- National Institute of Biology, Ljubljana, Slovenia
Dickeya fangzhongdai, an aggressive plant pathogen, causes symptoms on a variety of crops and ornamental plants including bleeding canker of Asian pear trees. Historical findings stress the need for a specific detection tool for D. fangzhongdai to prevent overlooking the pathogen or assigning it to general Dickeya spp. Therefore, a qualitative real-time PCR for specific detection of D. fangzhongdai has been developed and validated. The developed assay shows selectivity of 100%, diagnostic sensitivity of 76% and limit of detection with 95% confidence interval in plant matrices ranging from 311 to 2,275 cells/mL of plant extracts. The assay was successfully used in a retrospective survey of selected host plants of relevance to Europe and environmental niches relevant to D. fangzhongdai. Samples of potato tubers and plants, plants from the Malinae subtribe (apple, pear, quince, and Asian pear tree) and fresh surface water from Slovenia were analyzed. D. fangzhongdai was not detected in any plant samples, however, 12% of surface water samples were found to be positive.
1 Introduction
Dickeya fangzhongdai is one of the more recently described species of the pectinolytic genus Dickeya (Tian et al., 2016). This genus groups diverse bacterial isolates that cause soft-rot disease in a variety of plant species, including economically important crops and ornamental plants, and cause wilting, black leg, and soft-rot symptoms (Toth et al., 2011). Reports of soft-rot disease caused by the genus Dickeya have been limited to herbaceous plants, however, D. fangzhongdai was originally described as a causative agent of bleeding canker of Asian pear (Pyrus pyrifolia) in China (Tian et al., 2016). The pathogen description was later expanded to soft rot strains isolated from various plant species, prevalently in monocots (Alič et al., 2017a,2018), and isolates from surface water (Pritchard et al., 2013a; Alič et al., 2018). Since the description of the species, multiple reports of the pathogen have been made in various host plants (see Table 1), including affecting fruits of additional tree species (Jaffar et al., 2019). The species description has also been expanded to include strains isolated in the last century. Extension of the host range of Dickeya spp. to trees is a relatively new observation, however, it does not appear to be a unique characteristic of D. fangzhongdai, since it was also reported for D. dadantii (Ogoshi et al., 2019; Fujikawa et al., 2020). Both species predominantly affect fruit trees such as pear, apple, peach, and jackfruit trees (Tian et al., 2016; Ogoshi et al., 2019; Chen et al., 2020; Fujikawa et al., 2020; Choi et al., 2021). Based on the literature, the majority of the reported D. fangzhongdai strains and all tree infections have been reported in Asian countries (Table 1). There is little information regarding the economic damage and the extent of bacterial occurrence in host plants outside of Asia. The bacteria were reported as a causative agent of soft rot of orchids in commercial production in Europe (Alič et al., 2017a) and Canada (Zhou et al., 2021), which originated with material from Asia and Europe, and as the causative agent of soft rot of onions in USA (Ma et al., 2020). However, based on the outbreak reports, it has a more significant impact on agriculture in Asia. While data on losses is limited, several authors report outbreaks on various economically important plants such as orchids (Shen et al., 2019; Balamurugan et al., 2020; Wei et al., 2021; Chi et al., 2022), banana (Yang et al., 2022), onion (Tsai et al., 2019; Wei et al., 2021), jack fruit (Jaffar et al., 2019), Asian pear (Tian et al., 2016; Choi et al., 2021), and even staple food crops such as taro (Dobhal et al., 2020; Huang et al., 2021; Hugouvieux-Cotte-Pattat et al., 2022). Based on the reports, it can be surmised that D. fangzhongdai is well established in Asia.
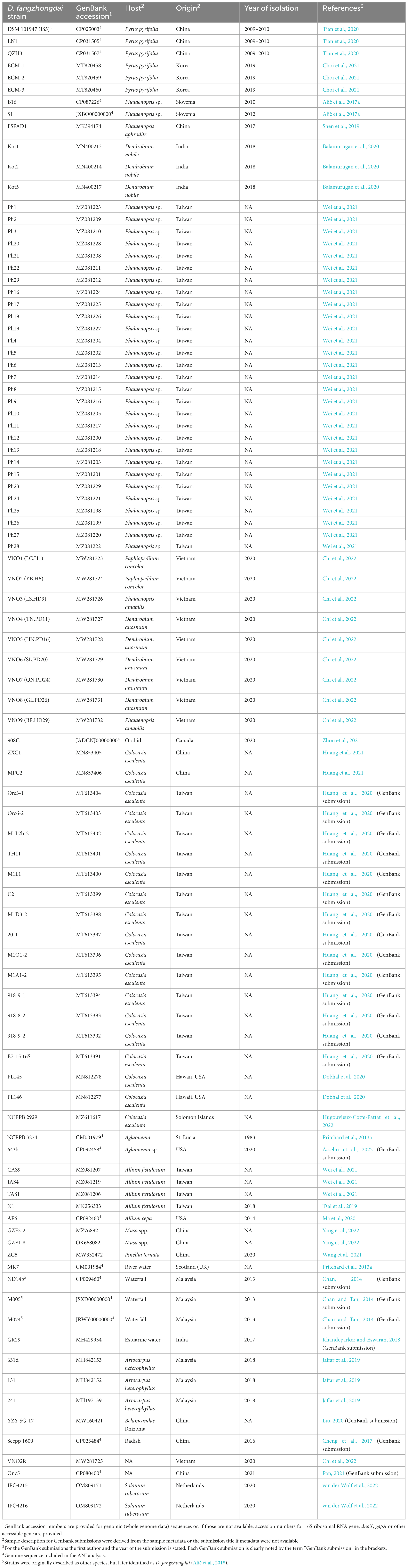
Table 1. Reports of D. fangzhongdai species in the literature to date. Most of the reported D. fangzhongdai strains were isolated in Asia.
There are currently no data on whether the strains causing bleeding canker can infect and cause soft rot symptoms of herbaceous plants in nature, however, based on inoculation experiments, pear isolates can cause symptoms on potato, tomato, cabbage, and orchids (Chen et al., 2020). The ability to persist in potato plants was also reported for the orchid pathogens D. fangzhongdai S1 and B16 (Alič et al., 2017a). In addition, D. fangzhongdai was isolated from field grown potato tubers in a 2020 study in the Netherlands. The isolates showed high virulence in a field bioassay, capable of causing blackleg to a similar extent as Pectobacterium brasiliense (van der Wolf et al., 2022). As yet, the species is not associated with diseases of food crops outside of Asia. However, the high aggressiveness of some D. fangzhongdai strains (Alič et al., 2017a; Chen et al., 2020; van der Wolf et al., 2022), the general adaptability of the genus Dickeya (Toth et al., 2011) and the plasticity of the species phenotype (Alič et al., 2018) call for caution.
Multiple D. fangzhongdai outbreaks (Tian et al., 2016; Jaffar et al., 2019; Shen et al., 2019; Tsai et al., 2019; Balamurugan et al., 2020; Ma et al., 2020; Choi et al., 2021; Zhou et al., 2021; Chi et al., 2022; Yang et al., 2022) in recent years suggest that the pathogen is present and spreading in Asia and also other continents. It was indicated before that trade plays can promote the spread of the Dickeya disease (Toth et al., 2011). Moreover, there is evidence that ornamental plants may represent potential routes for the introduction of additional Dickeya species and strains with a broad host range (Parkinson et al., 2009; Toth et al., 2011), as observed in D. solani spread to potato. Therefore, close consideration should be given to potential infection routes and the adaptability of these pathogens to other plant hosts and environments. Especially so for pathogens like D. fangzhongdai with a broad host range that predominantly consists of economically important plants with high trade rates (Hinsley et al., 2018). However, only adequately validated diagnostic tools enable timely identification of D. fangzhongdai infected plants to support prevention of its introduction and aid epidemiological investigations. Therefore, there is a great need for the development of specific and reliable diagnostic tools that enable monitoring the presence and spread of D. fangzhongdai. Two detection tests specific to D. fangzhongdai species were previously developed: a real-time PCR test described by Tian et al. (2020) specifically developed and tested on D. fangzhongdai strains from Asian pear trees, and a loop-mediated isothermal amplification (LAMP) test described by DeLude et al. (2022) that was comprehensively validated on taro, onion, and orchid matrices.
The aims of this study were to (i) develop a qualitative real-time PCR for specific detection of D. fangzhongdai in various plant matrices (and extend validation to novel matrices including potato and orchids) and ecological niches, (ii) to validate the developed test according to the guidelines of the European and Mediterranean Plant Protection Organization (EPPO) (European and Mediterranean Plant Protection organization, 2021), and compare its performance with the previously developed test described by Tian et al. (2020), and (iii) to use the newly developed test in a retrospective survey to assess the presence of these bacteria in potato plants with soft rot symptoms, and surface water in Slovenia.
2 Materials and methods
2.1 Selection of host plants and niches relevant for D. fangzhongdai survey
Based on previous experience with the occurrence of soft-rot and a literature search, an informed selection of potential target plants and environmental niches was made. The literature search was performed using Google Scholar, to find reports of novel D. fanzhongdai outbreaks and isolates. Further, the GenBank database (Sayers et al., 2022) was searched for D. fanzhongdai nucleotide sequences that have not yet been reported in any publication. Based on the search results, a table (Table 1) was created, summarizing the currently reported D. fangzhongdai strains, hosts, geographical origin, and year of isolation.
Average nucleotide identity (ANI) was calculated for all reported D. fangzhongdai isolates with publicly available genome sequences to confirm isolate identification using the Genome-based distance matrix calculator (Richter et al., 2016; Sayers et al., 2022).
