the new collections are new species in Lonicericola.
Lonicericola qujingensis D.Q. Dai, Wanas. & Wijayaw. sp. nov., (Figure 8)
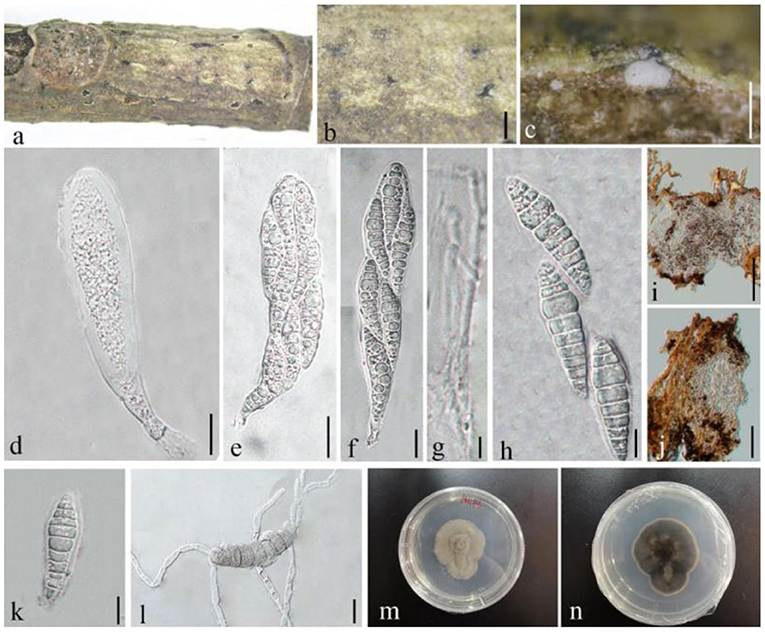
Figure 8. Lonicericola qujingensis (GMB1386, holotype). (a) Dead branch of M. grandiflora. (b) Immersed ascomata on host. (c) Vertical section of ascoma. (d–f) Asci. (g) Pseudoparaphyses. (h,k) Ascospores. (i,j) Peridium. (l) Germinated ascospore. (m,n) Seven-day-old colonies incubated at 28°C on PDA. Scale bars: (b) = 500, (c,j) = 50, (d–f) = 20, (g,h,k,l) = 10, and (i) = 100 μm.
Index Fungorum Registration Identifier IF 555252
Etymology: named after the locality from where it was collected, Qujing, Yunnan (China).
Saprobic dead branches of M. grandiflora, Sexual morph: Ascomata 250–300 × 100–160 μm ( = 284.2 × 129.7 μm, n = 10), black, scattered, solitary, immersed under host epidermis, slightly raised at maturity, globose to subglobose, uniloculate, glabrous, ostiolate, and papillate. Ostiole centrally located, oblong, with minute papilla, and filled with hyaline paraphyses. Peridium 10–20 μm wide, unequal thickness, composed of 2–3 layers, flattened to broad, brown to dark brown, composed of pseudoparenchymatous cells, and arranged in textura angularis to textura prismatica. Hamathecium composed of numerous, 1.5–3 μm wide, filamentous, and septate pseudoparaphyses. Asci 100–210 × 25–35 μm ( = 154.21 × 30.7 μm, n = 20), 8-spored, bitunicate, fissitunicate, broadly cylindrical to cylindric-clavate, subsessile to short pedicellate, with furcate to obtuse pedicel, and apically rounded. Ascospores 47–60 × 10–16 μm ( = 55.7 × 14.2 μm, n = 20), overlapping 2–3-seriate, hyaline, fusiform, 7–10-septate, constricted at the septa, smooth-walled, with small to large guttules, and surrounded by a mucilaginous sheath (14–18 μm diam.). Asexual morph: undetermined.
Culture characteristics
Ascospores germinating on PDA within 24 h and germ tubes produced from all sides. Colonies growing slowly on PDA, reaching 4 cm in diam. after 1 week at 28°C, dense, irregular, umbonate, surface smooth, with edge entire, cottony, white in the first week, white to gray from above; white at the margin and black in the center, and dark brown in the middle from below. Mycelium semi-immersed in PDA, with branches, septate, smooth-walled, and hyphae brown.
Material examined
China, Yunnan Province, Qujing, Qujing Normal University, 25°52′36.75″N, 103°74′46.73″E, 1,853.8 m, on dead branches of M. grandiflora L., 18 August 2020, Dong-Qin Dai and Mei-ling Zhu, Lin 46, (GMB1386, holotype), ex-type GMBCC1178; Ibid. 10 May 2021, Dong-Qin Dai and Ting-Ting Zhang, Lin 60, (GMB1046, paratype); ex-paratype GMBCC1037.
Notes
Currently, the genus comprises three species (including the new collection), and all species were reported from Yunnan, China. Interestingly, both Lonicericola hyaloseptispora and L. fuyuanensis have been reported from the same host family, i.e., Caprifoliaceae. Nevertheless, the new collection was made from decaying branches of M. grandiflora (Magnoliaceae). In our phylogenetic analyses (Figure 1), our new collection formed a distinct clade in Lonicericola s. str. with high bootstrap values (99% and 1 in ML and Bayesian analysis, respectively). This result is also supported by morphological characters (see the taxonomic key). Based on current data, we assume that Lonicericola species are restricted to subtropical regions in China but could be distributed in different host families.
The taxonomic key below can be used to distinguish the Lonicericola species based on ascospore and asci morphology.
1. Ascospores with only 3 septa …… L. fuyuanensis
1. Ascospores with more than 3 septa……2
2. Ascospores 37–49 × 8–12 μm, 8–9-septate…… L.hyaloseptispora*
2. Ascospores 47–60 × 10–16 μm, 7–10-septate…… L. qujingensis
*Phookamsak et al. (2019) did not provide ascospore dimensions; thus, we received them through personal communication with R. Phookamsak.
