- 1The Second Affiliated Hospital and Yuying Children’s Hospital, Wenzhou Medical University, Wenzhou, China
- 2Key Laboratory of Medical Genetics of Zhejiang Province, Key Laboratory of Laboratory Medicine, Ministry of Education, Wenzhou, China
- 3School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 4Medical Molecular Biology Laboratory, School of Medicine, Jinhua Polytechnic, Jinhua, China
Background: Pantoea species of the family Erwiniaceae are well-known plant pathogens and animal and human conditional pathogens. Due to the widespread and continuous use of antimicrobials, multidrug-resistant strains continue to emerge, making clinical treatment difficult; therefore, there is an increasing need to clarify the mechanisms of drug resistance.
Methods: A rabbit anal fecal sample was collected by a swab and the streak plate method was used to isolate single colonies. The standard agar dilution method was used to determine the minimum inhibitory concentrations (MICs) against antimicrobials. The complete genome sequence of the bacterium was obtained using Next-Generation Sequencing platforms. The potential resistance gene was annotated based on the Comprehensive Antibiotic Resistance Database (CARD) and verified by molecular cloning. The β-lactamase PSZ-1 was expressed via the pCold I expression vector and its enzyme kinetic parameters were analyzed. The genetic environment and evolutionary process of the novel resistance gene-related sequences were analyzed by bioinformatic methods.
Results: The isolate Pantoea endophytica X85 showed some degree of resistance to penicillins as well as cephalosporins. A novel AmpC resistance gene, designated blaPSZ-1 in this research, was identified to be encoded in the plasmid (pPEX85) of P. endophytica X85. BlaPSZ-1 showed resistance to penicillins and several first-, second-and third-generation cephalosporins as well as aztreonam, but it did not show resistance to the fourth-generation cephalosporins or carbapenems tested. Enzyme kinetic assays revealed that it could hydrolyze amoxicillin, penicillin G, cephalothin, and cefazolin, and its hydrolytic activity could be strongly inhibited by the inhibitor avibactam, which was generally consistent with antimicrobial susceptibility testing results. No hydrolytic activity was observed for third-generation cephalosporins or aztreonam.
Conclusion: In this study, a novel AmpC β-lactamase gene, designated blaPSZ-1, was characterized and it was encoded in the plasmid of the bacterium P. endophytica X85. It shows resistance to penicillins and several cephalosporins. The discovery of novel drug resistance mechanisms can help guide the scientific use of drugs in animal husbandry and clinical practice, effectively avoiding the abuse of antimicrobials and thus preventing the further development and spread of bacterial resistance.
Introduction
Pantoea species of the family Erwiniaceae are ubiquitous in the environment and are well-known plant pathogens (Walterson and Stavrinides, 2015). They have been isolated from contaminated soil, water, plants (as epiphytes or endophytes), seeds, dairy products, and from gastrointestinal tracts, blood, and urine of humans and animals (Fullerton et al., 2007; Dutkiewicz et al., 2016). They usually cause opportunistic infections, especially when the immune system is compromised; for example, they have been reported to cause blood infections in several newborn preterm infants (Habsah et al., 2005). The wider and even abused use of antimicrobials in clinical and agricultural farming caused the rapid emergence of bacterial resistance, and the emergence of multidrug resistance in human pathogens is considered to be a major public global health threat. Therefore, understanding the molecular mechanisms of bacterial drug resistance is crucial. Mechanisms of antimicrobial resistance can be mainly classified as follows: (i) modification of antibiotic molecules to inactivate them, such as the production of β-lactamases and aminoglycoside-modifying enzymes, (ii) reduction of antibiotic penetration (reduction of pore protein-mediated outer membrane permeability) and efflux mechanisms, and (iii) alteration of target sites (Munita and Arias, 2016). Among them, the presence of β-lactamases is a very common mechanism of drug resistance in pathogenic bacteria. β-lactamase is an enzyme that inactivates β-lactam antibiotics by hydrolyzing the β-lactam ring (Bush, 2018) and is classified by Ambler into class A to class D according to amino acid sequence homology (conserved and distinguished amino acid motifs; Bush and Jacoby, 2010). AmpC β-lactamases are commonly produced by many Enterobacteriaceae and a few other organisms and are encoded by bla genes located on bacterial chromosomes and, to a lesser extent, by plasmids. Organisms expressing AmpC enzymes are usually resistant to penicillins, β-lactamase inhibitors (clavulanic acid and tazobactam), and most cephalosporins, including cefoxitin, ceftriaxone, and cefotaxime (Bush et al., 1995). In addition, AmpC β-lactamases have a poor ability to hydrolyze the extended-spectrum cephalosporin cefepime and are readily inactivated by carbapenems. Notably, AmpC β-lactamases are strongly inactivated by avibactam (Jacoby, 2009).
Ampicillin, amoxicillin, and first-generation cephalosporins such as cefazolin and cephalothin are very strong inducers of AmpC enzymes and good substrates for hydrolysis. Cefoxitin and imipenem are also strong inducers but are more stable for hydrolysis. Cefotaxime, ceftriaxone, ceftazidime, cefepime, cefuroxime, piperacillin, and aztreonam are weak inducers and substrates but can also be hydrolyzed if sufficient enzyme expression is present. Thus, the minimum inhibitory concentrations (MICs) of weakly induced oxyimino β-lactams increase dramatically in the presence of AmpC overproduction (Jacoby, 2009).
In the present study, we described a novel AmpC enzyme gene, designated blaPSZ-1, which was harbored on a strain of the genus Pantoea isolated from a rabbit in a livestock farm in Wenzhou, China. Its enzymatic kinetic parameters were also determined. The identification of this novel resistance gene from an animal bacterium is of great value for our in-depth understanding of bacterial drug resistance and its dissemination pattern, as well as for the clinical treatment of infectious diseases caused by the related bacteria.
Materials and methods
Bacteria and plasmids
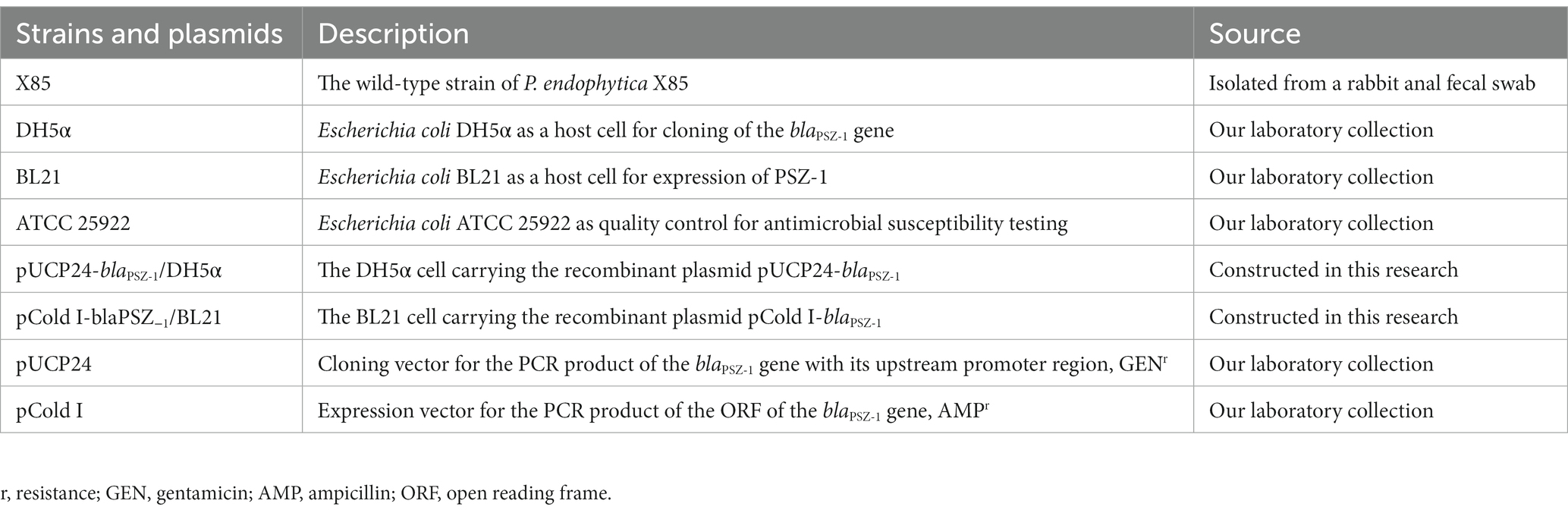
P. endophytica X85 carrying the novel drug-resistance gene blaPSZ-1 was isolated from the fecal swab of a rabbit at a livestock farm during a survey on the antimicrobial resistance of bacteria in Wenzhou, southern China. The genomes of the isolates were sequenced and their resistance profiles were determined. The relationship between the resistance genotypes and phenotypes was further analyzed. Species identification of the isolate was performed first by 16S rRNA gene homology and then by genome-wide average nucleotide identity (ANI) analyses. The bacterial strains and plasmids used in the study are listed in Table 1.