2.2 Bacterial strains
Bacterial strains used in the study are shown in Table 2. Dickeya spp. were grown overnight on Casamino acid-Peptone-Glucose (CPG; Schaad et al., 2001) medium at 28°C. Bacteria of other genera were grown overnight on yeast-extract peptone glucose agar (YPGA; EU, 1993), CPG or nutrient agar (NA; Schaad et al., 2001) medium at 25°C.
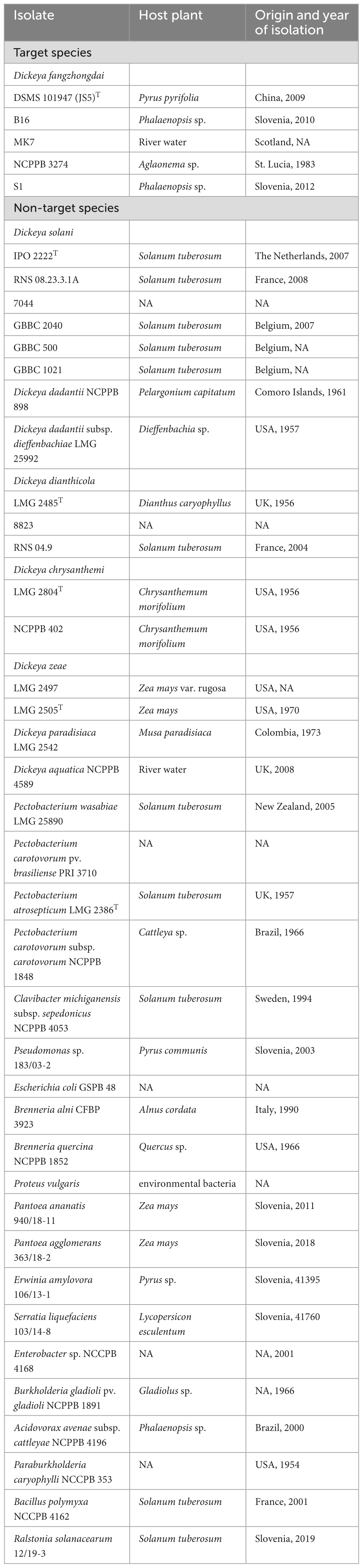
Table 2. Table of bacteria isolates used to determine analytical specificity of the developed real-time PCR test.
2.3 Samples and sample preparation
2.3.1 Analytical specificity
Suspensions of target and non-target bacteria (Table 2) were prepared from overnight cultures in 10 mM phosphate buffer (PB; 1.07 g Na2HPO4, 0.4 g NaH2PO4 × 2H2O per liter of water, pH 7.2) to an approximate concentration of 106 cells/mL (Densitometer DEN-1, Biosan). Inclusivity was determined on five D. fangzhongdai isolates from three different geographical regions (Asia, America, Europe) and 3 different host niches (herbaceous plants, trees, water). Exclusivity was determined on other Dickeya spp. (17 isolates), selected bacteria from Enterobacteriaceae family (15 strains), and bacteria colonizing the same host plant niches (5 isolates) as the target bacteria (Table 2).
2.3.2 Analytical sensitivity
A D. fangzhongdai B16 and JS5T bacterial suspension with concentration of 107 cells/mL was prepared in a 10 mM PB with 30% (V/V) glycerol. DNA was extracted and standard curves were prepared by 10-fold dilutions in TE buffer (Sigma-Aldrich, Merck, Germany) with the addition of salmon sperm DNA (25 μg/mL).
2.3.3 Diagnostic sensitivity
Plant extracts were prepared from relevant asymptomatic plants, namely orchids (genus Phalaenopsis), potato (Solanum tuberosum, cultivar Carrera), and apple (Malus domestica). For the preparation of Phalaenopsis extracts, leaf material was collected in July 2019 and surface sterilized with 70% ethanol. One gram of leaf tissue was macerated in 3.5 mL of sterile 10 mM phosphate buffered saline (10 mM PBS; 1.08 g Na2HPO4, 0.4 g NaH2PO4 × 2H2O, 8 g NaCl, 1 L distilled water, pH 7.2). The supernatant was separated from the plant tissue by pipetting. A field sample of an asymptomatic potato plant, cultivar Carrera, was collected in July 2016. The surface of the sampled plant stems was cleaned and surface sterilized with 70% ethanol. The asymptomatic potato stem was cut into smaller pieces (approximately 2 cm size) and covered with sterile 10 mM PBS buffer, vortexed, and incubated for several minutes (up to 20 min) at room temperature. The supernatant was separated from the plant tissue by pipetting. An apple extract was prepared from Malus domestica asymptomatic plant material collected in July 2018. The sample material consisted of twigs that were surface sterilized with 70% ethanol. Vascular tissue was scraped from sampled twigs and covered with sterile 10 mM PBS containing 0.1% Tween 20, vortexed, and incubated with shaking for 90 min at room temperature. The supernatant was then separated from the plant tissue and centrifuged at 1,500 g for 10 min, transferred to a new tube and centrifuged at 7,000 g for 20 min. The pellets were suspended in 2 mL of 10 mM PBS.
The health status of plant extracts was confirmed with real-time PCR analysis using generic Dickeya spp. assay (Pritchard et al., 2013b).
Surface water was sampled from the Pivka River in a western part of Slovenia in August 2017. Temperature and pH of the water at the time of sampling were 22°C and 7, respectively. One liter of water was aliquoted to 250 mL and centrifuged for 20 min at 10,000 g at 4–10°C. Pellets were resuspended in 1 mL of 10 mM PB buffer. The absence of Dickeya spp. in the surface water extract was confirmed by real-time PCR analysis using generic Dickeya spp. assay (Pritchard et al., 2013b).
Standard curves of Dickeya fangzhongdai B16 in plant extracts and surface water extract were prepared by mixing bacterial suspensions with aliquots of extracts to final concentrations ranging from 107 to 101 cells/mL of plant or surface water extract.
2.3.4 Retrospective survey
A retrospective survey was performed on the collection of DNA extracts from sample material selected as described in Section “2.1 Selection of host plants and niches relevant for D. fangzhongdai survey.” Potato samples, Malinae samples, and surface water samples previously obtained in diagnostic activity in the years 2017–2021 were included in the survey. Altogether, 278 plant samples were analyzed, consisting of 130 potato samples, 148 Malinae samples and 53 surface water samples.
2.3.4.1 Samples of potato plants and tubers
Samples of potato plants and tubers with soft rot symptoms were analyzed. The surface of the sampled plants was cleaned, and surface sterilized with 70% ethanol. Symptomatic material was covered with sterile 10 mM PBS buffer, vortexed, and incubated for several minutes (up to 20 min) at room temperature. The supernatant was separated from plant tissue. DNA was extracted as described in Section “2.3 Samples and sample preparation.” Extracted DNA was stored below −15°C until analysis. Potato samples comprised of potato plants with soft rot symptoms (119 samples) and potato tubers with soft rot (11 samples).
2.3.4.2 Malinae samples
Tree samples (from the Malinae subtribe) were surface sterilized with 70% ethanol. Vascular tissue was scraped from sampled twigs and covered with sterile 10 mM PBS containing 0.1% Tween 20, vortexed, and incubated for 90 min at room temperature. The supernatant was then separated from the plant tissue and centrifuged at 1,500 g for 10 min, transferred to a new tube and centrifuged at 7,000 g for 20 min. The pellets were suspended in 2 mL of 10 mM PBS. DNA was extracted as described in Section “2.4 DNA extraction and purification.” Extracted DNA was stored below −15°C until analysis. The Malinae samples were included apple samples (84 samples), pear samples (8 samples) and Asian pear sample (1 sample) with fire blight symptoms, and mixed Malinae samples (55 samples) sampled for latent testing to fire blight.
2.3.4.3 Surface water samples
Fifty diagnostic samples of surface water were collected from different freshwater. All samples were collected in summer, and the temperature and pH of the water at the sampling site were recorded (Supplementary Table 1). Samples were analyzed as follows: one liter of water was aliquoted to 250 mL and centrifuged for 20 min at 10,000 g at 4–10°C. Pellets were resuspended in collectively 1 mL of 10 mM PB buffer. DNA was extracted as described in Section “2.3 Samples and sample preparation.” Extracted DNA was stored below −15°C until analysis.
2.4 DNA extraction and purification
DNA was extracted from 100 μL aliquots of pure bacterial suspensions, spiked plant extracts, spiked surface water, field plant samples, and surface water samples using magnetic beads-based DNA extraction on QuickPick SML Plant DNA kits (BioNobile, Finland), according to Pirc et al. (2009), with the minor modification of using 440 μL lysate in the downstream purification.
DNA used for analytical specificity was extracted from 500 μL of pure bacterial suspension in PB buffer using heat lysis. Samples were incubated at 95°C for 10 min in a thermoblock, and then immediately put on ice for 3 min. After centrifugation for 1 min at 6,000 rpm supernatant was collected.
2.5 Real-time PCR assay design
A D. fangzhongdai specific real-time PCR assay was designed according to Alič et al. (2022). Unique diagnostic markers of D. fangzhongdai strains were identified by RUCS (Thomsen et al., 2017). A positive dataset comprised of 10 D. fangzhongdai genomic sequences was compared to a negative dataset of 39 Dickeya spp. genomic sequences, including D. solani, D. dadantii, D. dianthicola, D. chrysanthemi, D. undicola, D. aquatica, D. zeae, and D. paradisiaca (Supplementary Table 2). The complete genome sequence of D. fangzhongdai ND14b was selected as the positive reference genome.
