- National Fungal Culture Collection of India, Biodiversity and Palaeobiology Group, MACS' Agharkar Research Institute, Pune, India
In this study, a new species Fusarium indicum belonging to the Fusarium concolor species complex is established to accommodate an endophytic fungus isolated from Bambusa sp. and collected from Himachal Pradesh. The identity of this isolate was confirmed based on the asexual morphs, its cultural characteristics, and phylogenetic analyses. This isolate revealed out to be distinct by showing less similarity with described species in the genus Fusarium based on molecular sequence data, approximately 93.9% similarity based on translation elongation factor 1-alpha, and 94.2% similarity based on RNA polymerase II subunit. Furthermore, to increase knowledge about this novel species, whole-genome sequencing was carried out. The results displayed that Fusarium indicum NFCCI 5145 possesses a 40.2 Mb genome and 48.39% of GC content. Approximately 12,963 functional protein-coding genes were carefully predicted and annotated using different BLAST databases, such as Uniprot, Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), Pathogen Host Interactions (PHI), Clusters of Orthologous Groups (COG), and Carbohydrate-Active enzymes (CAZy). The orthologous proteins were identified using OrthoFinder and used for the phylogenetic analysis. ANIb confirmed that the isolate is closely related to the F. concolor species complex. It is known that Fusarium strains can produce a wide range of bioactive secondary metabolites. Therefore, in-depth mining for biosynthetic gene clusters for secondary metabolite biosynthesis of Fusarium indicum NFCCI 5145 was investigated using Antibiotics and Secondary Metabolites Analysis Shell (AntiSMASH) annotation. AntiSMASH results displayed that this isolate possesses 45 secondary metabolites of biosynthetic gene clusters (BGCs). These findings significantly improved our understanding of the strain Fusarium indicum NFCCI 5145 and its possible applications in different sectors including industry for the secondary metabolites and enzymes it can produce.
1 Introduction
The first report on the genus Fusarium was described by Link in 1809 (Link, 1809). Fusarium species are present in almost any ecosystem globally (Leslie and Summerell, 2006). They have been reported from nearly all bioclimatic regions of the world, including tropical and temperate grasslands, shrublands, forests, harsh desert, and alpine environments, soils associated with plants, organic debris, and any part of plants from plants’ deepest roots to highest flowers (Leslie and Summerell, 2006). Therefore, Fusarium species can colonize multiple habitats in almost all ecosystems worldwide (Young et al., 1978; Nelson et al., 1994; Arney et al., 1997). Fusarium is one of the most economically significant fungi, mostly known as a pathogen, capable of infecting essential agricultural and horticultural crops worldwide (Booth, 1971; Leslie and Summerell, 2006). Fusarium species are mainly responsible for wilts, blights, root rots, and cankers (Ingle, 2017). Its species can also occur as saprophytes, endophytes, parasites, pathogens of plants, and pathogens or mutualists of animals (Torbati et al., 2021).
Fusarium concolor was first established on Hordeum vulgare from Montevideo, Uruguay (Reinking, 1934) and later was reported from various hosts, such as wheat, banana, koa tree, Guarana, Hybanthus prunifolius, plant debris, and soil (Chambers, 1972; Saadabi, 2006; Murad and Baiti, 2017; Almeida et al., 2021). It has also been known to cause keratitis and fusariosis (Al-Hatmi et al., 2016). The Fusarium concolor species complex currently consists of four species, namely, F. anguioides, F. austroafricanum, F. bambusarum, and F. concolor. The isolates of F. anguioides have been reported from bamboo, Cordyline stricta, and Alocasia odora; Fusarium bambusarum have been reported from bamboo (Nelson et al., 1995; Wang et al., 2022). Fusarium austroafricanum was isolated as an endophyte of Kikuyu grass associated with putative mycotoxicosis of cattle (Jacobs-Venter et al., 2018). In the present study, an endophytic isolate was isolated from Bambusa sp. and was established as a new species of the genus Fusarium, falling into the Fusarium concolor species complex based on morphology and phylogeny. Furthermore, whole-genome sequencing was targeted to get insights into the genomic architecture of this newly discovered Fusarium species.