Antimicrobial susceptibility testing
Following the Clinical and Laboratory Standards Institute (CLSI) guidelines, MICs were determined on Mueller-Hinton (M-H) agar using the standard agar dilution method, and susceptibility patterns were explained in accordance with CLSI M100 (31st Edition, 2021). The reference strain used for quality control in this study was E. coli ATCC 25922. The antimicrobials tested in this research included 13 β-lactams, 4 aminoglycosides, and 2 β-lactamase inhibitors (Table 2). All antibiotics were for human clinical use and purchased from pharmacies and hospitals, and MIC values were the mean values of three independent measures.
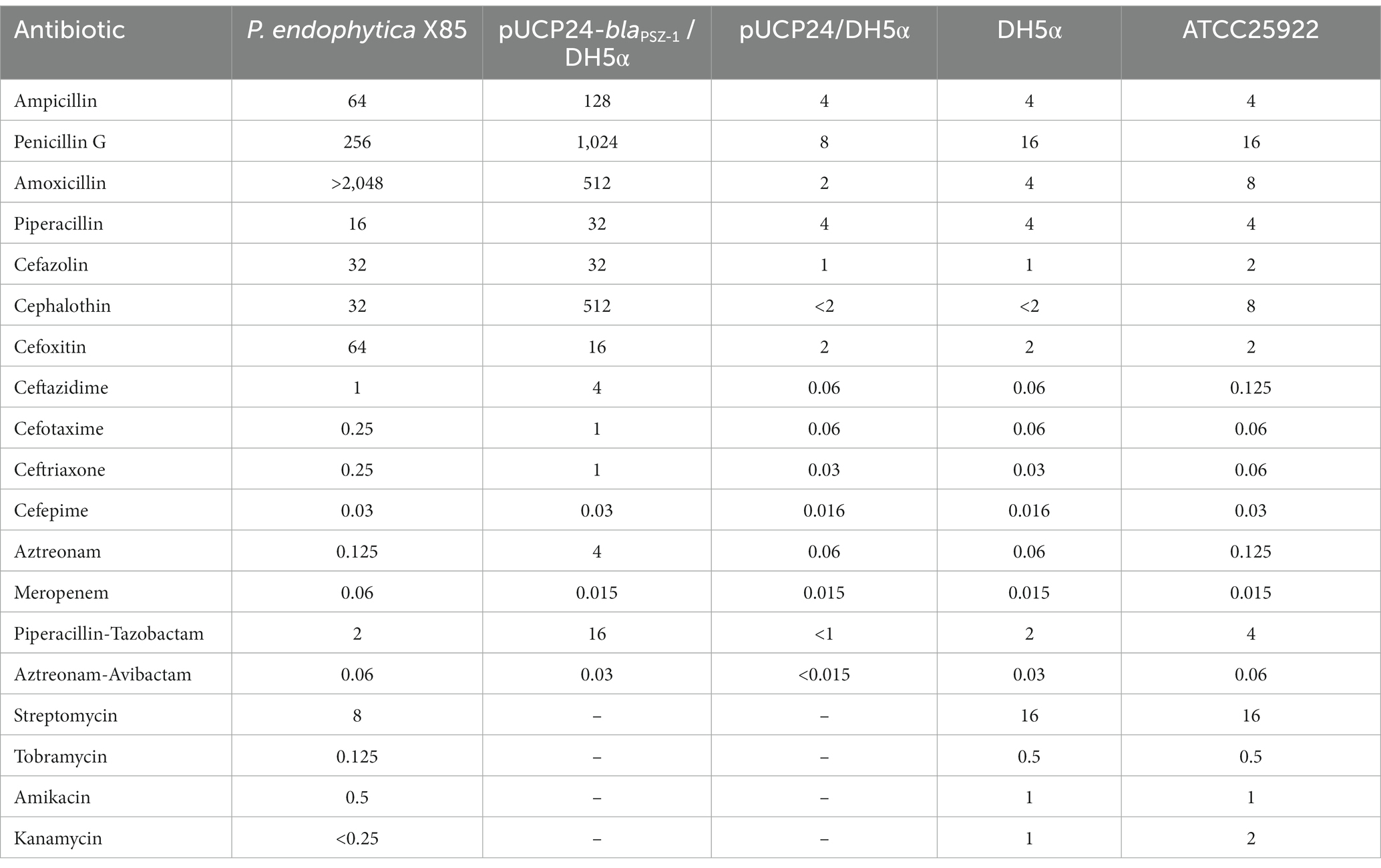
Table 2. Minimum inhibitory concentrations of antimicrobials for P. endophytica X85, the recombinant carrying blaPSZ-1, and the control strains (μg/mL).
Cloning and expression of the blaPSZ-1 gene and purification of PSZ-1
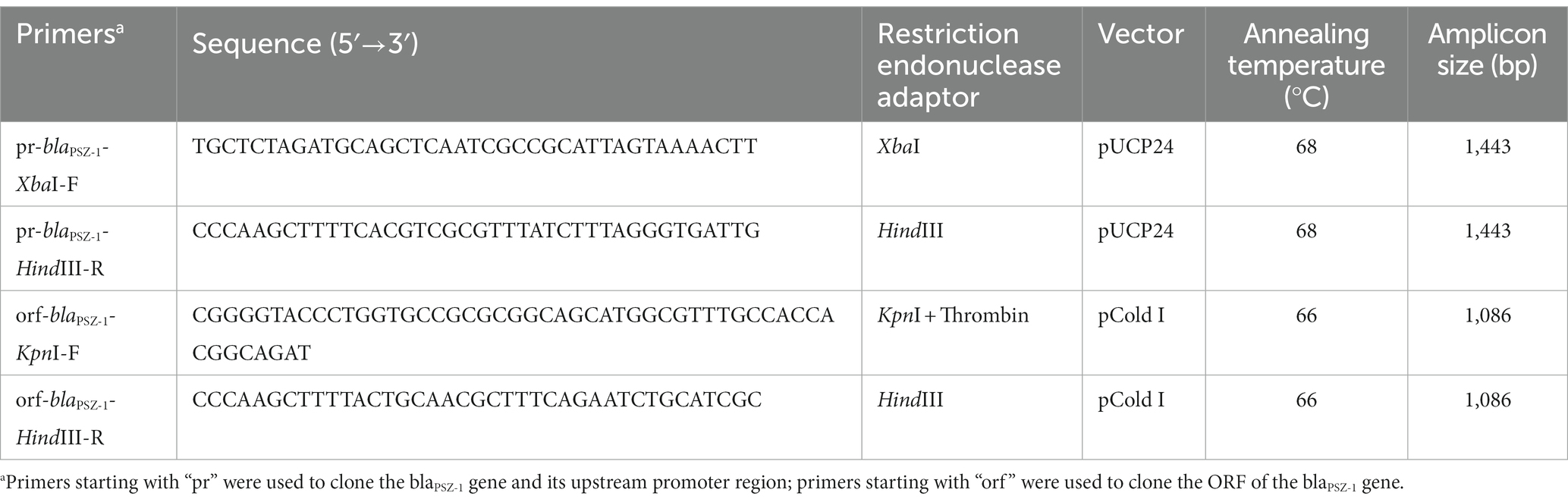
The blaPSZ-1 gene and its upstream promoter region were amplified using 2× Phanta® Max Master Mix (Nanjing Vazyme Biotech Co., Ltd., Nanjing, China), and P. endophytica X85 genomic DNA was extracted using the Generay Genomic DNA Miniprep kit (Shanghai Generay Biotech Co., Ltd., Shanghai, China) and used as the template for PCR. The primers with restriction endonuclease adaptors at the 5′-end (XbaI for the forward primer and HindIII for the reverse primer) are listed in Table 3. PCR products were digested with XbaI and HindIII and linked to the cloning vector pUCP24, which was also digested with XbaI and HindIII using a T4 DNA ligase cloning kit (Takara Bio, Inc., Dalian, China). The recombinant plasmid was transformed into E. coli DH5α by the calcium chloride method and the transformants were then screened on Luria-Bertani (LB) agar plates supplemented with gentamicin (40 μg/mL). Individual colonies were inoculated into LB mediums supplemented with the same antibiotic and cultured overnight, and then the inserts were verified by PCR and Sanger sequencing (Shanghai Sunny Biotechnology Co., Ltd., Shanghai, China).