Specificity of the identified unique sequences was confirmed by Blastn (Altschul et al., 1990) analysis against the whole GenBank database. Altogether, nine suitable unique sequences of sufficient length (above 100 bp) were identified. Primers and hydrolysis probes for real-time PCR were designed using Primer Express version 2.0 (Applied Biosystems). The quality of the designed assays was evaluated in silico by OligoAnalyzer Tool (IDT) and Blastn (Altschul et al., 1990), and experimentally.
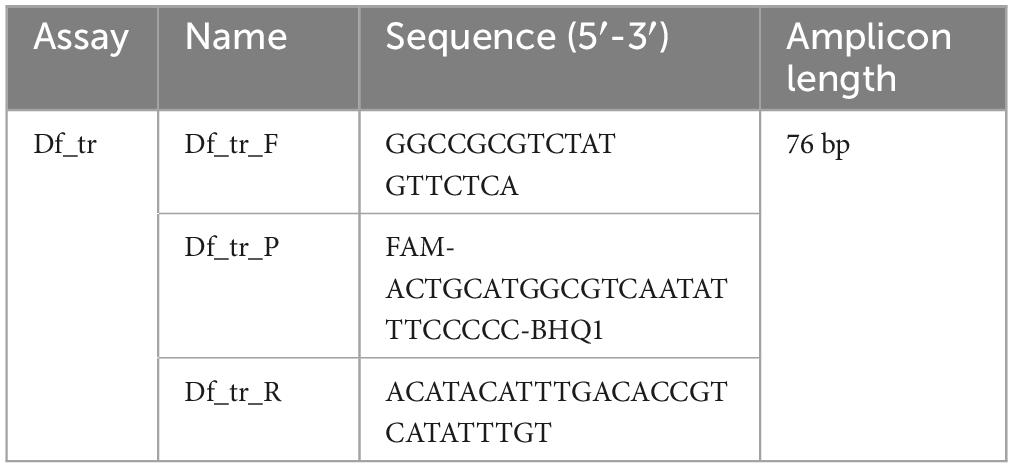
The optimal assay, assay Df_tr (Table 3; Supplementary Table 3), designed against a transcriptional regulator gene (Dickeya_fangzhongdai_ND14b.0976; GenBank locus tag LH89_04605), was selected for validation (Alič et al., 2019). Assays with poor performance or those targeting hypothetical genes or genes of extrachromosomal origin were omitted from further analysis.
2.6 Setup of the real-time PCR experiment
Real-time PCR reactions were performed on a QuantStudio 7 (Applied Biosystems, Thermo Fisher) using universal cycling conditions (2 min at 50°C, 10 min at 95°C, followed by 45 cycles of 15 s at 95°C and 1 min at 60°C, with 1.6°C/s ramping speed) according to the PCR Master Mix manufacturer’s recommendations. The reaction volume of 10 μL contained, in final concentrations: 1x TaqMan™ Universal PCR Master Mix (Applied Biosystems, Thermo Fisher), 900 nM primers (Eurofins), 200 nM probe (Eurofins), and 2 μL DNA. The QuantStudio™ real-time PCR Software 1.3 and 1.6 (Applied Biosystems, Thermo Fisher Scientific) were used for fluorescence acquisition and calculation of the threshold cycles (Cq). The baseline was set automatically, and the fluorescence threshold was set manually to intersect with the linear part of the amplification curves of all real-time PCR assays.
Analysis parameters in Df_tr validation procedure included the automatic baseline setting, and the fluorescence threshold set manually to 0.05.
Amplification of the plant endogenous sequence COX was used as an extraction and amplification control [COX; Weller et al. (2000), forward primer and probe and Mumford et al. (2004), reverse primer)]. real-time PCR assays for non-specific detection of Dickeya spp., assay ECH (Pritchard et al., 2013b) was used as a control for presence of Dickeya spp. The standard curves prepared by mixing target bacteria and plant extracts were used to determine analytical sensitivity of the novel assay and the real-time assay described by Tian et al. (2020). Fluorescence thresholds for those assays were manually set to 0.1, 0.1, and 0.06 for COX, ECH, and Df_tr assay, respectively. A reaction was interpreted as positive if it produced an amplification curve and a fluorescence signal that exceeded the threshold.
Positive amplification controls and negative amplification controls were included in every real-time PCR experiment for each assay.
2.7 Validation of D. fangzhongdai specific real-time PCR assays
2.7.1 Analytical specificity and selectivity
The analytical specificity of the real-time PCR assay was tested in silico by Blastn (Altschul et al., 1990) and experimentally by amplification of five target D. fangzhongdai strains and 37 non-target strains, including strains from eight different Dickeya genera (Table 2). Selectivity of the assay was tested on relevant plant matrixes, namely plant extracts from orchid plants, potato plants, and apple tree bark scrapings, free of disease symptoms.
2.7.2 Analytical and diagnostic sensitivity
Analytical sensitivity was determined in dilutions of DNA from pure cultures of D. fangzhongdai B16 and D. fangzhongdai JS5T, and diagnostic sensitivity was determined on standard curves of D. fangzhongdai B16 in plant extracts of Phalaenopsis, potato and apple, and surface water. Each standard curve was analyzed in triplicate. The following control systems were used to assure reliability of results and provide further information on method performance: (i) use of undiluted and diluted (1:10 in molecular grade water) DNA extracts from spiked plant extracts and surface water, and (ii) amplification of plant endogenous sequence as an extraction and amplification control (COX) (Weller et al., 2000; Mumford et al., 2004).
The limit of detection LOD95 was defined as the target amount giving positive results with 95% confidence and was calculated using drc package in R (Ritz and Strebig, 2016; R Core Team, 2021). The slope (k) of the linear regression line between logarithmic values of cell numbers (independent variable) and Cq values (dependent variable) was used to calculate the amplification efficiency, E = (10[-1/k])–1, where a value of one corresponds to 100% amplification efficiency (Pfaffl, 2001). The dynamic range, i.e., the range of concentrations for which Cq values were in linear relationship with logarithms of concentrations, was determined by visually exploring the slope across sections of the Cq values × log concentration plot.
Performance of the developed real-time PCR Df_tr assay was compared to real-time PCR assay described by Tian et al. (2020).
3 Results
3.1 Selection of host plants and niches relevant for D. fangzhongdai survey
Dickeya fangzhongdai isolates collected from reports in publications and the GenBank database are shown in Table 1. The majority of the species (87%) were reported from Asia. The reported isolates were predominantly isolated from soft rot symptoms on orchids (48%) and taro plants (20%). Six isolates (6%) were reported to be isolated from Asian pear trees, causing bleeding canker disease and 5 isolates (5%) were isolated from water sources. Overall, 80% of isolates were isolated from monocot plants, suggesting that D. fangzhongdai might have preference based on cotyledon types. Since D. fangzhongdai species description is relatively new, it is very likely that isolates found before the species description were assigned only to Dickeya spp. level, as was the case for NCPPB 3274. Therefore, the true list of D. fangzhongdai isolates is likely to be far more substantial. For example, it was indicated that Dickeya spp. isolates from several host plants described by Suharjo et al. (2014) correspond to D. fangzhongdai (Alič et al., 2017a,2018).
Dickeya fangzhongdai isolates with known whole genome sequence (Table 1) share above 96% average nucleotide identity (ANI) and above 86% coverage, regardless of the geographical origin or host.
Based on the literature search and previous experiences, the survey was focused on plants of agricultural importance (e.g., potato and members of the Malinae subtribe) and water samples. The latter give broader environment representation compared to individual plant samples.
3.2 Validation of the real-time PCR Df_tr assay
3.2.1 Analytical specificity and selectivity
The real-time PCR assay Df_tr, targeting a transcriptional regulator containing an amidase domain and an AraC-type DNA-binding HTH domain, was found to be specific for detection of D. fangzhongdai species. The assay exhibited 100% inclusivity (5/5 isolates) since all D. fangzhongdai isolates were reliably detected regardless of their host or geographical origin. Moreover, no cross reactivity with any of the tested non-target bacteria (37 isolates) was observed, meaning that the test is 100% exclusive to D. fangzhongdai.
Lastly, no non-specific amplification was obtained from healthy plant matrices, therefore selectivity of the assay was determined to be 100% on tested matrices, namely potato, orchid, and apple tree.
3.2.2 Analytical sensitivity
Analytical sensitivity was determined on DNA standard curves of two different D. fangzhongdai isolates, B16 and JS5T, from different environmental niches. The newly developed assay showed high analytical sensitivity (Table 4) with LOD95 below 104 cells/mL of plant extracts in all three matrices. The performance characteristics of the real-time PCR Df_tr assay were very similar to the real-time PCR described by Tian et al. (2020) (Supplementary Table 4). Both assays gave almost identical results tested on DNA standard curves.