Lentitheciaceae Y. Zhang ter, C.L. Schoch, J. Fourn., Crous & K.D. Hyde, Stud. Mycol. 64: 93 (2009)
Index Fungorum Registration Identifier IF 515470
Notes
Zhang et al. (2009) introduced this genus with Lentithecium, Katumotoa, and Keissleriella. Currently, Lentitheciaceae comprises 14 genera (Wijayawardene et al., 2022). Members of the family occur in both terrestrial and aquatic environments and are common as saprobes.
Phragmocamarosporium Wijayaw., Yong Wang & K.D. Hyde, Index Fungorum 370: 1 (2018)
Index Fungorum Registration Identifier IF 555365
Notes
Wijayawardene et al. (2015) introduced this genus with two species, P. hederae Wijayaw. et al. (from Hedera helix, Germany) and P. platani (type species, from Platanus sp., Guizhou, China). Wanasinghe et al. (2018) introduced the third species, which was inhabitant on spines of Rosa canina from Great Britain. However, according to the Index Fungorum (2022), the genus was invalidly published in Wijayawardene et al. (2015); thus later, the genus and all the species have been validated in Index Fungorum (2022).
Phragmocamarosporium magnoliae Wijayaw., D.Q. Dai & Wanas. sp. nov. (Figure 9)
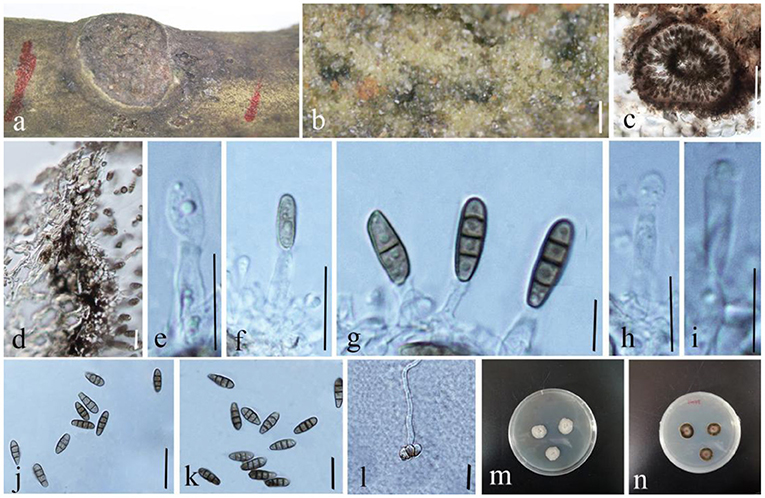
Figure 9. Phragmocamarosporium magnoliae (GMB1388, holotype). (a) Dead branch of M. grandiflora. (b) Appearance of conidiomata on host. (c) Vertical section of conidioma. (d) Pycnidium wall. (e–i) Developing and matured conidia attached to conidiogenous cells. (j,k) Conidia. (l) Germinated conidium. (m,n) Seven-day-old colonies incubated at 28°C on PDA. Scale bars: (b) = 100, (c) = 50, (d) = 20, (e,g–i) = 5, (f,l) = 15 μm, and (j) = 20 μm.
Index Fungorum Registration Identifier IF 555250
Etymology: named after the host genus from which it was collected, Magnolia.
Saprobic on branches of M. grandiflora. Sexual morph: undetermined. Asexual morph: Conidiomata 100–145 μm high, 80–105 μm in diam. ( = 122.2 × 88.9 μm, n = 20), pycnidial, immersed, black, gregarious to solitary, unilocular, globose to subglobose, and with a centrally located papillate ostiole. Pycnidial wall with outer 3–4 layers of dark brown cells of textura angularis, with inner layer of thin hyaline cells. Conidiophores reduced to conidiogenous cells. Conidiogenous cells simple to simple branch at the base, smooth, long, 12–17 × 3–6 μm ( = 13.5 × 4.5 μm, n = 20), phialidic, and hyaline. Conidia 13–17 × 4–6 μm ( = 14.5 × 5.35 μm, n = 20), medium brown, clavate or ellipsoid to sub-cylindrical, with obtuse apex and truncate base, straight to curved, 3(−4)-transverse septate, guttulate or eguttulate, and constricted at the septa.
Culture characteristics
Conidia germinating on PDA within 24 h and germ tubes produced from the middle. Colonies growing slowly on PDA, reaching 2.5 cm in diam. after 1 week at 28°C, circular, zonate, uneven margin, cottony, white from above, with thin mycelium, dark brown at the margin, and yellowish-brown at the center from below. Mycelium semi-immersed in PDA, with branches, septate, smooth-walled, and hyphae brown.
Material examined
China, Yunnan Province, Qujing, Qujing Normal University, 25°52′36.75″N, 103°74′46.73″E, 1,853.8 m, on dead branches of M. grandiflora L., 18 August 2020, Dong-Qin Dai and Mei-ling Zhu, Lin 48, (GMB1388, holotype), ex-type GMBCC1180; Ibid. 10 May 2021, Dong-Qin Dai and Ting-Ting Zhang, Lin 68, (GMB1048, paratype); ex-paratype GMBCC1041.
Notes
In the phylogenetic analyses (Figure 3), Phragmocamarosporium magnoliae groups with P. hederae and P. platani (Figure 8), but in morphology they are different (Table 4). Besides, P. hederae and P. platani are lacking sequences of ITS and tef1 loci. Species resolution of this subclade will be higher with more genes and more collections.