The corresponding primers (Table 3) were used to amplify the ORF of blaPSZ-1 containing the thrombin digestion site by PCR, which was inserted into the pCold I expression vector, and the recombinant plasmid (pCold I-blaPSZ-1) was transformed into E. coli BL21 competent cells. For the expression of the PSZ-1 protein, E. coli BL21 cells with verified recombinant plasmids were cultured overnight in LB mediums supplemented with ampicillin (100 μg/mL), diluted 100 times in fresh mediums, and incubated at 37°C. After incubation in an ice bath for more than 30 min, the OD600 of the culture reached 0.6 to 0.8 and protein expression was induced by the addition of 1 mM isopropyl-β-dithiogalactopyranoside (IPTG, Sigma Chemicals Co., St. Louis, MO, United States), and further incubated for approximately 20 h at 15–16°C. Cells were collected by centrifugation (5,000 × g, 10 min) at 4°C, resuspended in lysis buffer (20 mM Tris–HCl, 150 mM NaCl, 3 mM β-mercaptoethanol, 0.5% Nonidet-P-40; pH 8.0), and lysed by ultrasound. After the cell fragments were removed by centrifugation (12,000 × g, 30 min) at 4°C, the lysates were combined with pre-balanced nickel-nitrilotriacetic acid (Ni-NTA) agarose resin (Beyotime Biotechnology, Shanghai, China) at 4°C and shaken gently overnight. Then, the recombinant protein was purified by standard Ni-NTA affinity chromatography and digested with thrombin (Solarbio Science & Technology Co., Ltd., Beijing, China) at 37°C for 24 h to remove the His-tag. The purity of PSZ-1 protein was validated by SDS–PAGE with a 12% acrylamide separation gel and Coomassie blue G-250 staining, and the protein concentration was determined by a BCA protein assay kit (Thermo Fisher Scientific, Rockford, IL, United States).
Enzyme kinetic parameter analysis
Kinetic parameters of the purified β-lactamase PSZ-1 against β-lactam antimicrobials were determined at 37°C, in a 10 mM phosphate buffer (pH 7.4), and a final reaction volume of 200 μL on a Synergy™ Neo2 Multi-Mode Microplate Reader (BioTek Instruments, Inc., United States). The steady-state kinetic parameters kcat and Km were determined by non-linear regression of the initial reaction rates with the Michaelis–Menten equation in Prism (version 8.0.2) software (GraphPad Software, CA, United States).
The concentrations of the β-lactamase inhibitors avibactam and tazobactam, leading to a 50% reduction in the hydrolysis of nitrocefin (IC50), were measured after 5 min of preincubation of the enzymes with the inhibitors at 37°C, and nitrocefin (0.1 mM) was used as the substrate. The IC50 values were determined by non-linear regression analysis (GraphPad Prism, version 8.0.2) using log (inhibitor) vs. response (three parameters). Values are the average of three independent measures.
Whole-genome sequencing and sequence analysis
The total bacterial DNA of P. endophytica X85 was extracted from an individual colony subcultured in LB at 37°C for approximately 16 h by using the Generay Genomic DNA Miniprep kit (Shanghai Generay Biotech Co., Ltd., Shanghai, China). Genomic DNA was sequenced by both the Illumina HiSeq 2,500 and PacBio RS II platforms, with a read length of PE150 and 10 kb, respectively, and a sequencing depth of 150× for both (Shanghai Personal Biotechnology Co., Ltd., China). Unicycler v0.4.8 was used to hybrid assemble the PacBio long reads and confirm the cyclization of the whole-genome assembly (Wick et al., 2017). Pilon improves the quality of genomic assembly sketches by mapping Illumina short reads onto the assembly to correct possible incorrect assembly (Walker et al., 2014). Genes were predicted and annotated by using Prokka v1.14.6 (Seemann, 2014); furthermore, DIAMOND v2.0.11 was used to search the predicted proteins in the NCBI non-redundant protein databases with an e-value threshold of 1e-5 (Buchfink et al., 2021). The 16S rRNA homology analysis was performed by comparing the 16S rRNA sequences extracted from the Prokka-annotated genome of P. endophytica X85 with those in the 16S ribosomal RNA sequence database in NCBI. Drug resistance gene identifier v5.2.02 and CARD were used to annotate drug resistance genes (McArthur et al., 2013). FastANI was used to calculate the ANI (Jain et al., 2018) and dDDH was calculated through the Type Strain Genome Server (TYGS) online database1 (Meier-Kolthoff et al., 2022). Multiple sequence alignments of PSZ-1 and its relatives from the Beta-Lactamase Database2 and UniProt/Swiss-Prot database3 were performed by MAFFT v7.490 (Katoh and Standley, 2013) and then a msa R package was used to visualize the comparison results and embellish the generated visual figure (Bodenhofer et al., 2015). MEGA11 was used to construct a neighbor-joining (N-J) phylogenetic tree including PSZ-1 and other functionally characterized β-lactamases (Kumar et al., 2018). The generated tree was visualized using the online tool iTol4 (Letunic and Bork, 2007). The figure depicting the genetic environments around the blaPSZ-1 and blaPSZ-1-like genes was generated by clinker v0.0.24 (Gilchrist and Chooi, 2021). GView was used to construct the basic genomic characteristics and comparative genomes of P. endophytica X85 (Stothard et al., 2019). ProtParam5 was used to predict the molecular weight and pI value of PSZ-1 (Wilkins et al., 1999).
Accession numbers
The complete chromosome, plasmid, and blaPSZ-1 gene sequences of P. endophytica X85 have been submitted to GenBank, and the accession numbers are CP121108, CP121109, and OQ725878, respectively.
Results and discussion
Identification and genome characterization of the isolate Pantoea endophytica X85
The 16S rRNA gene homology analysis revealed that the 16S rRNA gene of the strain X85 had the highest homology with that of Pantoea endophytica 596 T (NR_178843.1); the coverage was 100.00%, and the identity was 100% (Supplementary Table S3). Furthermore, the ANI analysis between the genomes of P. endophytica X85 and all 863 genomes from Pantoea downloaded from the NCBI databases demonstrated that among them, 14 genomes with ≥95% ANI (the cutoff to define a classified bacterial species) were found (Jain et al., 2018), of which 3 were from the Pantoea endophytica species (Pantoea endophytica 596 T, Pantoea endophytica HN-23, and Pantoea endophytica RIT-836 with ANIs of 98.29, 98.28 and 95.68%, respectively), and the remaining 11 genomes were all species-undetermined ones of genus Pantoea (Supplementary Table S1). The digital DNA–DNA hybridization (dDDH) analysis results obtained by TYGS showed that P. endophytica X85 shared the highest identity (86.00%) with Pantoea endophytica 596 T (GCA_002858935.1), which was higher than the cutoff (70%) to classify a bacterial species (Goris et al., 2007; Supplementary Table S4). Therefore, based on the results mentioned above, the isolate X85 was finally designated P. endophytica X85.