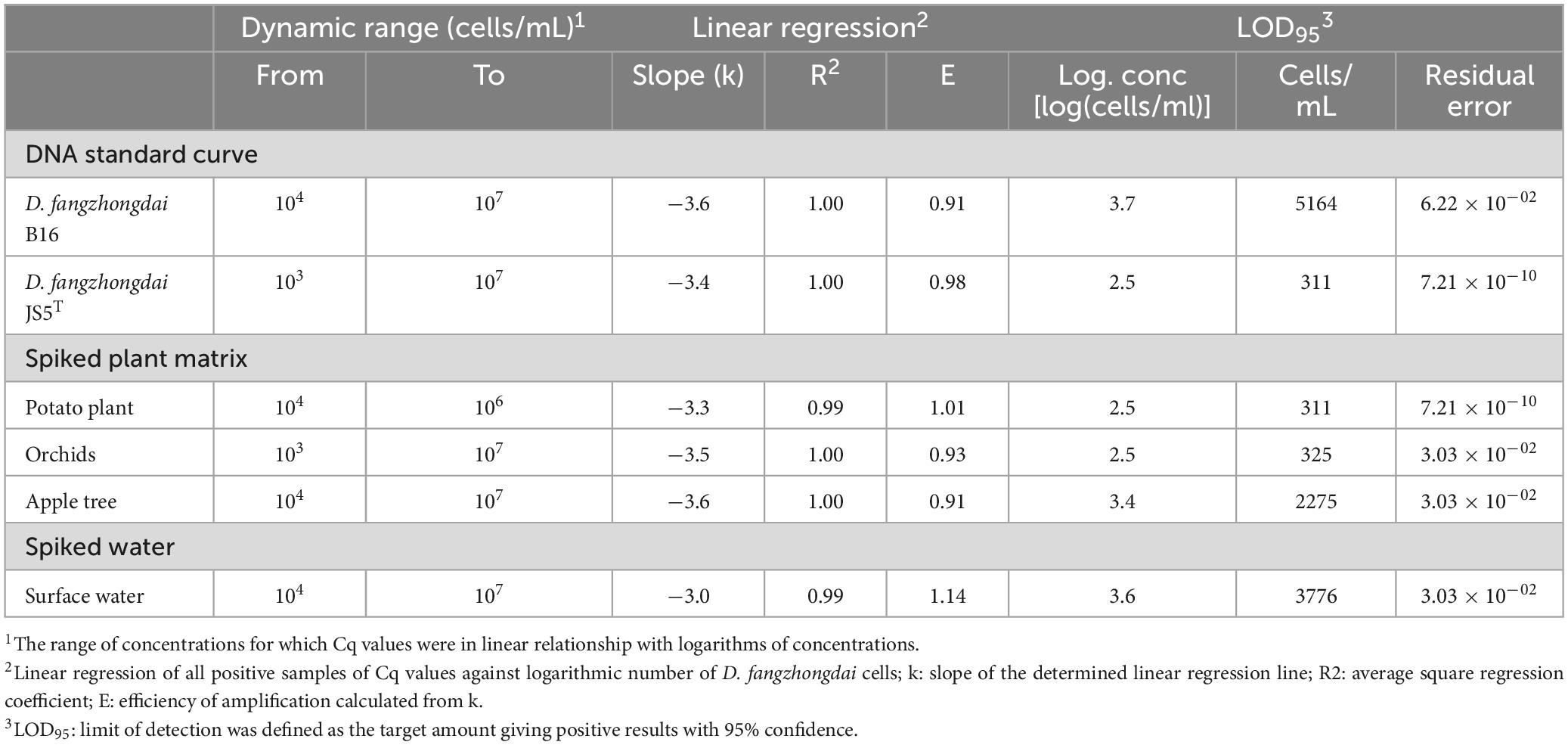
Table 4. Performance characteristics of real-time PCR Df_tr assays evaluated on bacterial suspension, spiked plant matrices and spiked surface water.
3.2.3 Diagnostic sensitivity
Diagnostic sensitivity was determined on spiked plant matrices and surface water. No inhibition of amplification could be observed in any of the matrices tested; however, the sensitivity of the assay varied from matrix to matrix, suggesting a matrix effect on DNA extraction procedure (Figure 1). Of all the plant matrices tested, sensitivity was most affected by the apple tree matrix, with an LOD95 of 2,275 cells/mL (Supplementary Figure 1) compared to potato and orchid matrices with LOD95 in the range of 311 – 325 cell/mL of sample. The diagnostic sensitivity of the assay was slightly lower in surface water than in plant matrices. The LOD95 was of 3,776 cells/mL of sample (Supplementary Figure 1). However, the performance characteristics of the matrices tested were not significantly different (Table 4). The Cq values were consistently below 37 at the detection limit where all parallel reactions were positive for all samples tested. Inter-run repeatability was high for all the samples and matrices tested, with coefficients of variation of Cq values below 7% within the dynamic range. The performance characteristics of the real-time PCR Df_tr assay were better compared to performance characteristics of the real-time PCR described by Tian et al. (2020) in all spiked matrices (Supplementary Table 4).
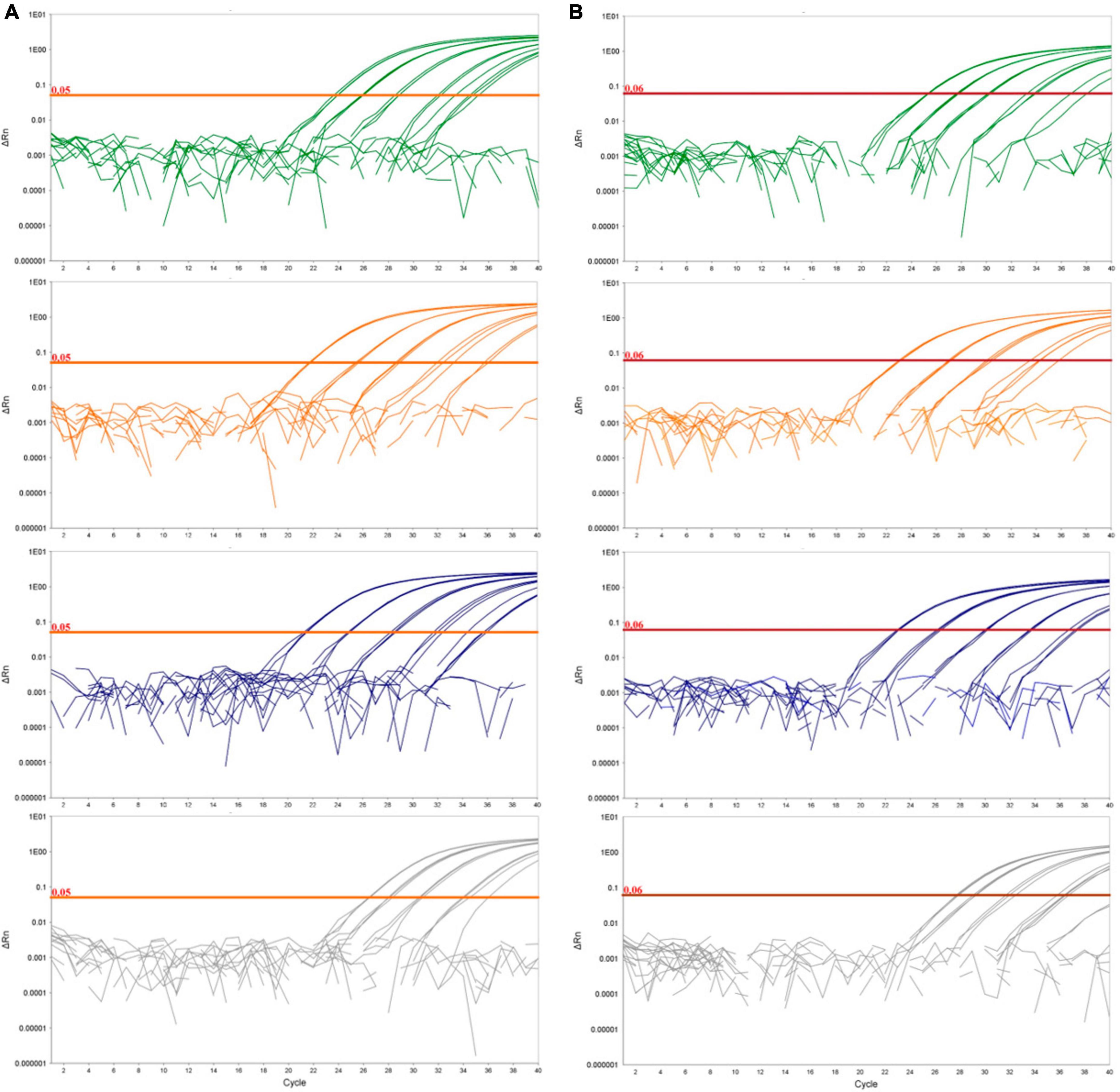
Figure 1. Logarithmic amplification curves of D. fangzhongdai DNA extracted from spiked plant matrices and spiked surface water for the novel real-time PCR assay Df_tr (A) and assay described by Tian et al. (2020) (B). The curves show bacterial standard curves prepared in potato matrix (green curves), in apple matrix (orange curves), in orchid matrix (blue curves) and in surface water (gray curves). The threshold line for the real-time PCR assay Df_tr is shown in orange and for Tian et al. (2020) in red.
The greatest difference in sensitivity between assays was observed in the plant matrices. The LOD95 of the Df_tr assay was 311 and 325 cells/mL, compared to the real-time PCR described by Tian et al. (2020), which had LOD95 of 2,275 and 2,438 cells/mL, for potato and orchid matrices, respectively. Nonetheless, the difference in sensitivity was less pronounced for the apple tree matrix (LOD95 of 2,275 for the Df_tr assay and 3,776 for the real-time PCR described by Tian et al. (2020). A similar difference in sensitivity was observed in surface water. The Df_tr assay showed higher sensitivity, with LOD95 of 3,776 cells/mL than the real-time PCR described by Tian et al. (2020) with LOD95 of 15,241 cells/mL (Figure 2). Overall, the sensitivity of Df_tr assay was better than the real-time PCR described by Tian et al. (2020) in plant matrices and surface water (Figure 2). In addition, fluorescence (ΔRn; Figure 1) was consistently higher in the Df_tr assay compared to real-time PCR described by Tian et al. (2020).