2 Materials and methods
2.1 Collection, isolation, and morphological characterization of endophytic Fusarium indicum
Healthy leaves of Bambusa sp. collected from Panchrukhi, Palampur, District Kangra of Himachal Pradesh, India, on 4 October 2021 were placed in sterile polythene bags and transported carefully to the laboratory. The surface adherents were removed after thorough washing under tap water. Then, bigger leaves were chopped into small pieces and subjected to surface sterilization following a modified method by Dobranic et al. (1995). Concisely, bamboo leaves were first dipped in 70% ethanol for 5 s, followed by 4% sodium hypochlorite for 90 s, and later rinsed with sterile water for 10 s (four times). These surface sterilized leaves were cut into small pieces using a sterilized sharp blade and inoculated on potato dextrose agar (PDA) plates. These plates were kept at 25°C until any vegetative growth appeared from the inoculated tissues. Individual colonies arising from inoculated tissues were transferred to fresh PDA plates by hyphal tipping and allowed to grow (Bills and Polishook, 1992). Furthermore, pure culture was raised using a single spore isolation technique. Colony characteristics of this isolate were studied on PDA and synthetic nutrient agar (SNA). Methuen’s Handbook of Color was referred to recording the colors of the colonies on different agar media (Kornerup and Wanscher, 1978). Microscopic structures of the isolates were recorded from pure culture using staining cum-mounting medium, lactophenol cotton blue, under a Carl Zeiss Image Analyzer 2 (Germany) microscope. Measurements and photomicrographs of the fungal structures were recorded using Axiovision Rel 4.8 software and Digi-Cam attached with Carl Zeiss Image Analyzer 2 microscope. The holotype specimen is deposited and accessioned in Ajrekar Mycological Herbarium (AMH 10381), and ex-type pure culture is deposited and accessioned in the National Fungal Culture Collection of India (NFCCI 5145).
2.2 DNA extraction, PCR amplification, and DNA sequencing
Genomic DNA was extracted from pure colonies raised from single spore isolation on PDA Petri plates. After approximately 1 week of incubation, DNA extraction was performed by a simple, easy, and rapid DNA extraction protocol using FastPrep®24 tissue homogenizer (MP Biomedicals GmbH, Eschwege, Germany; Aamir et al., 2015). The amplification and sequencing of tef-1α, rpb2, and LSU gene regions were carried out. The primers involved in amplification and sequencing were EF-1 (5′ ATGGGTAAGGARGACAAGAC 3′) and EF-2 (5′ GGARGTACCAGTSATCATG 3′) for translation elongation factor 1-alpha (tef1-α; O’Donnell et al., 1998), LR-OR (5′ ACCCGCTGAACTTAAGC 3′; Vilgalys and Sun, 1994), LR-7 (5′ TACTACCACCAAGATCT 3′; Vilgalys and Hester, 1990) for 28S large subunit of the nrDNA (LSU), and fRPB2-5f (5′ GAYGAYMGWGATCAYTTYGG 3′) and fRPB2-7cR (5′ CCCATRGCTTGYTTRCCCAT 3′) for RNA polymerase second largest subunit (rpb2; Liu et al., 1999).
PCR was carried out in a 25 μL reaction using 12.5 μL 2x Invitrogen Platinum SuperFi PCR Mastermix, 2 μL template DNA (10–20 ng), 1.5 μL 10 pmol primer, 5 μL 5x GC enhancer, and H2O (Sterile Ultra-Pure Water, Sigma, St. Louis, MO, United States), with the total volume made to 25 μL. The conditions of the thermocycling involved those as follows: For tef1-α gene region, an initial denaturation of 5 min at 94°C, 30 cycles of 45 s at 94°C, 30 s at 57°C, and 1 min at 72°C followed by a final 7-min extension at 72°C; 5 min denaturation at 94°C, 35 cycles of 1 min at 94°C, 50 s at 52°C, and 1.2 min at 72°C, with a final 8 min extension at 72°C for LSU; and 5 min denaturation at 95°C, 35 cycles of 45 s at 95°C, 1 min at 52°C, and 1.5 min at 72°C, with a final 10 min extension at 72°C for rpb2.