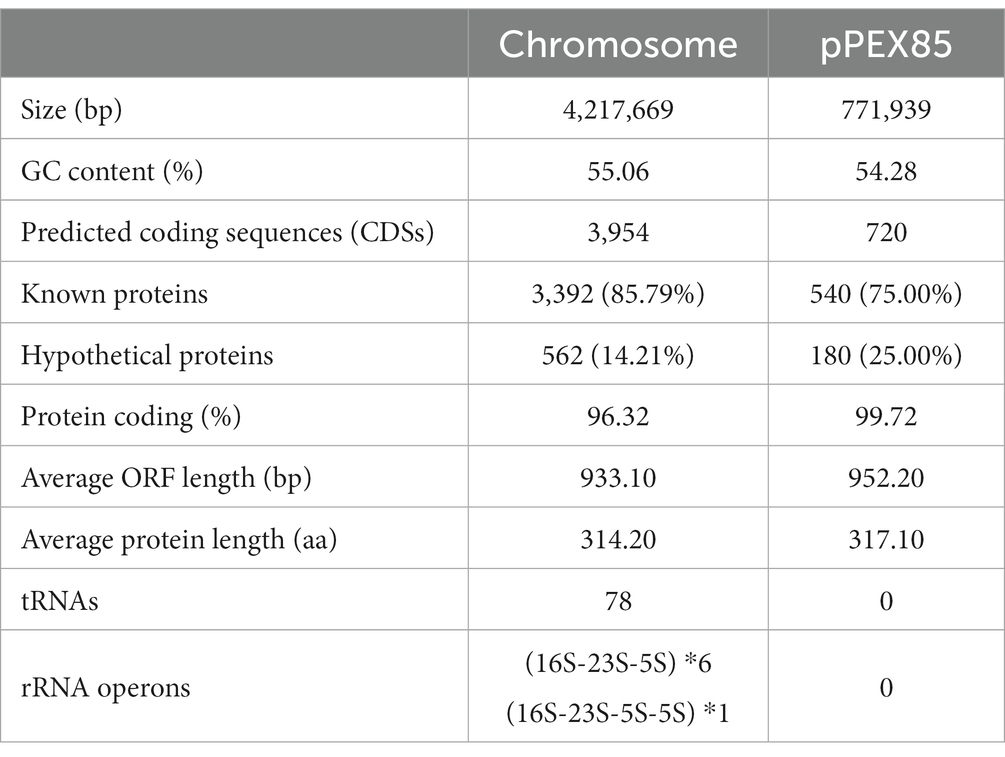
The complete genome of P. endophytica X85 consisted of a chromosome and a plasmid named pPEX85. The size of the chromosome was approximately 4.22 Mb and encoded approximately 4,954 ORFs with an average GC content of 55.06%. The length of the plasmid was 771,939 bp, encoding approximately 720 ORFs, of which 540 (75.0%) were predicted to encode function-known proteins (Table 4).
The resistance profile of Pantoea endophytica X85
Of the 17 antimicrobials tested, including 13 β-lactams and 4 aminoglycosides, the wild-type P. endophytica X85 exhibited the highest MIC level for amoxicillin (> 2048 μg/mL) and higher MIC levels for penicillin G (256 μg/mL), cefoxitin (64 μg/mL), ampicillin (64 μg/mL), cefazolin (32 μg/mL), and cephalothin (32 μg/mL) (Table 2). When analyzing the relationship between the drug resistance phenotype and genotype, especially for β-lactam antibiotics, we found that there was no functionally characterized β-lactam resistance gene annotated from the whole genome sequence. However, it had six predicted β-lactamase genes annotated in the genome, of which one gene showed the highest amino acid similarity of 75.13% (a coverage of 93.0% and an identity of 80.79%) with the function-characterized β-lactamase gene blaERH-1, which is described to confer resistance to some penicillin and cephalosporin antibiotics (Naas et al., 2004; Supplementary Table S2). The blaERH-1 homologous gene of this research was finally designated blaPSZ-1.
Resistance function characterization of the novel β-lactam resistance gene blaPSZ-1
To verify the resistance function of the gene, we cloned the ORF of blaPSZ-1 and its promoter region into the clone vector pUCP24, and the recombinant plasmid was transformed into E. coli DH5α competent cells. The transformant (pUCP24-blaPSZ-1/DH5α) conferred resistance to ampicillin, penicillin G, amoxicillin, cefazolin, cephalothin, cefoxitin, ceftazidime, cefotaxime, ceftriaxone, and aztreonam but not meropenem (Table 2). Compared with the control strain (pUCP24/DH5α), the MIC levels of the recombinant harboring blaPSZ-1 increased more than 8-fold for most β-lactam antibiotics, especially cephalothin (>256-fold), amoxicillin (≥256-fold), penicillin G (128-fold), ceftazidime (64-fold), aztreonam (64-fold), ampicillin (32-fold), cefazolin (32-fold), and ceftriaxone (32-fold) (Table 2). However, the MIC level of carbapenem meropenem was not different for the recombinant strain compared to the control strain. Tazobactam, a classical class A β-lactamase inhibitor, had a poor inhibitory effect on the resistance activity of blaPSZ-1, whereas the MIC level of blaPSZ-1 against aztreonam was significantly reduced in the presence of avibactam. When comparing the antimicrobial resistance spectrum of blaERH-1 (ERH-1 shared the highest amino acid sequence similarity with PSZ-1) and blaPSZ-1, blaERH-1 did not show resistance to the second-generation cephalosporin cefoxitin, whereas blaPSZ-1 did. However, blaERH-1 demonstrated resistance to carbapenems (Naas et al., 2004) but blaPSZ-1 did not.
Kinetic parameters of PSZ-1
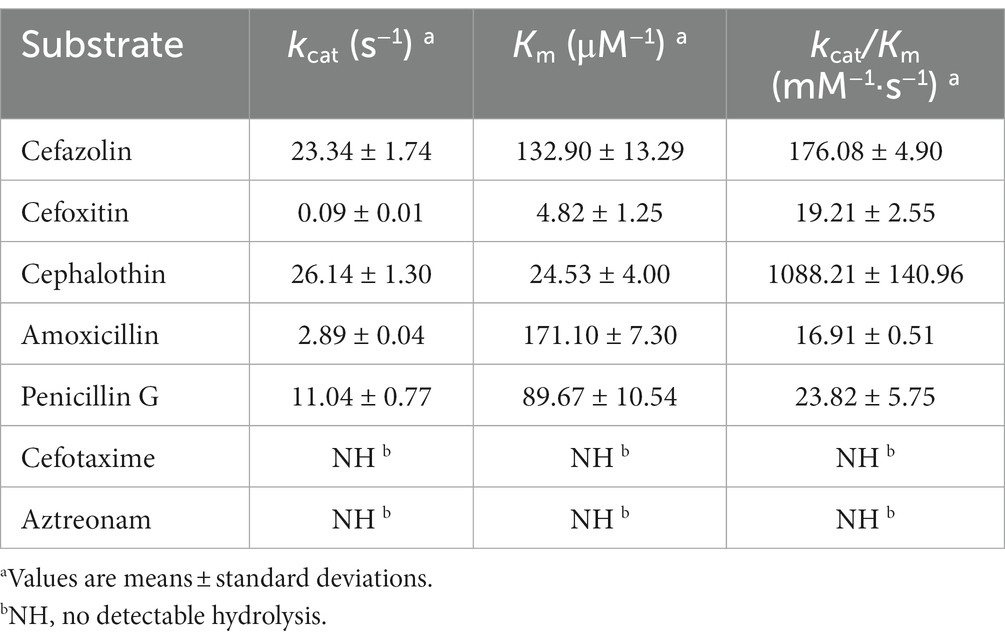
The length of the blaPSZ-1 gene was 1,086 bp, and it encoded an AmpC β-lactamase of 361 amino acids. The predicted molecular weight of mature β-lactamase PSZ-1 was 39.68 kDa, and the pI was 9.14. The kinetic parameters of the purified PSZ-1 demonstrated different degrees of hydrolytic activities against penicillins and narrow-spectrum cephalosporins. Among them, the strongest hydrolytic activity (kcat/Km 1,088.21 ± 140.96 mM−1·s−1) was observed for cephalothin. PSZ-1 showed moderate hydrolytic activities for penicillin G (kcat/Km 176.08 ± 4.90 mM−1·s−1) and the first-generation cephalosporin cefazolin (kcat/Km 123.82 ± 5.75 mM−1·s−1). However, PSZ-1 showed almost no hydrolytic activity against the third-generation cephalosporin cefotaxime or monoamide ring beta-lactam class antibiotic aztreonam (Table 5). Notably, the kinetic hydrolytic activity results of the enzyme were not completely consistent with the MIC levels of the recombinant (pUCP24-blaPSZ-1/DH5α) compared with the control strain. The hydrolytic activity of the PSZ-1 β-lactamase for cefotaxime or aztreonam was not detectable, which contrasted with the increased MIC levels (increased MIC levels of 16-and 64-fold, respectively) of the recombinant with cloned blaPSZ-1 to the two antimicrobials. This discrepancy between in vivo and in vitro results might be attributed to the fact that, despite several attempts, for weak substrates (e.g., cefotaxime or aztreonam), β-lactamase production may be excessively induced in vivo (even up to 260-fold), whereas in vitro experiments were unable to enrich a sufficient amount of β-lactamase production for us to observe the detectable hydrolysis of the enzyme against the antibiotics (Lakaye et al., 1999). A similar phenomenon was reported in a study on the AmpC enzyme CDA-1 (Ammenouche et al., 2014). When comparing the hydrolytic activity of PSZ-1 with CDA-1, which is another AmpC β-lactamase that shares an amino acid similarity of 65.65% with PSZ-1, the highest among the β-lactamases that had the kinetic parameters characterized, PSZ-1 showed lower hydrolytic activity than CDA-1 toward cefoxitin (kcat/Km of 19.21 vs. 840 mM−1·s−1) and cephalothin (kcat/Km of 1,088.21 vs. 14,500 mM−1·s−1). In accordance with the kinetic parameters, the MIC result of the recombinant strain with the cloned blaPSZ-1 against cefoxitin was also significantly lower than that of blaCDA-1 (increased 8-fold vs. >256-fold). However, the MIC values of blaPSZ-1 and blaCDA-1 to cephalothin did not seem to vary much, and both of them increased >256-fold to cephalothin compared to the controls.