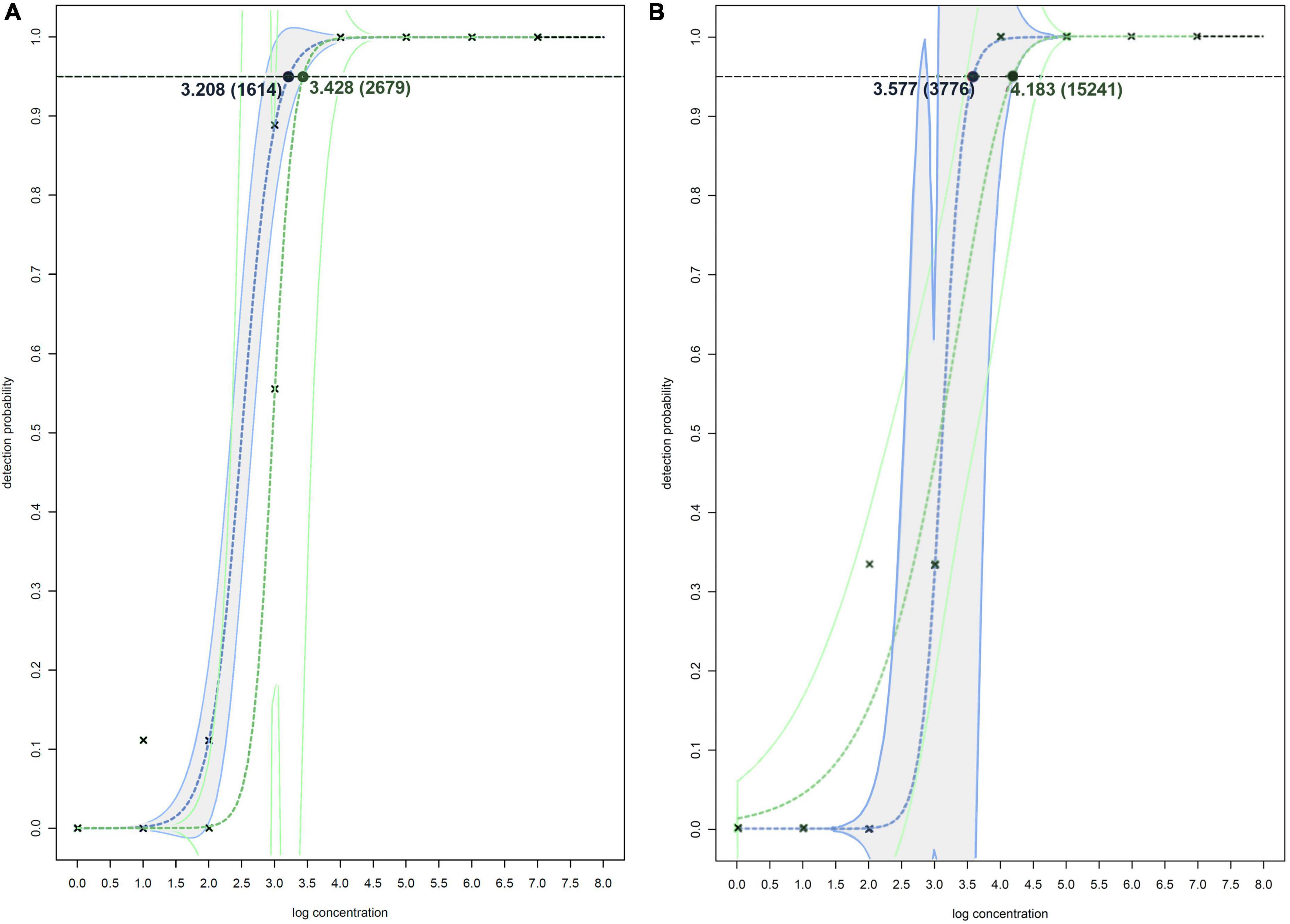
Figure 2. Non-linear modeling of probability of detection on spiked plant matrixes (A) and spiked surface water (B) for Df_tr real-time PCR assay (shown in blue and gray) and real-time PCR assay described by Tian et al. (2020) (shown in green). The concentrations shown are expressed as log(cells/mL of plant extract), and in the brackets as cells/mL of plant extracts. The model used for both assays on spiked plant matrixes (A) is two-parameter log-logistic function (LL.2), and models used on spiked surface water (B) is log-logistic function (LL.2) for Df_tr real-time PCR assay and two-parameter Weibull function (W2.2) for real-time PCR assay described by Tian et al. (2020). The dotted line denotes 95% probability of detection.
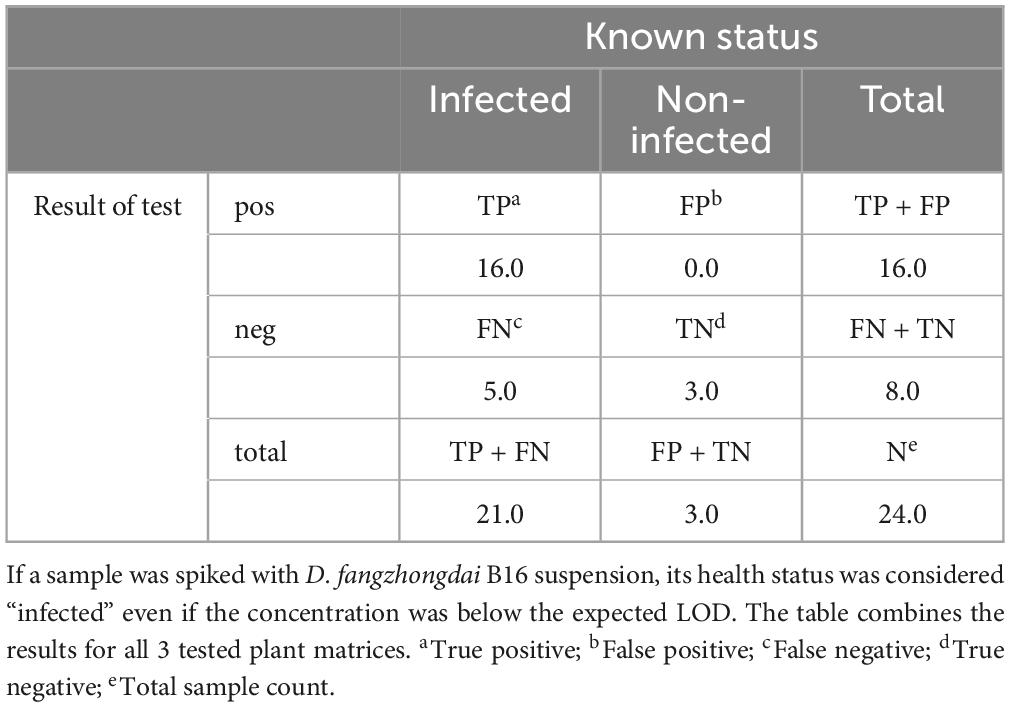
No false positives were observed for Df_tr. As expected, only samples with a bacterial concentration below LOD95 gave false negative results when compared with their known health status (Table 5). Accuracy of the test based on spiked samples was 79% and diagnostic sensitivity 76%.
Samples of potato plants and tubers, and samples of Malinae trees were tested for a general presence of Dickeya spp. and D. fangzhongdai strains. The retrospective assay did not confirm the presence of D. fangzhongdai in any of the tested plant samples. However, 12% (16/130 samples) of tested samples with soft rot symptoms were positive using a non-specific Dickeya spp. real-time assay (Pritchard et al., 2013b), indicating the presence of other Dickeya species in potato. Similarly, no D. fangzhongdai nor other Dickeya spp. were detected in any of the samples of Malinae members. The Malinae samples were collected from trees that are of economic importance in the Slovenian environment, therefore the majority of the samples represent the genera Pyrus and Malus. The general presence of Dickeya spp. was confirmed in samples of surface water using real-time PCR. Dickeya spp. were detected in 70% (35 out of 50) surface water samples in low concentrations (32 ≤ Cq ≤ 39). In 6 of these samples, we also detected D. fangzhongdai, in 4 samples from August 2018 and 2 samples from August 2021. In all samples, concentrations were relatively low (34 ≤ Cq ≤ 38; Figure 3), close to the limit of detection of the assay (LOD95 of 3,776 cells/mL of sample water extract; Figure 2). The sample was considered positive if at least one reaction produced a signal above threshold and a characteristic amplification curve was present.