Phragmocamarosporium qujingensis D.Q. Dai, Wanas. & Wijayaw. sp. nov., (Figure 10)
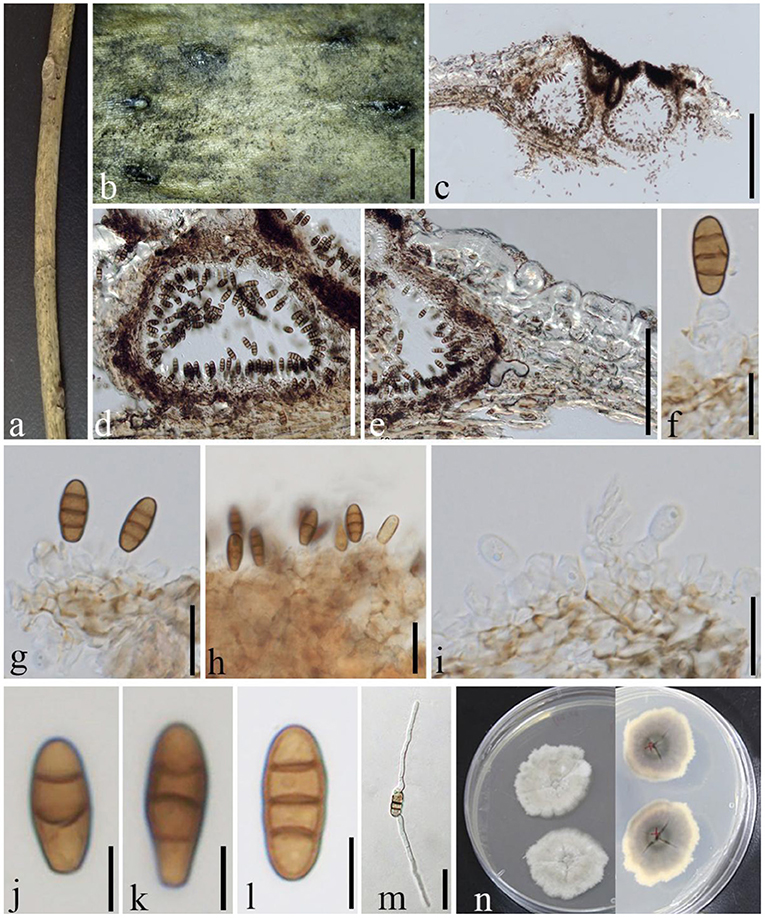
Figure 10. Phragmocamarosporium qujingensis (GMB1384, holotype). (a) Dead branch of M. grandiflora. (b) Appearance of ascomata on host. (c,d) Vertical sections of conidiomata. (e) Peridium. (f–i) Conidia attached to conidiogenous cells. (j–l) Conidia. (m) Germinated conidium. (n) Seven-day-old colonies incubated at 28°C on PDA. Scale bars: (b) = 1,000, (c) = 200, (d) = 100, (e) = 100, (f,g,i) = 10, (h,m) = 15, and (j–l) = 5 μm.
Index Fungorum Identifier IF 555251
Etymology: named after the locality from where it was collected, Qujing, Yunnan (China).
Saprobic on dead branches of M. grandiflora. Sexual morph: undetermined. Asexual morph: Conidiomata 100–300 μm high, 100–150 μm diam. ( = 142 × 122 μm, n = 10), pycnidial, immersed, semi immersed at maturity, black, gregarious to solitary, unilocular, globose to subglobose, and with a centrally located papillate ostiole. Pycnidial wall with outer 3–4 layers of dark brown cells of textura angularis and with inner layer of thin hyaline cells. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 6–11 × 2–5 μm ( = 8.4 × 3.3 μm, n = 20), simple to simple branch at the base, smooth, long, phialidic, and hyaline. Conidia 10–14 × 3–6 μm ( = 11.5 × 4.4 μm, n = 30), medium brown, clavate or ellipsoid to subcylindrical, with obtuse apex and truncate base, straight to curved, 3(−4) transverse septate, guttulate or eguttulate, constricted at the septa.
Culture characteristics
Conidia germinating on PDA within 24 h and germ tubes produced from both sides. Colonies growing slowly on PDA, reaching 3 cm in 1 week at 28°C, dense, irregular, uneven margin, white in the first week, gray from above, light yellow at the margin and dark gray at the center, and with light gray in the middle from below.
Material examined
China, Yunnan Province, Qujing, Qujing Normal University, 25°52′36.75″N, 103°74′46.73″E, 1,853.8 m, on dead branches of M. grandiflora L., 18 August 2020, Dong-Qin Dai and Mei-ling Zhu, Lin 43-1, (GMB1384, holotype), ex-type GMBCC1176; Ibid. 10 June 2021, Dong-Qin Dai and Ting-Ting Zhang, Lin 101, (GMB1066, paratype); ex-paratype GMBCC1044.
Notes
Phragmocamarosporium qujingensis is morphologically and phylogenetically distinct from other species (Figure 3; Table 4).
Longiostiolaceae Phukhams., Doilom, & K.D. Hyde, Fungal Diversity 102: 43 (2020)
Shearia Petr., Annls mycol. 22(1/2): 180 (1924)
Index Fungorum Registration Identifier IF 9914
Notes
Petrak (1924) introduced this genus, which is typified by Shearia magnoliae (Shear) Petr. (Basionym: Camarosporium magnoliae Shear). However, Sutton (1980) regarded that Stegonsporium formosum Ell. & Ev. 1863 as the older name for this taxon. Thus, S. formosa (Ell. & Ev.) Petrak was regarded as the correct name for the type of species (Sutton, 1980). Wanasinghe et al. (2020) re-collected Shearia formosa from Magnolia denudate and M. soulangeana and designated the neotype. All collections were made from Kunming, Yunnan. In this study, we report Shearia formosa from M. grandiflora as a new host record from China (Wanasinghe et al., 2020; Farr and Rossman, 2022).