When analyzing the inhibitory effect of the β-lactamase inhibitors on PSZ-1, avibactam had a strong inhibitory effect on PSZ-1 (IC50: 0.0555 μM), whereas tazobactam had a weaker inhibitory effect (IC50: 7.96 μM) when the concentration of PSZ-1 was 0.00214 μM and the nitrocefin substrate concentration was 100 μM. This result is consistent with the properties of AmpC β-lactamases for inhibitors.
Evolution and structure analysis of PSZ-1 and blaPSZ-1 related sequences
The evolutionary relationship analysis revealed that PSZ-1 formed a new branch in the phylogenetic tree of the function-characterized AmpC β-lactamases (Figure 1). Sequence comparison analysis between PSZ-1 and these function-characterized β-lactamases in the Beta-Lactamase DataBase and UniProt/Swiss-Prot database together revealed that PSZ-1 shared higher amino acid sequence similarities with ERH-1 (75.13%), CDA-1 (65.65%), EC-152 (65.65%), EC-2045 (65.37%), EC-96 (65.10%), EC-329 (65.10%), EC-106 (64.82%), and EC-P49 (64.82%; Figure 2). Within the deduced amino acid sequence of the protein, a serine-valine-serine-lysine tetrad (S-V-S-K) with the conserved and characteristic serine and lysine amino acid residues of β-lactamases possessing a serine active site was found at positions 65 to 68. Additionally, three motifs characteristic of class C β-lactamases (cephalosporinases) were also found: YAN (tryptophan-alanine-asparagine) at positions 151 to 153, DAEX (aspartic acid-alanine-glutamic acid-xaa) at positions 218 to 221, and KTG (lysine-threonine-glycine) at positions 315 to 317 (Joris et al., 1988; Mack et al., 2020; Figure 2).
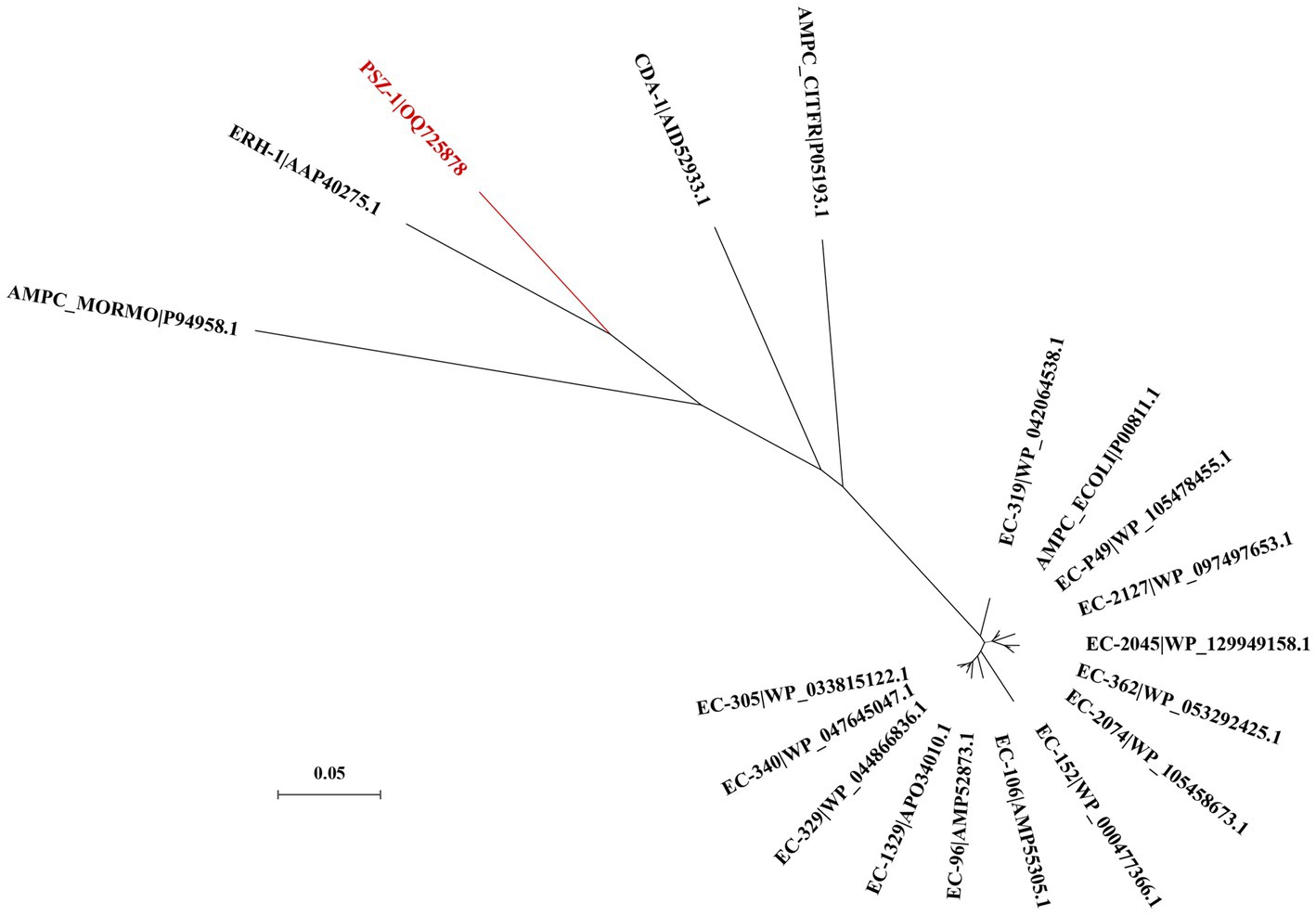
Figure 1. A phylogenetic tree showing the relationship of PSZ-1 (OQ725878) with other functionally characterized β-lactamases. PSZ-1 is highlighted in red. Other β-lactamases include ERH-1 (AAP40275.1), AMPC_ECOLI (P00811.1), EC-P49 (WP_105478455.1), EC-152 (WP_000477366.1), AMPC_MORMO (P94958.1), EC-2045 (WP_129949158.1), EC-96 (AMP52873.1), EC-329 (WP_044866836.1), EC-106 (AMP55305.1), AMPC_CITFR (P05193.1), and CDA-1 (AID52933.1).