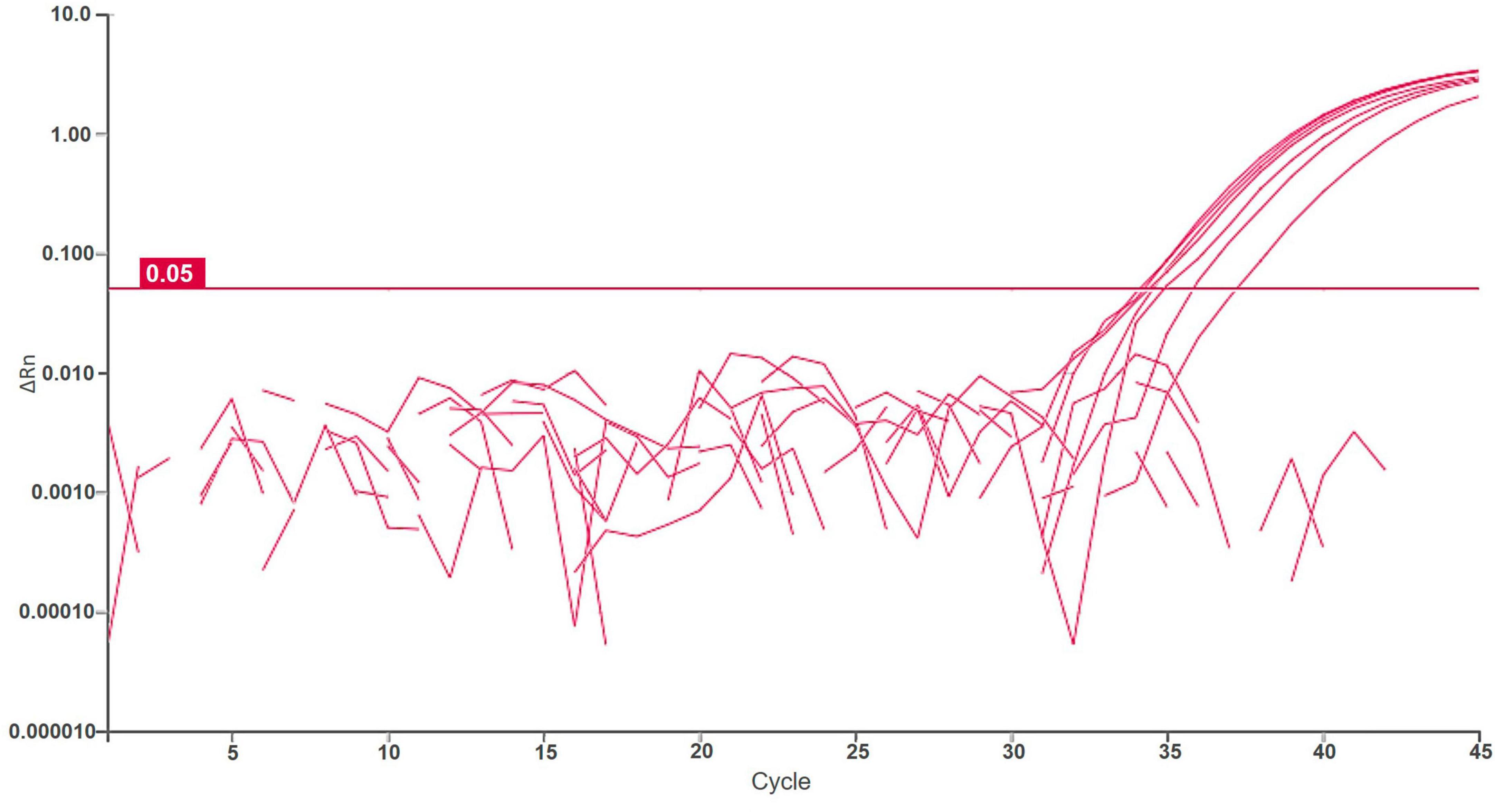
Figure 3. Logarithmic amplification curves of positive surface water samples of real-time PCR Df_tr. Threshold line is shown in magenta. Each sample was analyzed in duplicates, and it was considered positive if at least one reaction was positive.
Positive water samples were from different freshwater sources and of different types (Table 6). The samples were collected from different parts of Slovenia, but most samples had slightly acidic pH of 6 (4 of 6 samples). The temperature of the water ranged from 17°C to 28.5°C, measured at the sampling site.
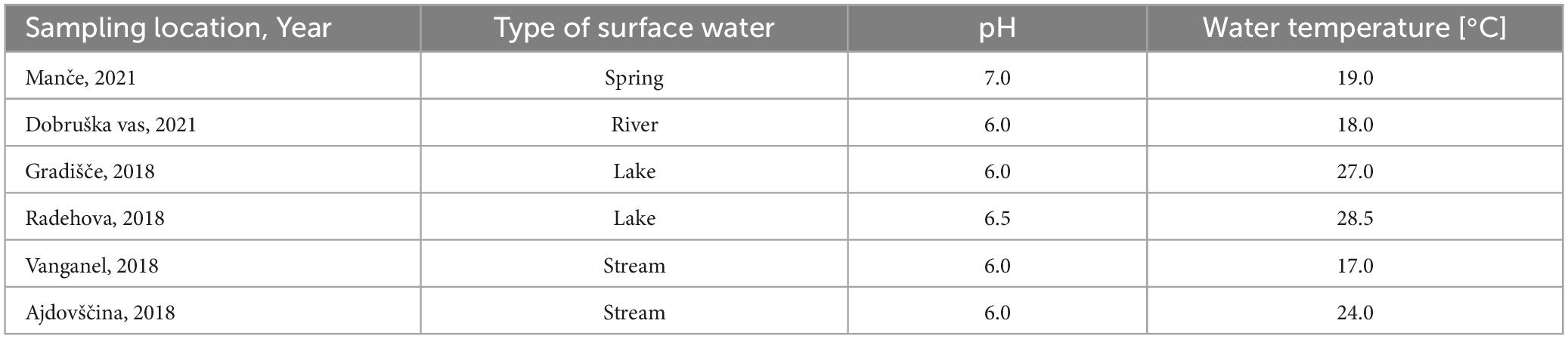
Table 6. Metadata of the water samples positive presence of Dickeya spp. (real-time PCR ECH) and D. fangzhongdai (real-time PCR Df_tr).
4 Discussion
In this study, a real-time PCR assay was developed for specific detection of D. fangzhongdai, along with its validation in matrices of orchids, potatoes, and Malinae members. The assay was used in a retrospective survey of relevant ecological niches in Slovene environments.
Designed assay exhibited very good performance characteristics in the validation, which proves its suitability for the detection of D. fangzhongdai with 100% inclusivity and exclusivity and good analytical and diagnostic sensitivity. Diagnostic sensitivity of the test ranged from 1 to 10 cells per reaction (LOD95), showing that the sensitivity of the assays is close or equal to the theoretical sensitivity of the method (Kralik and Ricchi, 2017). No inhibition of the real-time PCR reaction could be observed in any of the tested plant matrices and the reaction efficiency was close to optimal. Compared to the real-time assay described by Tian et al. (2020), the new test exhibited better diagnostic sensitivity in samples that contained plant matrices, showing better suitability for diagnostic purposes.
Dickeya fangzhongdai is the first known member of Dickeya spp. that causes disease not only on herbaceous plants, but also on trees. The majority of Dickeya fangzhongdai isolates originate from Asia, and few occurrences of D. fangzhongdai have been reported in Europe or America. This species has not yet been found to be associated with any significant plant disease in Europe’s open environment. It has been isolated from asymptomatic potato tubers in the Netherlands, but never from symptomatic plants in farmers’ fields in Europe (van der Wolf et al., 2022). Water sources seem to be an alternative habitat for Dickeya spp., as three species, namely D. aquatica, D. undicola and D. lacustris, are limited to water habitats and many others, including D. fangzhongdai, were also isolated from various water sources (Pritchard et al., 2013a; Parkinson et al., 2014; Alič et al., 2018; Hugouvieux-Cotte-Pattat et al., 2019; Oulghazi et al., 2019). Furthermore, presence of D. fangzhongdai was indirectly confirmed in wastewaters in Slovenia by isolation of D. fangzhongdai specific bacteriophages (Alič et al., 2017b). The results of the screening test performed in this study correspond with the described findings. However, the importance of water as an ecological niche is not yet understood. It is not known whether water presents a transmission source or only transient ecological niche that the bacteria is able to persist in. Nevertheless, virulence genes and genes involved in virulence regulation are also conserved in isolates from water (Alič et al., 2019).
In the screening test of potato plants and tubers with and without soft rot symptoms, and trees from the Malinae subtribe, D. fangzhongdai could not be detected in the tested samples. In Asian pear trees, bark tissue has been shown to be affected by D. fangzhongdai, therefore vascular tissue from twigs was selected as sample material for Malinae samples (Tian et al., 2016; Chen et al., 2020). However, it is not known which tissue would be most suitable for testing asymptomatic trees for the presence of D. fangzhongdai. Sixty-five of the surface water samples tested in this study were positive for presence of Dickeya spp., and 11% of those samples also contained D. fangzhongdai. D. fangzhongdai was present at low concentration, and the limit of detection (LOD95) of the assay in surface water is 3,776 cells/mL sample water extract. Other Dickeya spp. was detected in some potato samples with soft rot symptoms, however its prevalence is approximately 5 times lower compared to water samples. Based on the results, Dickeya spp. including D. fangzhongdai have not yet entered the agricultural environment but is present at low concentrations in some water sources in Slovenia.
Repeated reports of D. fangzhongdai outbreaks in Asia indicate that the pathogen is posing a threat to cultivation of various crops, ornamental plants and trees (Tian et al., 2016; Zhang et al., 2018; Jaffar et al., 2019; Shen et al., 2019; Tsai et al., 2019; Balamurugan et al., 2020; Choi et al., 2021; Huang et al., 2021; Wang et al., 2021). There have been no reports of D. fangzhongdai outbreaks or infections of plants in the open environment in Europe, however due to lack of specific testing for D. fangzhongdai species, isolates can be overlooked or assigned to Dickeya spp. The newly developed real-time PCR is reliable, sensitive and adequately validated, and therefore a suitable detection test for D. fangzhongdai detection, identification, and monitoring. Based on the results of the retrospective survey, D. fangzhongdai seems to be present in some water sources in Slovene environment. Presence of D. fangzhongdai was not confirmed in any tested plant species, however, its persistence cannot be excluded from hosts that were not included in this study. Specific testing for D. fangzhongdai presence and accordingly implementing preventive measures, is currently the only mechanism to prevent establishment of the species in new environments and environments in which the species had been sporadically detected.