Shearia formosa (Ellis and Everh.) Petr., Sydowia 15 (1–6): 216 (1962), (Figure 11)
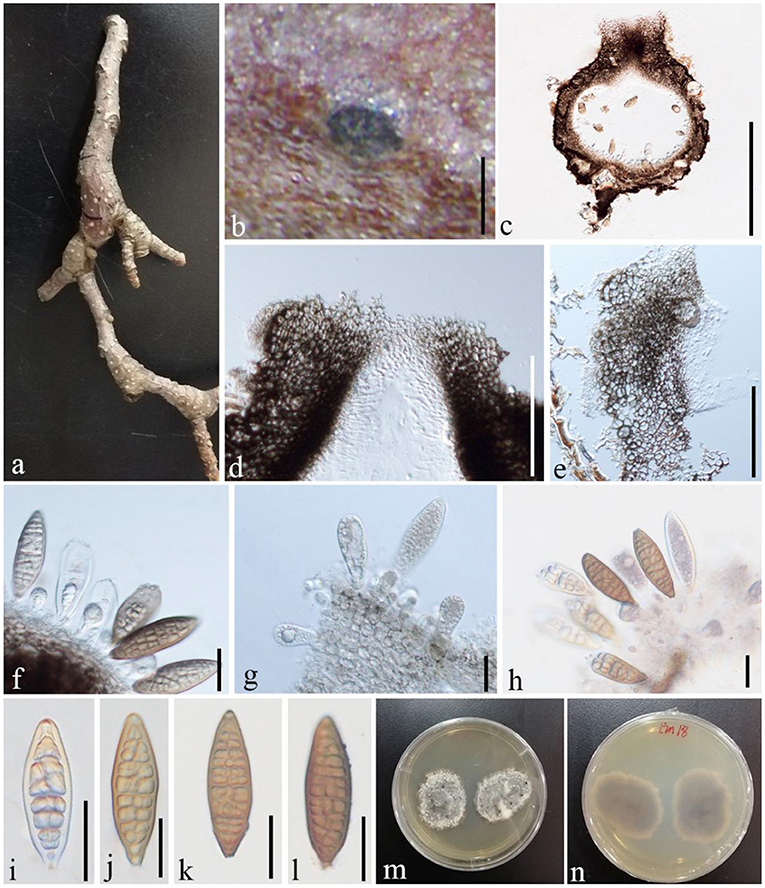
Figure 11. S. formosa (GMB1379, a new host record on M. grandiflora from China). (a) M. grandiflora branch. (b) Appearance of ascomata on host. (c) Vertical sections of conidioma. (d) Osti-ole opening. (e) Peridium. (f–h) Conidia and conidiogenous cells. (i–l) Conidia. (m,n) Seven-day-old colonies incubated at 28°C on PDA. Scale bars: (b) = 300, (c) = 430, (d) = 100, (e) = 55, (f) = 25, (g) = 20, (h) = 25, and (i–l) = 20 μm.
Basionym: Stegonsporium formosa Ellis & Everh., Bull. Torrey bot. Club 10(7): 76 (1883)
Index Fungorum Registration Identifier IF 339263
Saprobic on dead twigs of M. grandiflora. Sexual morph: undetermined. Asexual morph: Conidiomata 500–870 μm high, 530–1,000 μm diam. ( = 635.5 × 763.3 μm, n = 20), pseudostromatic, solitary, immersed, globose, unilocular, dark brown, central, papillate ostiole, and circular. Conidiomata wall 15–35 μm wide at the base, 30–80 μm wide at the sides and ostiole region, outer layer composed of thick-walled, very dark brown occluded cells, and lateral and basal walls composed of dark brown cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 7–14 × 6–8 μm ( = 11.2 × 7.3 μm, n = 10), holoblastic, annellidic, doliiform or cylindrical, discrete, inde-terminate, hyaline, and smooth-walled. Conidia 70–95 × 22–30 μm ( = 82 × 25 μm, n = 28), light brown to medium brown, fusiform, base truncate, apex obtuse, with several transverse and lateral distosepta, continuous, smooth and thick-walled, initially enveloped in a gelatinous sheath, depressed at the apex, and at maturity remaining as a basal lateral sheath.
Culture characteristics
Ascospores germinating on PDA within 24 h and germ tubes produced from both sides. Colonies growing slowly on PDA, reaching 3 cm in 1 week at 28°C, effuse, velvety to hairy, oval, uneven margin, white in the first week, white at the margin and gray at the center from above, and white at the margin and dark brown at the center from below.
Material examined
China, Yunnan Province, Qujing Normal University, 25°52′36.75″N, 103°74′46.73″E, 1,853.8 m, on branches of M. grandiflora L., 7 November 2019, Dong-Qin Dai and Mei-Ling Zhu, Lin 18, GMB1379 (new host record from China), culture GMBCC1172.