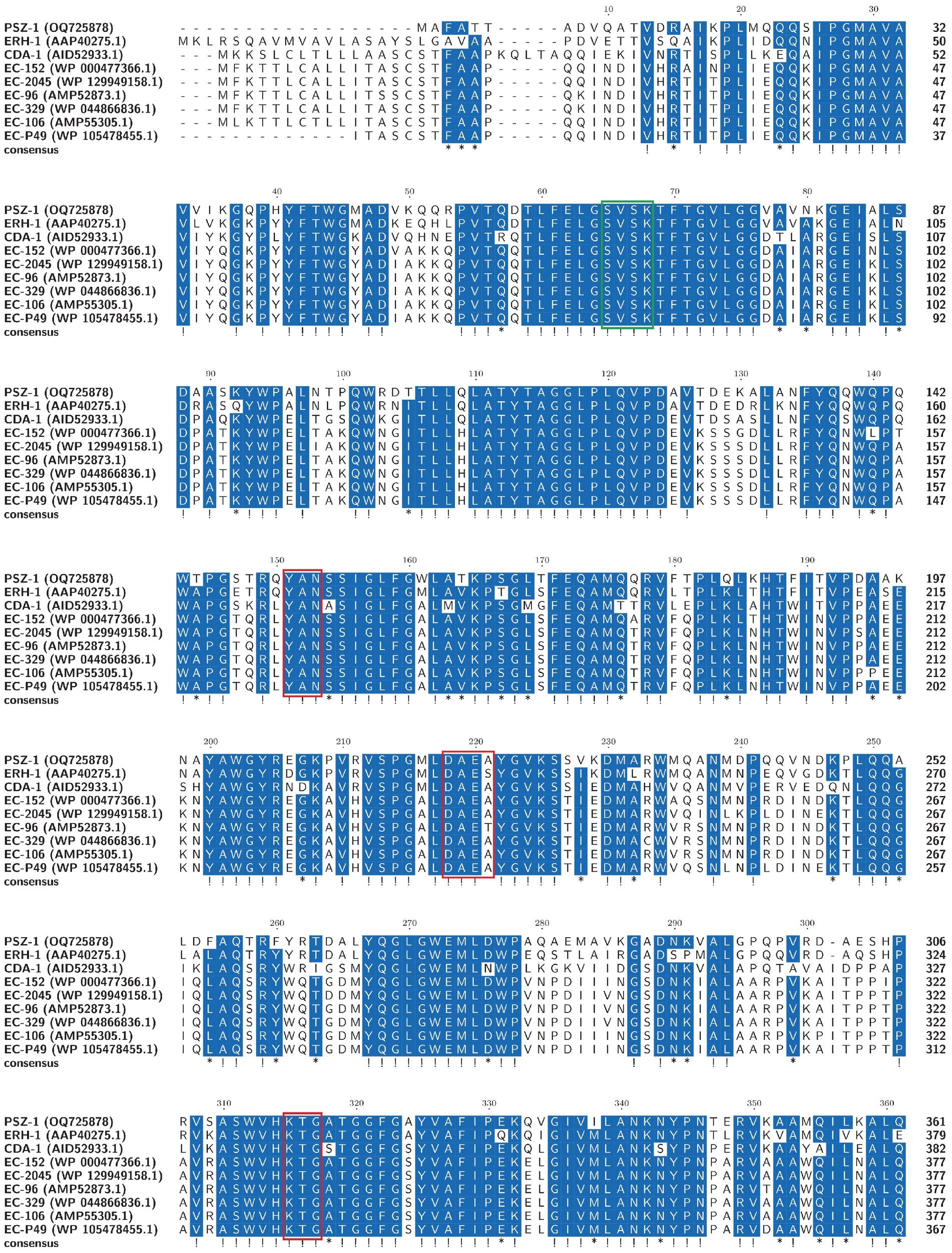
Figure 2. Multiple alignments of the deduced amino acid sequences of PSZ-1 and its close relatives. Exclamations indicate fully conserved residues, asterisks indicate strong similar residues, and gaps are represented using hyphens. The numbers on the right represent the corresponding sequence length. Three conserved motifs of class C β-lactamases are boxed in red. The β-lactamase characteristic serine active site is boxed in green.
To analyze the structure of the blaPSZ-1-related sequences, comparative analyses of the blaPSZ-1-encoding plasmid and the gene context were carried out. We found that five sequences sharing ≥45.0% similarities with P. endophytica X85 plasmid (CP121109) were present in the NCBI nucleotide database, all of which were complete plasmids from the genus Pantoea. Among them, the plasmid from Pantoea sp. SOD02 (CP102605, 926,844 bp in length approximately 180 kb larger than pPEX85) shared significantly higher similarities, of 62.20%, with pPEX85, while the other four, namely, the plasmid from Pantoea dispersa YSD_J2 (CP074351.1, 710,238 bp in length nearly the same size as pPEX85), the plasmid from P. dispersa Lsch (CP082347.1, 689,940 bp in length approximately 80 kb smaller than pPEX85), the unnamed plasmid from P. dispersa AHKW2b (CP082342.1, 653,898 bp in length approximately 120 kb smaller than pPEX85), and the plasmid from Pantoea sp. SO10 (NZ_CP040096, 744,154 bp in length, nearly the same size as pPEX85), shared similar sequence similarities of, respectively, 49.80, 49.70, 49.50, and 48.30% with pPEX85 (Figure 3). These sequences did not contain the blaPSZ-1 gene, except for Pantoea sp. SO10 and Pantoea sp. SOD02.
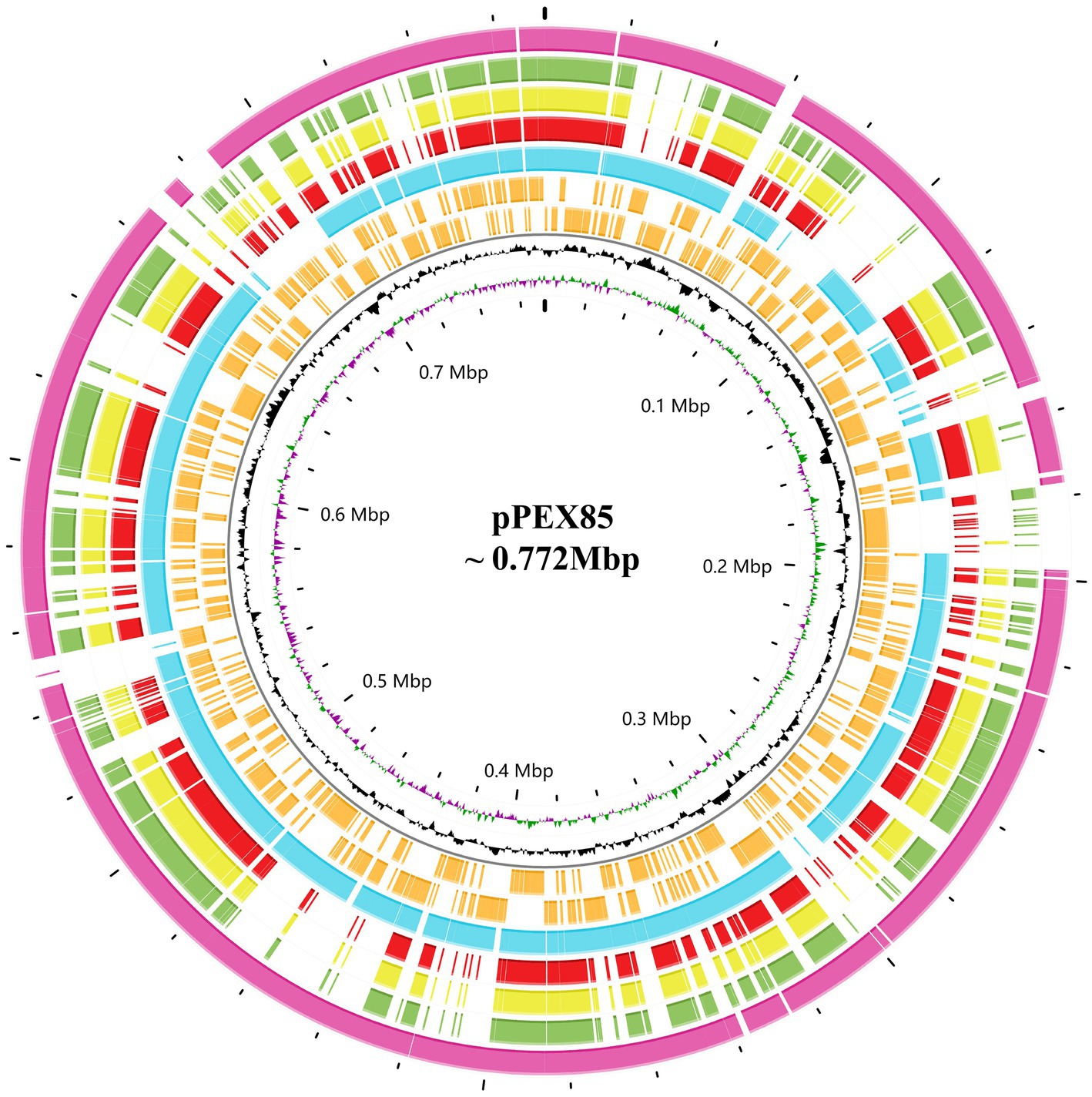
Figure 3. Genomic comparison of P. endophytica X85 plasmid with its close relatives. From outside to inside. Circles 1 to 5 are homologous regions of Pantoea sp. SOD02 plasmid (CP102605), P. dispersa YSD_J2 plasmid (CP074351.1), Pantoea dispersa Lsch plasmid (CP082347.1), unnamed P. dispersa AHKW2b plasmid (CP082342.1), and Pantoea sp. SO10 plasmid (NZ_CP040096). They are compared to P. endophytica X85 plasmid with unmatched regions left blank. Circles 6 and 7 are the genes encoded in the forward and reverse strands, respectively, circles 8 and 9 represent the GC content and GC skew, respectively, and circle 10 shows the scale in Mb.