The PCR amplicons were purified with a FavorPrep™ PCR purification kit as per the manufacturer’s instructions. Purified PCR products of all marker genes were checked on 1.2% agarose gel electrophoresis stained with 0.5 μg/mL ethidium bromide and were further subjected to a sequencing PCR using a BigDye®Terminator v3.1 Cycle Sequencing Kit, as per the manufacturer’s instructions. In brief, the sequencing PCR of 20 μL included 4 μL of 5× sequencing buffer, 2 μL of BigDye™ Terminator premix, 4 μL of primer (5 pmol), and 4 μL of the purified amplicon and H2O (Sterile Ultra-Pure Water, Sigma), with the volume made to 20 μL. Thermal cycling conditions consisted of an initial denaturing at 96°C for 3 min, followed by 30 cycles of 94°C for 10 s, 50°C for 40 s, and 60°C for 4 min. The BigDye® terminators and salts were removed using the BigDye Xterminator® Purification Kit (Thermo Fisher Scientific, Waltham, MA, United States) as per the manufacturer’s instructions. After performing cycle sequencing with BigDye™ terminators, 80 μL of SAM™ solution and 20 μL of XTerminator™ solution were added to each tube. The mixture was vortexed for 30 min and then centrifuged at 10,000 rpm for 30 s. The supernatant was transferred to a 96-well microplate, and the module was selected and run was set up. The sequence was elucidated on Applied Biosystems SeqStudio Genetic Analyzer (Applied Biosystems, Foster City, CA, United States). Sequences obtained were submitted in NCBI GenBank [accession numbers OM032811 (tef-1α), OM032812 (rpb2), and OM025235 (LSU)].
2.3 Phylogenetic analysis
To determine the phylogenetic status of this isolate, tef-1α and rpb2 gene regions were used to compare the present isolate with already known authentic strains in the genus Fusarium. The sequences of the related authentic strains were retrieved from NCBI. A total of 83 isolates of the genus Fusarium were used in the phylogenetic analysis and were aligned along with the sequences of Fusarium indicum NFCCI 5145. Geejayessia zealandica CBS 111.93 and Geejayessia cicatricum CBS 125549 were selected as the outgroup taxa. The strains which were used in making phylogenetic tree, along with their accession numbers and other related details, are presented in Supplementary Table 1. Each gene region was aligned individually with MAFFT v. 6.864b (Katoh and Standley, 2013). The alignments were checked and adjusted manually using AliView (Larsson, 2014). Furthermore, alignments were concatenated and processed for the phylogenetic analyses. The best substitution model was figured using ModelFinder (Kalyaanamoorthy et al., 2017). Additionally, Windows version IQ-tree tool v.1.6.11 (Nguyen et al., 2015) was used to reconstruct the phylogenetic tree. The reliability of the tree branches was assessed and tested on the basis of 1,000 ultrafast bootstrap support replicates (UFBoot) and the SH-like approximate likelihood ratio test (SH-like aLRT) with 1,000 replicates. The constructed phylogenetic tree was visualized in FigTree v.1.4.4.