Since the beginning of the 21st century the most detrimental Dickeya spp. for in European agriculture was D. solani. In 2012 it was listed among the 10 most important bacterial pathogens because of its sudden clonal spread and impact on the potato industry under higher temperatures (Mansfield et al., 2012). The pathogen was first isolated in 2005 and then in 2009 (Sławiak et al., 2009) but was recognized as a species only in 2014 (van der Wolf et al., 2014). However, in more recent studies it was shown that D. solani was present in potato more than a decade before the first reported outbreak. The early strains are genetically very close to the epidemic clones isolated during the 2000s outbreaks. Potentially aggressive D. solani strains in potato seeds were already present in the last century (Pédron et al., 2021), therefore it does not seem that genetics played an exclusive role in promoting pathogenicity, but rather an additional factor to the environmental conditions. Based on the reports, D. fangzhongdai seems to be as or even more aggressive than D. solani (Alič et al., 2017a; van der Wolf et al., 2022). Even if there is currently no association of D. fangzhongdai with any host in the open European environment, it is highly likely that the pathogen will not have a problem finding a host in favorable environmental conditions. Previous experience with D. solani showed that sporadic detection of such an aggressive pathogen in the environment might be a warning sign supporting the need for specific surveying of D. fangzhongdai.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
TD and ŠA: conceptualization, methodology, validation, and funding acquisition. ŠA and KB: formal analysis, investigation, and data curation. ŠA, KB, and TD: resources. ŠA: writing—original draft preparation and visualization. TD: writing—review and editing, supervision, and project administration. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financed by the Slovenian Research and Innovation Agency (contract numbers: P4-0165 and Z1-3190) and by the Euphresco Phytosanitary ERA-NET project on taxonomy and epidemiology of Pectobacterium and Dickeya spp. in Europe, North America, and South Africa (2020-F-336). The plant material used in this study was collected by the Administration of the Republic of Slovenia for Food Safety, Veterinary and Plant Protection, of the Ministry of Agriculture and Environment and Phytosanitary Inspectorate.
Acknowledgments
We thank Prof. Ian Toth and Dr. Sonia Humphries of the James Hutton Institute for the Dickeya sp. MK7 strain; Dr. Manca Pirc, Neža Turnšek, Lidija Matičič, Špela Prijatelj Novak, and Aleš Blatnik of the NIB for diagnostic samples preparation and technical assistance, and Dr. Carissa Robyn Bleker for critical reading and English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1249955/full#supplementary-material
References
Alič, Š., Dermastia, M., Burger, J., Dickinson, M., Pietersen, G., Pietersen, G., et al. (2022). Genome-informed design of a LAMP assay for the specific setection of the strain of ‘Candidatus Phytoplasma asteris’ phytoplasma occurring in grapevines in South Africa. Plant Dis. 106, 2927–2939. doi: 10.1094/PDIS-10-21-2312-RE
Alič, Š., Naglič, T., Tušek-Žnidarič, M., Peterka, M., Ravnikar, M., and Dreo, T. (2017a). Putative new species of the genus Dickeya as major soft rot pathogens in Phalaenopsis orchid production. Plant Pathol. 66, 1357–1368. doi: 10.1111/ppa.12677
Alič, Š., Naglič, T., Tušek-Žnidarič, M., Ravnikar, M., Rački, N., Peterka, M., et al. (2017b). Newly Isolated bacteriophages from the Podoviridae, Siphoviridae, and Myoviridae families have variable effects on putative novel Dickeya spp. Front. Microbiol. 8:1870. doi: 10.3389/fmicb.2017.01870
Alič, Š., Pédron, J., Dreo, T., and Van Gijsegem, F. (2019). Genomic characterisation of the new Dickeya fangzhongdai species regrouping plant pathogens and environmental isolates. BMC Genomics 20:34. doi: 10.1186/s12864-018-5332-3
Alič, Š., Van Gijsegem, F., Pédron, J., Ravnikar, M., and Dreo, T. (2018). Diversity within the novel Dickeya fangzhongdai sp., isolated from infected orchids, water and pears. Plant Pathol. 67, 1612–1620. doi: 10.1111/ppa.12866
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Asselin, J. E., Karp, M., Rychlik, P., Daughtrey, M., Filiatrault, M., and Stodghill, P. (2022). Genomic Comparison of Dickeya fangzhongdai Strains Isolated from Different Hosts and Pathogenicity in Onion (Allium cepa) Bulbs. Bethesda, MD: GenBank Direct Submission. s
Balamurugan, A., Kumar, A., Sakthivel, K., Ashajyothi, M., Sahu, K. P., and Karthikeyan, M. (2020). Characterization of Dickeya fangzhongdai causing bacterial soft rot disease on Dendrobium nobile in India. Eur. J. Plant Pathol. 158, 773–780.
Chan, K. G. (2014). Dickeya fangzhongdai strain ND14b chromosome. Bethesda, MD: GenBank Direct Submission.
Chan, K. G., and Tan, W. S. (2014). Genome sequence of Dickeya solani M005. Bethesda, MD: GenBank Direct Submission.
Chen, B., Tian, Y., Zhao, Y., Wang, J., Xu, Z., Li, X., et al. (2020). Bleeding canker of pears caused by Dickeya fangzhongdai: symptoms, etiology and biology. J. Integr. Agric. 19, 889–897. doi: 10.1016/S2095-3119(19)62882-0
Cheng, Y., Yan, Z., Gao, C., Yu, Y., Li, Z., Zen, L., et al. (2017). Complete genome sequence of the emerging pathogen for Southern Economic Crops, Dickeya Secpp 1600. Changsha: Institute of Bast Fiber Crops and Center of Southern Economic CropsGenBank Direct Submission.
Chi, N. M., Anh, D. T. K., Hung, T. X., Nhung, N. P., BaO, H. Q., Toan, D. V., et al. (2022). Soft rot disease caused by Dickeya fangzhongdai in epiphytic orchids in Vietnam. Can. J. Plant Pathol. 44, 386–399. doi: 10.1080/07060661.2021.1998226
Choi, E. D., Kim, Y., Lee, Y., Jeong, M.-H., Kim, K. H., Song, J. H., et al. (2021). First report of bleeding canker of pear tree trunks caused by Dickeya fangzhongdai in Korea. Plant Dis. 105, 2238. doi: 10.1094/PDIS-09-20-1948-PDN
DeLude, A., Wells, R., Boomla, S., Chuang, S.-C., Urena, F., Shipman, A., et al. (2022). Loop-mediated isothermal amplification (LAMP) assay for specific and rapid detection of Dickeya fangzhongdai targeting a unique genomic region. Sci. Rep. 12:19193. doi: 10.1038/s41598-022-22023-4
Dobhal, S., Boluk, G., Babler, B., Stulberg, M. J., Rascoe, J., Nakhla, M. K., et al. (2020). Comparative genomics reveals signature regions used to develop a robust and sensitive multiplex TaqMan real-time qPCR assay to detect the genus Dickeya and Dickeya dianthicola. J. Appl. Microbiol. 128, 1703–1719. doi: 10.1111/jam.14579
EU (1993). Council directive 93/85 of 4 October 1993 on the control of potato ring rot. Official J. Eur. Union L259, 1–25.
European and Mediterranean Plant Protection organization (2021). PM 7/98 (5) Specific requirements for laboratories preparing accreditation for a plant pest diagnostic activity. EPPO Bull. 51, 468–498. doi: 10.1111/epp.12780
Fujikawa, T., Hatomi, H., and Ota, N. (2020). Draft genome sequences of seven strains of Dickeya dadantii, a quick decline-causing pathogen in fruit zrees, isolated in Japan. Microbiol. Resour. Announc. 9:e00609-20. doi: 10.1128/MRA.00609-20
Hinsley, A., de Boer, H. J., Fay, M. F., Gale, S. W., Gardiner, L. M., Gunasekara, R. S., et al. (2018). A review of the trade in orchids and its implications for conservation. Bot. J. Linn. Soc. 186, 435–455. doi: 10.1093/botlinnean/box083
Huang, C. J., Lin, P. R., and Ting, C. L. (2020). Bacterial soft rot of taro in Taiwan. Bethesda, MD: GenBank Direct Submission.
Huang, S., Chen, Z., Hu, M., Xue, Y., Liao, L., and Zhang, L. (2021). First report of bacterial soft rot disease on taro caused by Dickeya fangzhongdai in China. Plant Dis. 105:3737. doi: 10.1094/PDIS-10-20-2225-PDN
Hugouvieux-Cotte-Pattat, N., Jacot-des-Combes, C., and Briolay, J. (2019). Dickeya lacustris sp. nov., a water-living pectinolytic bacterium isolated from lakes in France. Int. J. Syst. Evol. Microbiol. 69, 721–726. doi: 10.1099/ijsem.0.003208
Hugouvieux-Cotte-Pattat, N., Royer, M., Gueguen, E., Le Guen, P., Süssmuth, R. D., Reverchon, S., et al. (2022). Specificity and genetic polymorphism in the Vfm quorum sensing system of plant pathogenic bacteria of the genus Dickeya. Environ. Microbiol. 24, 1467–1483. doi: 10.1111/1462-2920.15889
Jaffar, N. S., Osman, M. S., and Koyube, M. N. K. (2019). New bacterial fruit rot disease of jackfruit caused by Dickeya fangzhongdai in Malaysia. Malays. J. Microbiol. 15, 214–219. doi: 10.21161/mjm.191547
Khandeparker, D. L., and Eswaran, R. (2018). Dickeya fangzhongdai strain GR29 16S ribosomal RNA gene. Bethesda, MD: GenBank Direct Submission.
Kralik, P., and Ricchi, M. (2017). A basic guide to real-time PCR in microbial diagnostics: definitions, parameters, and everything. Front. Microbiol. 8:108. doi: 10.3389/fmicb.2017.00108
Liu, D. (2020). First report of Dickeya fangzhongdai causing soft rot of belamcandae Rhizoma in China State. Bethesda, MD: GenBank Direct Submission.