Notes
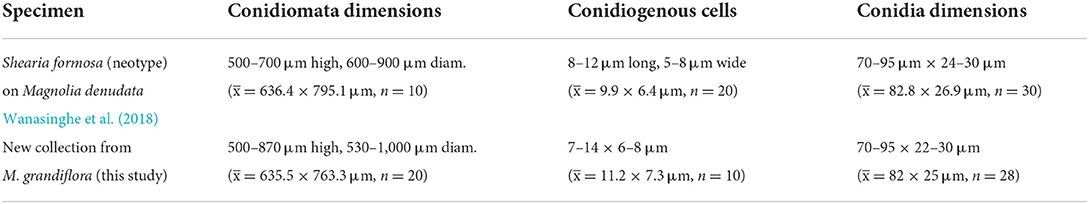
In conidial morphology and dimensions (in mean values), the new collection resembles the neotype of Shearia formosa (which was reported on Magnolia denudate) but is distinct from conidiomatal and conidiogenous cell characters (Wanasinghe et al., 2020) (Table 5). However, in phylogenetic analyses, the new collection accommodated in Shearia s. str. and clustered with MFLUCC 20–0019, the ex-neotype (Figure 1). Hence, we confirm our collection as Shearia formosa. S. formosa has previously been reported on M. grandiflora from the United States (Miller, 1990; Schubert, 1991; Farr and Rossman, 2022). However, according to our knowledge, S. formosa has not been reported on M. grandiflora from China. Hence, this is the first host record of S. formosa on M. grandiflora from China.
Discussion
Tropical and subtropical regions are rich in biodiversity. Several studies concluded that some regions in Asia have not been properly studied; thus, a large number of fungal species are yet to be discovered (Hyde et al., 2018). In China, the southwestern region has higher biodiversity including higher floral, faunal, and microbial diversity (Xu et al., 2017). The Guizhou and Yunnan provinces are important in this region as a large number of research studies confirmed rich fungal diversity (Farr and Rossman, 2022).
In southwest China (i.e., Guizhou and Yunnan Provinces), Magnolia species are widely used in gardening (as an ornamental plant), horticulture, and Chinese traditional medicine. In this study, we focused on M. grandiflora in Qujing Normal University Garden, Qujing city, Yunnan province. We recognized that this species has been widely used in Qujing for gardening purposes. Thus, we selected the university garden as a preliminary collecting site to assess the fungal diversity of M. grandiflora.
Botryosphaeriaceae species are common in southwest China and other regions as well (e.g., Wijayawardene et al., 2016; Xiao et al., 2021). Our new collections of Botryosphaeriaceae taxa from M. grandiflora resided in Botryosphaeria sensu stricto and Diplodia sensu stricto (Figure 1). Among the taxa, one species is confirmed as B. dothidea while two other strains are confirmed as D. mutila and D. seriata. This is the first report of both Diplodia species on M. grandiflora. Botryosphaeria dothidea has been reported as a pathogen of broad range of hosts in China, including gardening plants (e.g., causal agent of trunk and extended up to branches of Acer platanoides fide; Wang et al., 2015) and agricultural crops (e.g., causal agent of apple ring rot of apple fide; Tang et al., 2012). According to Farr and Rossman (2022), Botryosphaeria dothidea has not been reported from Magnolia species from China; thus this is the first report. Nevertheless, Botryosphaeria dothidea was reported as a pathogen of M. grandiflora from Serbia (Zlatkovic et al., 2018). However, none of the new collections have been observed associated with any diseased symptoms such as cankers or leaf spots. Besides, in a recent genomic study by Yan et al. (2018), they predicted that some Botryosphaeriaceae species (e.g., Lasiodiplodia theobromae) could be opportunistic pathogens of woody plants with changes in the environment. Hence, it is essential to expand the sample number to confirm whether their life modes are adversely impacted by M. grandiflora populations in Qujing.
Angustimassarina populi was introduced as a saprobe of dead branches of Populus sp. from Italy (Thambugala et al., 2015). The genus Angustimassarina comprises twelve species epithets (Index Fungorum 2022) including A. populi, and all species have been reported from Europe. In this study, we reported Angustimassarina populi, which occurred on M. grandiflora from Qujing, Yunnan. This is the first report of a member of Angustimassarina reported outside Europe. Moreover, this is the first report of Angustimassarina populi from China and on M. grandiflora and, thus, the first country and host records, respectively. This collection confirms that Angustimassarina species could have a broader distribution and, apparently, are not host-specific. It is necessary to promote biogeographic studies of this type of genus, which was previously reported only in one geographic region but recently found in other countries. Based on this result, we predict that more novel species can be reported from China as Angustimassarina was originally reported in temperate countries, i.e., Italy.
The novel species of Lonicericola, L. qujingensis is the third member of the genus. Interestingly, all the species have been reported only from Yunnan Province, China. However, previous species (i.e., L. hyaloseptispora and L. fuyuanensis) have been reported from the host family Caprifoliaceae. Since the new collection was made from decaying branches of M. grandiflora, we predict that members of Lonicericola could occur in a broad range of host families. However, geographical distribution is not clear and thus needs further collections from other regions in Yunnan.
Currently, the genus Phragmocamarosporium comprises three species that were reported from Germany, The United Kingdom, and Guizhou Province, China. In this study, we introduce two more species of Phragmocamarosporium from M. grandiflora viz., Phragmocamarosporium magnoliae and P. qujingensis. Phragmocamarosporium platani, the type of species of Phragmocamarosporium, was reported from Platanus species, in Guizhou. Species resolution in the subclade in which Phragmocamarosporium magnoliae, P. hedeare, and P. platani are included is not clear as the latter species are lacking ITS and protein loci in the GenBank (Wijayawardene et al., 2015). Hence, here, we used morphological characteristics to differentiate the species as a supporting factor. Besides, we predict that the Guizhou-Yunnan region could be harboring more Phragmocamarosporium species. Moreover, the strain named KUMCC 18-0165 (of Phragmocamarosporium hederae) in the GenBank must represent a novel lineage in Phragmocamarosporium s. str. (Figures 1, 10; Table 4). Wanasinghe et al. (2020) reported S. formosa on Magnolia denudate from Yunnan, China. In this study, we report S. formosa on M. grandiflora for the first time.