To analyze the genetic environment of blaPSZ-1, we first searched the NCBI nucleotide database with the blaPSZ-1 gene as a query and collected sequences carrying a blaPSZ-1-like gene that shared a nucleotide sequence similarity higher than 75.0% with blaPSZ-1. Among them, only the sequences longer than 20 kb with a blaPSZ-1-like gene at the center were kept, and finally, a total of four sequences were left for further analysis. When the five 20 kb sequences (including one of the present study) were analyzed, it was found that two of them shared >75.0% nucleotide sequence similarities with the 20 kb sequence of P. endophytica X85, whereas the similarities between the sequence of P. endophytica X85 and any of the remaining two were less than 50.0%.
Among the five sequences, three (including one of this study) were from the same genus Pantoea, and only one of the three, namely, the one in the present study, has been classified as a definite species. The 20 kb sequence from Pantoea sp. SO10 plasmid (NZ_CP040096) was particularly similar (99% coverage and 97.94% identity) to that of the present study, while the 20 kb sequence from Pantoea sp. SOD02 plasmid (CP102605) showed a lower similarity (88% coverage and 87.30% identity) with the sequence in this study. The ANI between P. endophytica X85 and Pantoea sp. SO10 (GCA_005281435.1) was 98.29%, and the two might belong to the same species, whereas the ANI between P. endophytica X85 and Pantoea sp. SOD02 (GCA_024707505.1), or between Pantoea sp. SO10 (GCA_005281435.1) and Pantoea sp. SOD02 (GCA_024707505.1) was less than 95.0 (93.49 and 93.51%, respectively). This result indicated that Pantoea sp. SOD02 was not of the same species as either of the other two.
The other two 20 kb sequences were from the genus Erwinia: Erwinia rhapontici BY21311 (NZ_CP085627) and Erwinia rhapontici MAFF 311155 (NZ_AP024333), and the upstream and downstream regions of the blaPSZ-1-like genes were almost completely different from those of P. endophytica X85, except for the ampR-ampC fragment. No mobile genetic element was identified in these 20 kb sequences. The upstream regions of all five sequences, including the one from the present study, had an ampR gene that encoded an AmpR protein (Figure 4). AmpR activates AmpC by binding anhydrous forms of cell wall precursor muropeptides, which are believed to act as cofactors for AmpC induction (Girlich et al., 2000). These findings suggested that the sequences from phylogenetically closer species had higher sequence identity and were more conserved in the related species.
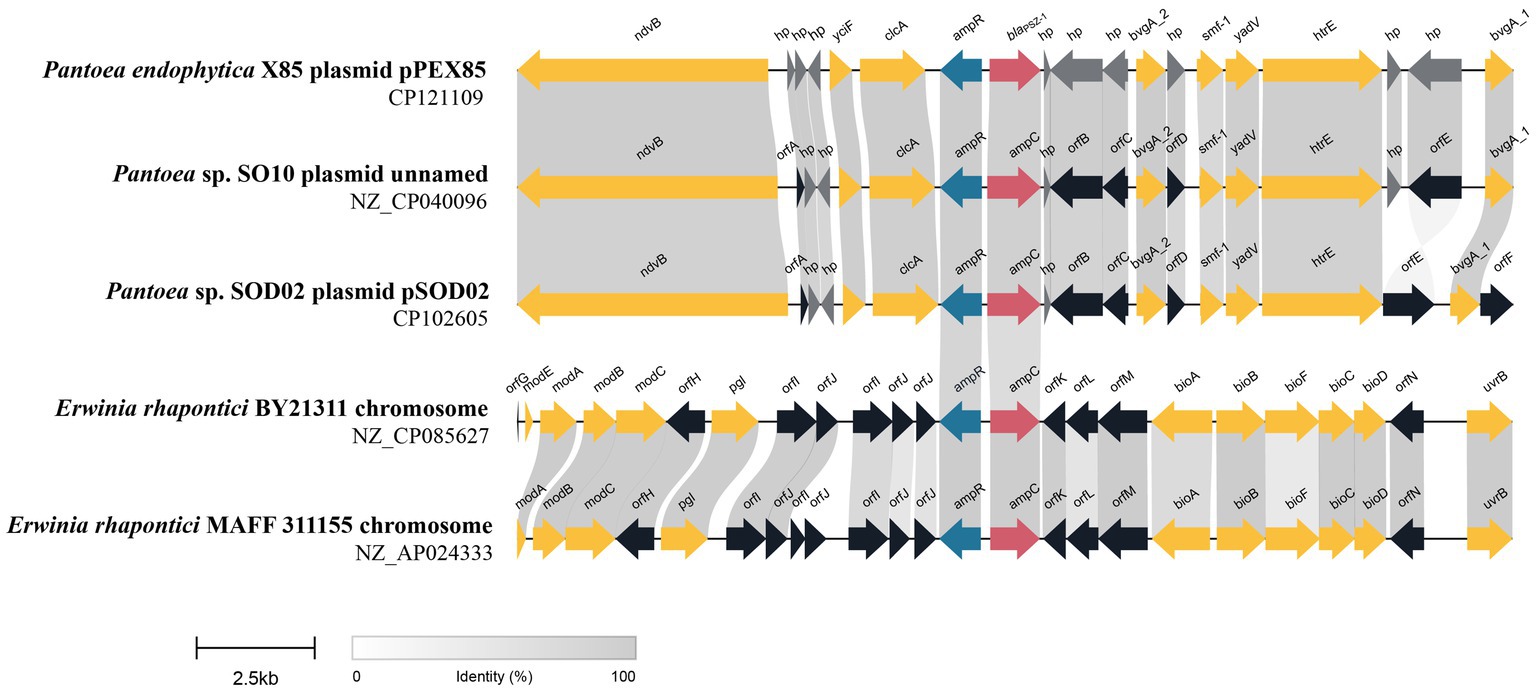
Figure 4. Genetic environment of the blaPSZ-1 and blaPSZ-1-like genes. The direction of genes is shown via an arrow. The blaPSZ-1 gene and putative ampC (blaPSZ-1-like) genes are colored red and the other genes are colored based on gene function classification. Those without direct names are depicted as orfA, YnfU family zinc-binding protein; orfB, MFS transporter; orfC, TetR/AcrR family transcriptional regulator; orfD, Hpt domain-containing protein; orfE, fimbrial protein; orfF, transporter substrate-binding domain-containing protein; orfG, molybdenum-dependent transcriptional regulator; orfH, pyridoxal phosphatase; orfI, DUF3289 family protein; orfJ, DUF943 family protein; orfK, kinase inhibitor; orfL, type 1 glutamine amidotransferase domain-containing protein; orfM, NADP-dependent oxidoreductase; and orfN, ATP-binding cassette domain-containing protein. The predicted hypothetical proteins (hp) are in dark gray. Regions with ≥80% amino acid identities are colored light gray.