Ma, X., Bonasera, J. M., Asselin, J. A. E., Beer, S. V., and Swingle, B. (2020). First report of Dickeya fangzhongdai causing soft rot of onion in New York state. Plant Dis. 104:1251. doi: 10.1094/PDIS-09-19-1940-PDN
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology: top 10 plant pathogenic bacteria. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Mumford, R. A., Skelton, A. L., Boonham, N., Posthuma, K. I., Kirby, M. J., and Adams, A. N. (2004). The improved detection of strawberry crinkle virus using real-time RT-PCR (TAQMAN®–). Acta Hortic. 656, 81–86. doi: 10.17660/ActaHortic.2004.656.11
Ogoshi, C., Monteiro, F. P., Becker, W. F., Kvitschal, M. V., Cardoza, Y. F., Zanin, J. G., et al. (2019). First report of Dickeya dadantii causing a new disease of apple trees in Brazil. New Dis. Rep. 39:8. doi: 10.5197/j.2044-0588.2019.039.008
Oulghazi, S., Pédron, J., Cigna, J., Lau, Y. Y., Moumni, M., Van Gijsegem, F., et al. (2019). Dickeya undicola sp. nov., a novel species for pectinolytic isolates from surface waters in Europe and Asia. Int. J. Syst. Evol. Microbiol. 69, 2440–2444. doi: 10.1099/ijsem.0.003497
Pan, L. (2021). Dickeya fangzhongdai strain Onc5 chromosome. Bethesda, MD: GenBank Direct Submission.
Parkinson, N., DeVos, P., Pirhonen, M., and Elphinstone, J. (2014). Dickeya aquatica sp. nov., isolated from waterways. Int. J. Syst. Evol. Microbiol. 64, 2264–2266. doi: 10.1099/ijs.0.058693-0
Parkinson, N., Stead, D., Bew, J., Heeney, J., Tsror (Lahkim), L., and Elphinstone, J. (2009). Dickeya species relatedness and clade structure determined by comparison of recA sequences. Int. J. Syst. Evol. Microbiol. 59, 2388–2393. doi: 10.1099/ijs.0.009258-0
Pédron, J., Schaerer, S., Kellenberger, I., and Van Gijsegem, F. (2021). early emergence of Dickeya solani revealed by analysis of Dickeya diversity of potato blackleg and soft rot causing pathogens in Switzerland. Microorganisms 9:1187. doi: 10.3390/microorganisms9061187
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pirc, M., Ravnikar, M., Tomlinson, J., and Dreo, T. (2009). Improved fireblight diagnostics using quantitative real-time PCR detection of Erwinia amylovora chromosomal DNA. Plant Pathol. 58, 872–881. doi: 10.1111/j.1365-3059.2009.02083.x
Pritchard, L., Humphris, S., Saddler, G. S., Elphinstone, J. G., Pirhonen, M., and Toth, I. K. (2013a). Draft genome sequences of 17 isolates of the plant pathogenic bacterium Dickeya. Genome Announc. 1:e00978-13. doi: 10.1128/genomeA.00978-13
Pritchard, L., Humphris, S., Saddler, G. S., Parkinson, N. M., Bertrand, V., Elphinstone, J. G., et al. (2013b). Detection of phytopathogens of the genus Dickeya using a PCR primer prediction pipeline for draft bacterial genome sequences: Dickeya diagnostics from draft bacterial genome sequences. Plant Pathol. 62, 587–596. doi: 10.1111/j.1365-3059.2012.02678.x
R Core Team (2021). R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
Richter, M., Rosselló-Móra, R., Oliver Glöckner, F., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931.
Ritz, C., and Strebig, J. C. (2016). Analysis of dose-response curves. Available online at: https://cran.r-project.org/web/packages/drc/drc.pdf (accessed September 4, 2019).
Rodriguez-R, L. M., and Konstantinidis, K. T. (2014). Bypassing cultivation to identify bacterial species: culture-independent genomic approaches identify credibly distinct clusters, avoid cultivation bias, and provide true insights into microbial species. Microbe Mag. 9, 111–118. doi: 10.1128/microbe.9.111.1
Sayers, E. W., Bolton, E. E., Brister, J. R., Canese, K., Chan, J., Comeau, D. C., et al. (2022). Database resources from the National Center for Biotechnology Information. Nucleic Acids Res. 50, D20-D26. doi: 10.1093/nar/gkab1112
Schaad, N. W., Jones, J. B., and Chun, W. (2001). Laboratory guide for identification ofplant pathogenic bacteria, 3rd Edn. St. Paul, MN: American Phytopathological Society Press.
Shen, Y., Lv, W. G., Du, Y. H., Zhang, Y. X., and Li, H. P. (2019). First report of Dickeya fangzhongdai causing soft rot of Phalaenopsis aphrodite in China. Plant Dis. 103, 2665–2665. doi: 10.1094/PDIS-01-19-0234-PDN
Sławiak, M., Łojkowska, E., and van der Wolf, J. M. (2009). First report of bacterial soft rot on potato caused by Dickeya sp. (syn. Erwinia chrysanthemi) in Poland. Plant Pathol. 58, 794–794. doi: 10.1111/j.1365-3059.2009.02028.x
Suharjo, R., Sawada, H., and Takikawa, Y. (2014). Phylogenetic study of Japanese Dickeya spp. and development of new rapid identification methods using PCR–RFLP. J. Gen. Plant Pathol. 80, 237–254. doi: 10.1007/s10327-014-0511-9
Thomsen, M. C. F., Hasman, H., Westh, H., Kaya, H., and Lund, O. (2017). RUCS: rapid identification of PCR primers for unique core sequences. Bioinformatics 33, 3917–3921. doi: 10.1093/bioinformatics/btx526
Tian, Y., Zhao, Y., Chen, B., Chen, S., Zeng, R., Hu, B., et al. (2020). Real-time PCR assay for detection of Dickeya fangzhongdai causing bleeding canker of pear disease in China. J. Integr. Agric. 19, 898–905. doi: 10.1016/S2095-3119(19)62881-9
Tian, Y., Zhao, Y., Yuan, X., Yi, J., Fan, J., Xu, Z., et al. (2016). Dickeya fangzhongdai sp. nov., a plant-pathogenic bacterium isolated from pear trees (Pyrus pyrifolia). Int. J. Syst. Evol. Microbiol. 66, 2831–2835. doi: 10.1099/ijsem.0.001060
Toth, I. K., van der Wolf, J. M., Saddler, G., Lojkowska, E., Hélias, V., Pirhonen, M., et al. (2011). Dickeya species: an emerging problem for potato production in Europe: Dickeya spp. on potato in Europe. Plant Pathol. 60, 385–399. doi: 10.1111/j.1365-3059.2011.02427.x
Tsai, W.-A., Lin, P.-R., and Huang, C.-J. (2019). First report of Dickeya fangzhongdai causing soft rot disease of Welsh onion in Taiwan. J. Plant Pathol. 101, 797–798. doi: 10.1007/s42161-019-00264-z
van der Wolf, J. M., Nijhuis, E. H., Kowalewska, M. J., Saddler, G. S., Parkinson, N., Elphinstone, J. G., et al. (2014). Dickeya solani sp. nov., a pectinolytic plant-pathogenic bacterium isolated from potato (Solanum tuberosum). Int. J. Syst. Evol. Microbiol. 64, 768–774. doi: 10.1099/ijs.0.052944-0
van der Wolf, J., Krijger, M., Mendes, O., Kurm, V., and Gros, J. (2022). Natural infections of potato plants grown from Minitubers with blackleg-causing soft rot Pectobacteriaceae. Microorganisms 10:2504. doi: 10.3390/microorganisms10122504
Wang, F., Tang, T., Mao, T., Guo, J., Guo, X., Duan, Y., et al. (2021). Occurrence of Dickeya fangzhongdai causing soft rot of Banxia (Pinellia ternata) in China. Plant Dis. 105:3288. doi: 10.1094/PDIS-01-21-0030-PDN
Wei, X., Deng, W., and Chu, C. (2021). Phylogenetic and phenotypic analyses on Dickeya spp. isolated from different host plants in Taiwan. J. Phytopathol. 169, 678–691. doi: 10.1111/jph.13038
Weller, S. A., Elphinstone, J. G., Smith, N. C., Boonham, N., and Stead, D. E. (2000). Detection of Ralstonia solanacearum strains with a quantitative multiplex real-time fluorogenic PCR (TaqMan) Assay. Appl. Environ. Microbiol. 66, 2853–2858. doi: 10.1128/AEM.66.7.2853-2858.2000
Yang, D., Du, C. J., Ye, Y. F., Pan, L. F., Zhang, J., and Fu, G. (2022). First report of Dickeya fangzhongdai causing peduncle soft rot of banana in China. Plant Dis. 106:1980. doi: 10.1094/PDIS-11-21-2513-PDN
Zhang, J., Hu, J., Shen, H., Zhang, Y., Sun, D., Pu, X., et al. (2018). Genomic analysis of the Phalaenopsis pathogen Dickeya sp. PA1, representing the emerging species Dickeya fangzhongdai. BMC Genomics 19:782. doi: 10.1186/s12864-018-5154-3
Keywords: molecular testing, diagnostics, plant pathogen, real-time PCR, Dickeya, survey, water
Citation: Alič Š, Bačnik K and Dreo T (2024) Retrospective survey of Dickeya fangzhongdai using a novel validated real-time PCR assay. Front. Microbiol. 14:1249955. doi: 10.3389/fmicb.2023.1249955
Received: 29 June 2023; Accepted: 27 December 2023;
Published: 13 February 2024.
Edited by:
Mohammad Arif, University of Hawai’i at Manoa, United StatesReviewed by:
Emilio Marco Stefani, University of Modena and Reggio Emilia, ItalyGamze Boluk-Sari, Ministry of Agriculture and Forestry, Türkiye
Copyright © 2024 Alič, Bačnik and Dreo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tanja Dreo, tanja.dreo@nib.si
 Špela Alič
Špela Alič Katarina Bačnik
Katarina Bačnik Tanja Dreo
Tanja Dreo