Our findings suggest that it is essential to check for fungal diversity on extensively studied host genera that occur in biodiversity-rich regions. Hence, we suggest expanding future studies on extensively studied host genera that occur in Yunnan such as Eucalyptus, Magnolia, and Quercus. Besides, these host genera could be species-rich and thus could harbor different fungal taxa. Hence, precise host identification is also important in this type of broad future study. This is essential to reveal hidden fungal diversity in biodiversity hotspots.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
NW and D-QD designed the study, performed the morphological study, and wrote the manuscript. DW and ST performed the phylogenetic analyses. JK, H-HC, and D-QD reviewed and edited the manuscript. T-TZ, G-QZ, and M-LZ made the plates. L-SH performed the fungal DNA extraction. All authors approved the final version of the manuscript.
Funding
The research was supported by the National Natural Science Foundation of China (Nos. NSFC 31860620, 31950410558, 31760013, and 32150410362), High-Level Talent Recruitment Plan of Yunnan Province (Young Talents Program and High-End Foreign Experts Program), CAS President's International Fellowship Initiative (Grant No: 2021FYB0005), and Postdoctoral Fund from Human Resources and Social Security Bureau of Yunnan Province, and it was partially supported by Chiang Mai University, Thailand.
Acknowledgments
The authors thank R. Phookamsak for providing the spore dimensions of the holotype of Lonicericola hyaloseptispora.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.954680/full#supplementary-material
References
Barr, M. E. (1972). Preliminary studies on the Dothideales in temperate North America. Contrib. Univ. Mich. Herb. 9, 523–638.
Calabon, M. S., Jones, E. B., Boonmee, S., Doilom, M., Lumyong, S., and Hyde, K. D. (2021). Five novel freshwater ascomycetes indicate high undiscovered diversity in lotic habitats in Thailand. J. Fungi. 7, 117. doi: 10.3390/jof7020117
Cesati, V., and De Notaris, G. (1863). Schema di classificazione degle sferiacei italici aschigeri piu' o meno appartenenti al genere Sphaeria nell'antico significato attribuitoglide Persono. Commentario della Società Crittogamologica Italiana 1, 177–420.
Chaiwan, N., Wanasinghe, D. N., Mapook, A., Jayawardena, R. S., Norphanphoun, C., and Hyde, K. D. (2020). Novel species of Pestalotiopsis fungi on Dracaena from Thailand. Mycology 11, 306–315. doi: 10.1080/21501203.2020.1801873
Chomnunti, P., Hongsanan, S., Aguirre-Hudson, B., Tian, Q., Persoh, D., Dhami, M. K., et al. (2014). The sooty moulds. Fungal Divers 66, 1–36. doi: 10.1007/s13225-014-0278-5
Dai, D. Q., Phookamsak, R., Wijayawardene, N. N., Li, W. J., Bhat, D. J., Xu, J. C., et al. (2017). Bambusicolous fungi. Fungal Divers 82, 1–105. doi: 10.1007/s13225-016-0367-8
Dai, D. Q., Wijayawardene, N. N., Tang, L. Z., Liu, C., Han, L. H., Chu, H. L., et al. (2019). Rubroshiraia gen. nov., a second hypocrellin-producing genus in Shiraiaceae (Pleosporales). MycoKeys 58, 1–26. doi: 10.3897/mycokeys.58.36723
Díaz, G. A., Latorre, B. A., Ferrada, E. E., and Lolas, M. (2018). Identification and characterization of Diplodia mutila, D. seriata, Phacidiopycnis washingtonensis and Phacidium lacerum obtained from apple (Malus x domestica) fruit rot in Maule Region, Chile. Eur. J. Plant Pathol. 153, 1259–1273. doi: 10.1007/s10658-018-01640-8
Dissanayake, A. J., Camporesi, E., Hyde, K. D., Yan, J. Y., and Li, X. H. (2017). Saprobic Botryosphaeriaceae, including Dothiorella italica sp. nov., associated with urban and forest trees in Italy. Mycosphere 8, 1157–1176. doi: 10.5943/mycosphere/8/2/7
Farr, D. F., and Rossman, A. Y. (2022). Fungal Databases, U.S. National Fungus Collections, ARS, USDA. Available online at: https://nt.arsgrin.gov/fungaldatabases/ (accessed March 20, 2022).
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acid Symp. Ser. 41, 95–98.
Hawksworth, D. L., and Lücking, R. (2017). Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 5, 79–95. doi: 10.1128/microbiolspec.FUNK-0052-2016
Hyde, K. D., Jeewon, R., Chen, Y. J., Bhunjun, C. S., Calabon, M. S., Jiang, H. B., et al. (2020). The numbers of fungi: is the descriptive curve flattening? Fungal Divers 103, 219–271. doi: 10.1007/s13225-020-00458-2
Hyde, K. D., Norphanphoun, C., Chen, J., Dissanayake, A. J., Doilom, M., Hongsanan, S., et al. (2018). Thailand's amazing diversity: up to 96% of fungi in northern Thailand may be novel. Fungal Divers 93, 215–239. doi: 10.1007/s13225-018-0415-7
Hyde, K. D., Tennakoon, D. S., Jeewon, R., Bhat, D. J., Maharachchikumbura, S. S., Rossi, W., et al. (2019). Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers 96, 1–242. doi: 10.1007/s13225-019-00429-2
Index Fungorum (2022). Available online at: http://www.indexfungorum.org/names/Names.asp (accessed March 20, 2022).