Conclusion
In this study, we described a novel AmpC β-lactamase gene, designated blaPSZ-1, from an animal isolate P. endophytica X85, which shares the highest amino acid similarity (75.13%) with the function-characterized AmpC enzyme ERH-1; it shows resistance to penicillins and narrow-spectrum cephalosporins. The enzyme PSZ-1 also demonstrated hydrolytic activities against these antimicrobials, and the hydrolytic activity was strongly inhibited by the β-lactamase inhibitor avibactam. Research on the discovery of novel drug-resistance genes and their resistance mechanisms can help guide the scientific use of drugs in animal husbandry and animal/human clinical practice.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
This study used strains obtained from an anal swab of a rabbit in an animal farm in Wenzhou, China. The owner of the farm was written informed of the study and expressed approval for sampling of animals. The studies involving human participants and animals were reviewed and approved by the Animal Welfare and Ethics Committee of Wenzhou Medical University, Zhejiang Province, China (Protocol number: wydw2021-0323).
Author contributions
KL, HZ, QB, and LL: conceived and designed the experiments. JZ, YZ, YS, NL, GZ, JL, and TZ: performed the experiments. JZ, XZ, QL, and XL: data analysis and interpretation. JZ, QB, and LL: drafting of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Science & Technology Project of Wenzhou City, China (N20210001), the Science & Technology Project of Jinhua City, China (2022-2-013, 2022-4-017), and Zhejiang Provincial Natural Science Foundation of China (LY19C060002 and LQ17H190001).
Acknowledgments
The authors would like to acknowledge all study participants and individuals who contributed to this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1222703/full#supplementary-material
Footnotes
References
Ammenouche, N., Dupont, H., and Mammeri, H. (2014). Characterization of a novel AmpC β-lactamase produced by a carbapenem-resistant Cedecea davisae clinical isolate. Antimicrob. Agents Chemother. 58, 6942–6945. doi: 10.1128/AAC.03237-14
Bodenhofer, U., Bonatesta, E., Horejš-Kainrath, C., and Hochreiter, S. (2015). Msa: an R package for multiple sequence alignment. Bioinformatics 31, 3997–3999. doi: 10.1093/bioinformatics/btv494
Buchfink, B., Reuter, K., and Drost, H.-G. (2021). Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368. doi: 10.1038/s41592-021-01101-x
Bush, K. (2018). Past and present perspectives on β-lactamases. Antimicrob. Agents Chemother. 62, e01076–e01018. doi: 10.1128/AAC.01076-18
Bush, K., and Jacoby, G. A. (2010). Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 54, 969–976. doi: 10.1128/AAC.01009-09
Bush, K., Jacoby, G. A., and Medeiros, A. A. (1995). A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39, 1211–1233. doi: 10.1128/AAC.39.6.1211
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M. K., Golec, M., Skórska, C., Góra-Florek, A., et al. (2016). Pantoea agglomerans: a mysterious bacterium of evil and good. Part II. Deleterious effects: dust-borne endotoxins and allergens – focus on grain dust, other agricultural dusts and wood dust. Ann. Agric. Environ. Med. 23, 6–29. doi: 10.5604/12321966.1196848
Fullerton, D. G., Lwin, A. A., and Lal, S. (2007). Pantoea agglomerans liver abscess presenting with a painful thigh. Eur. J. Gastroenterol. Hepatol. 19, 433–435. doi: 10.1097/MEG.0b013e3280ad4414
Gilchrist, C. L. M., and Chooi, Y.-H. (2021). Clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics 37, 2473–2475. doi: 10.1093/bioinformatics/btab007
Girlich, D., Naas, T., Bellais, S., Poirel, L., Karim, A., and Nordmann, P. (2000). Biochemical-genetic characterization and regulation of expression of an ACC-1-like chromosome-borne cephalosporinase from Hafnia alvei. Antimicrob. Agents Chemother. 44, 1470–1478. doi: 10.1128/AAC.44.6.1470-1478.2000
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Habsah, H., Zeehaida, M., Rostenberghe, H. V., Noraida, R., Pauzi, W. I. W., Fatimah, I., et al. (2005). An outbreak of Pantoea spp. in a neonatal intensive care unit secondary to contaminated parenteral nutrition. J. Hosp. Infect. 61, 213–218. doi: 10.1016/j.jhin.2005.01.004
Jacoby, G. A. (2009). AmpC β-lactamases. Clin. Microbiol. Rev. 22, 161–182. doi: 10.1128/CMR.00036-08
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Joris, B., Ghuysen, J. M., Dive, G., Renard, A., Dideberg, O., Charlier, P., et al. (1988). The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 dd-peptidase family. Biochem. J. 250, 313–324. doi: 10.1042/bj2500313
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lakaye, B., Dubus, A., Lepage, S., Groslambert, S., and Frère, J.-M. (1999). When drug inactivation renders the target irrelevant to antibiotic resistance: a case story with β-lactams. Mol. Microbiol. 31, 89–101. doi: 10.1046/j.1365-2958.1999.01150.x
Letunic, I., and Bork, P. (2007). Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Mack, A. R., Barnes, M. D., Taracila, M. A., Hujer, A. M., Hujer, K. M., Cabot, G., et al. (2020). A standard numbering scheme for class C β-lactamases. Antimicrob. Agents Chemother. 64, e01841–e01819. doi: 10.1128/AAC.01841-19
McArthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. doi: 10.1128/AAC.00419-13
Meier-Kolthoff, J. P., Carbasse, J. S., Peinado-Olarte, R. L., and Göker, M. (2022). TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 50, D801–D807. doi: 10.1093/nar/gkab902
Munita, J. M., and Arias, C. A. (2016). Mechanisms of antibiotic resistance. Microbiol. Spectr. 4, 1–24. doi: 10.1128/microbiolspec.VMBF-0016-2015
Naas, T., Aubert, D., Vimont, S., and Nordmann, P. (2004). Identification of a chromosome-borne class C β-lactamase from Erwinia rhapontici. J. Antimicrob. Chemother. 54, 932–935. doi: 10.1093/jac/dkh446
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Stothard, P., Grant, J. R., and Van Domselaar, G. (2019). Visualizing and comparing circular genomes using the CGView family of tools. Brief. Bioinform. 20, 1576–1582. doi: 10.1093/bib/bbx081
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Walterson, A. M., and Stavrinides, J. (2015). Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 39, 968–984. doi: 10.1093/femsre/fuv027
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Keywords: Pantoea , resistance mechanism, β-lactamase gene, blaPSZ-1 , kinetic parameter
Citation: Zhao J, Zhang Y, Sha Y, Lin N, Zhang G, Lu J, Zhu T, Zhang X, Li Q, Zhang H, Lin X, Li K, Bao Q and Lin L (2023) BlaPSZ-1, a novel AmpC gene identified from a Pantoea isolate. Front. Microbiol. 14:1222703. doi: 10.3389/fmicb.2023.1222703
Edited by:
Biao Tang, Zhejiang Academy of Agricultural Sciences, ChinaReviewed by:
Nomeda Kuisiene, Vilnius University, LithuaniaNikolaos Strepis, Erasmus Medical Center, Netherlands
Copyright © 2023 Zhao, Zhang, Sha, Lin, Zhang, Lu, Zhu, Zhang, Li, Zhang, Lin, Li, Bao and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hailin Zhang, zhlwz97@hotmail.com; Qiyu Bao, baoqy@genomics.cn; Li Lin, linli97@126.com
 Jingxuan Zhao
Jingxuan Zhao Yuan Zhang
Yuan Zhang Yuning Sha
Yuning Sha Naru Lin2,3
Naru Lin2,3 Guozhi Zhang
Guozhi Zhang Junwan Lu
Junwan Lu Tingting Zhu
Tingting Zhu Xueya Zhang
Xueya Zhang Qiaoling Li
Qiaoling Li Hailin Zhang
Hailin Zhang Xi Lin
Xi Lin Kewei Li
Kewei Li Qiyu Bao
Qiyu Bao Li Lin
Li Lin