Katoh, K., Rozewicki, J., and Yamada, K. D. (2017). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Liu, J., Xiao, J., Zhou, J., Cai, G., Li, X., and Lu, J. (2019). Alternaria alternata causing leaf spot on Magnolia grandiflora in China. Plant Dis. 103, 2672. doi: 10.1094/PDIS-04-19-0828-PDN
Liu, Y., Whelen, S., and Hall, B. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Lu, L., Tibpromma, S., Karunarathna, S., Thiyagaraja, V., Xu, J., Jayawardena, R. S., et al. (2021). Taxonomic and phylogenic appraisal of a novel species and a new record of Stictidaceae from coffee in Yunnan Province, China. Phytotaxa 14, 111–124. doi: 10.11646/phytotaxa.528.2.4
Luo, Z. L., Hyde, K. D., Bhat, D. J., Jeewon, R., Maharachchikumbura, S. S., Bao, D. F., et al. (2018). Morphological and molecular taxonomy of novel species Pleurotheciaceae from freshwater habitats in Yunnan, China. Mycol. Prog. 17, 511–530. doi: 10.1007/s11557-018-1377-6
Miller, J. W. (1990). Bureau of plant pathology. Triology Techn. Rep. Div. Plant Indust. Florida 29, 6.
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees,” in Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010, p. 1–8.
Montagne, J. P. F. C. (1834). Notice sur les plantes cryptogames récemment découvertes en France, contenant aussi l'indication précise des localités de quelques espèces les plus rares de la flore française. Annales des Sciences Naturelles Botanique. 1, 295–307.
Nylander, J. A. A., Wilgenbusch, J. C., Warren, D. L., and Swofford, D. L. (2008). AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24, 581–583. doi: 10.1093/bioinformatics/btm388
Phillips, A. J. L., Alves, A., Abdollahzadeh, J., Slippers, B., Wingfield, M. J., Groenewald, J. Z., et al. (2013). The Botryosphaeriaceae: genera and species known from culture. Stud. Mycol. 76, 51–167. doi: 10.3114/sim0021
Phookamsak, R., Hyde, K. D., Jeewon, R., Bhat, D. J., Gareth Jones, E. B., Maharachchikumbura, S. S. N., et al. (2019). Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers 95, 1–273. doi: 10.1007/s13225-019-00421-w
Rajeshkumar, K. C., Verma, R. K., Boonmee, S. A., Chandrasiri, S., Hyde, K. D., Ashtekar, N. I., et al. (2021). Paradictyocheirospora tectonae, a novel genus in the family Dictyosporiaceae from India. Phytotaxa 30, 259–271. doi: 10.11646/phytotaxa.509.3.1
Rambaut, A. (2012). FigTree version 1.4.0. Available online at: http://tree.bio.ed.ac.uk/software/figtree/ (accessed March 20, 2022).
Rehner, S. (2001). Primers for Elongation Factor 1-α (EF1-α). Available online at: http://ocid.NACSE.ORG/research/deephyphae/EF1primer.pdf/ (accessed March 20, 2022).
Ren, G. C., Wanasinghe, D. N., Monkai, J., Mortimer, P. E., Hyde, K. D., Xu, J. C., et al. (2021). Novel saprobic Hermatomyces species (Hermatomycetaceae, Pleosporales) from China (Yunnan Province) and Thailand. MycoKeys 82, 57. doi: 10.3897/mycokeys.82.67973
Schubert, T. S. (1991). Bureau of plant pathology. Tri-ology Techn. Rep. Div. Plant Indust. Florida. 30, 5–6.
Slippers, B., Fourie, G., Crous, P. W., Coutinho, T. A., Wingfield, B. D., Carnegie, A. J., et al. (2004). Speciation and distribution of Botryosphaeria spp. on native and introduced Eucalyptus trees in Australia and South Africa. Stud. Mycol. 50, 343–358.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sutton, B. C. (1980). The Coelomycetes. fungi imperfecti with pycnidia, acervuli and stromata. Kew, Surrey: Commonwealth Mycological Institute.
Tanaka, K., Hirayama, K., Yonezawa, H., Sato, G., Toriyabe, A., Kudo, H., et al. (2015). Revision of the massarineae (Pleosporales, Dothideomycetes). Stud. Mycol. 1, 75–136. doi: 10.1016/j.simyco.2015.10.002
Tang, W., Ding, Z., Zhou, Z. Q., Wang, Y. Z., and Guo, L. Y. (2012). Phylogenetic and pathogenic analyses show that the causal agent of apple ring rot in China is Botryosphaeria dothidea. Plant Dis. 96:486–496. doi: 10.1094/PDIS-08-11-0635
Thambugala, K. M., Hyde, K. D., Tanaka, K., Tian, Q., Wanasinghe, D. N., Ariyawansa, H. A., et al. (2015). Towards a natural classification and backbone tree for Lophiostomataceae, Floricolaceae, and Amorosiaceae fam. nov. Fungal Divers 74, 199–266. doi: 10.1007/s13225-015-0348-3
Tibpromma, S., Hyde, K. D., Jeewon, R., Maharachchikumbura, S. S. N., Liu, J. K., Bhat, D. J., et al. (2017). Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 83, 1–261. doi: 10.1007/s13225-017-0378-0
Vilgalys, R., and Sun, B. L. (1994). Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. PNAS 91, 4599–4603. doi: 10.1073/pnas.91.10.4599
Wanasinghe, D. N., Phukhamsakda, C., Hyde, K. D., Jeewon, R., Lee, H. B., Gareth Jones, E. B., et al. (2018). Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers 89, 1–236. doi: 10.1007/s13225-018-0395-7
Wanasinghe, D. N., Wijayawardene, N. N., Xu, J., Cheewangkoon, R., and Mortimer, P. E. (2020). Taxonomic novelties in Magnolia-associated pleosporalean fungi in the Kunming Botanical Gardens (Yunnan, China). PLoS ONE 13, e0235855. doi: 10.1371/journal.pone.0235855
Wang, K., Cai, L., and Yao, Y. (2021). Overview of nomenclature novelties of fungi in the world and China (2020). Biodivers. Sci. 29, 1064. doi: 10.17520/biods.2021202
Wang, X., Li, Y. X., Dong, H. X., Jia, X. Z., and Zhang, X. Y. (2015). First report of Botryosphaeria dothidea causing canker of Acer platanoides in China. Plant Dis. 99, 1857. doi: 10.1094/PDIS-03-15-0265-PDN
White, T. J., Bruns, T., Lee, S. J., and Taylor, J. L. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Prot. Guide Methods Appl. 18, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wijayawardene, N. N., Dissanayake, L. S., Dai, D. Q., Li, Q. R., Xiao, Y., Wen, T. C., et al. (2021). Yunnan-Guizhou Plateau: a mycological hotspot. Phytotaxa 523, 1–31. doi: 10.11646/phytotaxa.523.1.1
Wijayawardene, N. N., Hyde, K. D., Bhat, D. J., Goonasekara, I. D., Nadeeshan, D., Camporesi, E., et al. (2015). Additions to brown spored coelomycetous taxa in Massarinae, Pleosporales: introducing Phragmocamarosporium gen. nov. and Suttonomyces gen. nov. Cryptogamie Mycol. 36, 213–224. doi: 10.7872/crym/v36.iss2.2015.213
Wijayawardene, N. N., Hyde, K. D., Dai, D. Q., Sánchez-García, M., Goto, B. T., Saxena, R. K., et al. (2022). Outline of Fungi and fungus-like taxa - 2021. Mycosphere 13, 53–453. doi: 10.5943/mycosphere/13/1/2
Wijayawardene, N. N., Hyde, K. D., Wanasinghe, D. N., Papizadeh, M., Goonasekara, I. D., Camporesi, E., et al. (2016). Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Divers 77, 1–316. doi: 10.1007/s13225-016-0360-2
Wu, N., Dissanayake, A. J., Manawasinghe, I. S., Rathnayaka, A. R., Liu, J. K., Phillips, A. J. L., et al. (2021). https://botryosphaeriales.org/, an up-to-date classification and account of taxa of Botryosphaeriales. Database. 2021, baab061. doi: 10.1093/database/baab061
Xiao, X. E., Wang, W., Crous, P. W., Wang, H. K., Jiao, C., Huang, F., et al. (2021). Species of Botryosphaeriaceae associated with citrus branch diseases in China. Persoonia Mol. Phyl. Evol. Fungi 47, 106–135. doi: 10.3767/persoonia.2021.47.03
Xu, Y., Shen, Z., Ying, L., Wang, Z., Huang, J., Zang, R., et al. (2017). Hotspot analyses indicate significant conservation gaps for evergreen broadleaved woody plants in China. Sci. Rep. 7, 1–10. doi: 10.1038/s41598-017-02098-0
Yan, J. Y., Zhao, W. S., Chen, Z., Xing, Q. K., Zhang, W., Chethana, K. T., et al. (2018). Comparative genome and transcriptome analyses reveal adaptations to opportunistic infections in woody plant degrading pathogens of Botryosphaeriaceae. DNA Res. 25, 87–102. doi: 10.1093/dnares/dsx040
Yasanthika, E., Dissanayake, L. S., Wanasinghe, D. N., Karunarathna, S. C., Mortimer, P. E., Samarakoon, B. C., et al. (2020). Lonicericola fuyuanensis (Parabambusicolaceae) a new terrestrial pleosporalean ascomycete from Yunnan Province, China. Phytotaxa 27, 103–113. doi: 10.11646/phytotaxa.446.2.3
Zhang, Y., Schoch, C. L., Fournier, J., Crous, P. W., Gruyter, J., de Woudenberg, J. H. C., et al. (2009). Multilocus phylogeny of Pleosporales: a taxonomic, ecological and evolutionary reevaluation. Stud. Mycol. 64, 85–102. doi: 10.3114/sim.2009.64.04
Keywords: Ascomycota, Botryosphaeriales, diversity, phylogeny, Pleosporales, taxonomy
Citation: Wijayawardene NN, Dai D-Q, Zhu M-L, Wanasinghe DN, Kumla J, Zhang G-Q, Zhang T-T, Han L-S, Tibpromma S and Chen H-H (2022) Fungi associated with dead branches of Magnolia grandiflora: A case study from Qujing, China. Front. Microbiol. 13:954680. doi: 10.3389/fmicb.2022.954680
Received: 27 May 2022; Accepted: 30 June 2022;
Published: 04 August 2022.
Edited by:
Mohammad Arif, University of Hawaii at Manoa, United StatesReviewed by:
Danushka Sandaruwan Tennakoon, Chiang Mai University, ThailandTolgor Bau, Jilin Agriculture University, China
Hamid Mohammadi, Shahid Bahonar University of Kerman, Iran
Abolfazl Narmani, University of Tabriz, Iran
Kaivan Karimi, Iranian Research Institute of Plant Protection (IRIPP), Iran
Copyright © 2022 Wijayawardene, Dai, Zhu, Wanasinghe, Kumla, Zhang, Zhang, Han, Tibpromma and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong-Qin Dai, cicidaidongqin@gmail.com; Huan-Huan Chen, huanhuanc5337@gmail.com
†These authors have contributed equally to this work
 Nalin N. Wijayawardene
Nalin N. Wijayawardene Dong-Qin Dai
Dong-Qin Dai Mei-Ling Zhu1
Mei-Ling Zhu1 Dhanushka N. Wanasinghe
Dhanushka N. Wanasinghe Jaturong Kumla
Jaturong Kumla Saowaluck Tibpromma
Saowaluck Tibpromma Huan-Huan Chen
Huan-Huan Chen