2.4 High molecular weight DNA extraction for whole-genome sequencing
In total, 100 mg of the fungal mass was crushed using a mortar-pestle in liquid N2. The powder was placed in a 2 mL sterile Eppendorf tube. Overall, 1 mL of pre-heated CTAB buffer [20 mM EDTA, 100 mM Tris HCl, and 1.4 M NaCl and CTAB 2%] was added along with 20 μL of β-mercaptoethanol and 1 mg of polyvinylpyrrolidone (PVP). The mixture was mixed properly and incubated at 65°C ± 2°C for at least 30 min. An equal amount of phenol:chloroform:isoamyl alcohol (25:24:1, v/v) was added and mixed well, and centrifuged at 10,000 rpm for 10 min. The upper aqueous layer was transferred to a fresh 1.5 mL Eppendorf tube and followed by the addition of an equal volume of chloroform:isoamyl alcohol (24:1, v/v) and mixed well and then centrifuged at 10,000 rpm for 10 min. The upper aqueous layer was transferred to a fresh 1.5 mL Eppendorf tube, and an equal volume of isopropanol was added and incubated under cold conditions at −20°C for 20 min. Furthermore, centrifugation was carried out at 10,000 rpm for 10 min at 4°C. The supernatant was removed, and the pellet was carefully washed using 500 μL of 70% ethanol. Again, centrifugation was carried out at 10,000 rpm for 5 min at 4°C. The supernatant was discarded, and the pellet was dried. The pellet was dissolved in 70 μL of 1× TE buffer. In total, 1 μL of RNase A solution (20 mg mL−1) was added and later incubated at 37°C for at least 30 min. The integrity was evaluated by 1% agarose gel electrophoresis, and purity was accessed by a NanoDrop™ 1,000 Spectrophotometer (Thermo Fisher Scientific).
2.5 Library preparation and sequencing
Library construction was done using the QIASeq FX DNA library preparation protocol (Cat#180475) as per the manufacturer’s instructions. In total, 50 ng of Qubit quantified DNA was enzymatically fragmented, end-repaired, and A-tailed in the one-tube reaction using the FX enzyme mix provided in the QIASeq FX DNA kit. The end-repaired and adenylated fragments were subjected to adapter ligation, whereby the index-incorporated Illumina adapter was ligated to generate sequencing libraries. These libraries were subjected to 6 cycles of Indexing PCR [initial denaturation at 98°C for 2 min and cycling (98°C for 20 s, 60°C for 30 s, and 72°C for 30 s) and final extension at 72°C for 1 min] to enrich the adapter-tagged fragments. Finally, for purification of the amplified libraries, JetSeq Beads were used (Bio-68031). Furthermore, quantification of the sequencing library was carried out by a Qubit fluorometer (Thermo Fisher Scientific, MA, United States). The sequencing of the libraries was performed on an Illumina NovaSeq 6000 sequencer (Illumina, San Diego, CA) for 150 bp paired-end chemistry, according to the manufacturer’s procedure.
2.6 Gene prediction and annotation
Libraries were paired-end sequenced using an Illumina NovaSeq 6000 sequencer. After sequencing, the paired-end raw data were kept in FASTQ format. Fastp (0.20.1) tool was used to remove low-quality reads, adapters, and polyG tails from FastQ files.1 To get the draft genome sequence, St. Petersburg genome assembler SPAdes version 3.11.1 was used for assembly.2 The genome diagram of Fusarium indicum NFCCI 5145 was constructed using Circos version 0.69-9 (Krzywinski et al., 2009). Augustus 3.4.0 tool was used for performing gene prediction.3 The gene function and metabolic pathway data available in the already existing databases were used to carry out the function annotation, and BLAST searches were performed against various databases such as NR (NCBI non-redundant protein sequences), Uniprot, COG (Cluster of Orthologous Groups of proteins), KEGG (Kyoto Encyclopedia of Genes and Genomes), CAZy (Carbohydrate-Active Enzymes Database; Lombard et al., 2014), and PHI (Pathogen Host Interactions Database; Urban et al., 2020). Secondary metabolite biosynthetic gene clusters of Fusarium indicum NFCCI 5145 were analyzed using antiSMASH fungal 6.1.1 (Blin et al., 2021). Tandem Repeats Finder Program (trf) version 4.09.1 was used to predict the number of repeats4 (Benson, 1999).
2.7 Comparative genomics and phylogenetic analysis
Orthologous proteins were identified using OrthoFinder version 2.5.4 (Emms and Kelly, 2019), and the results were used to build a species tree, visualized using FigTree. Fusarium species used for the study of the orthologous proteins and phylogenetic tree construction are presented in Table 1. The analysis of average nucleotide identity (ANI) was performed using the pyani script and ANIb as an algorithm for the alignment (Pritchard et al., 2016). Fusarium species used in the analysis of ANI are presented in Table 2.
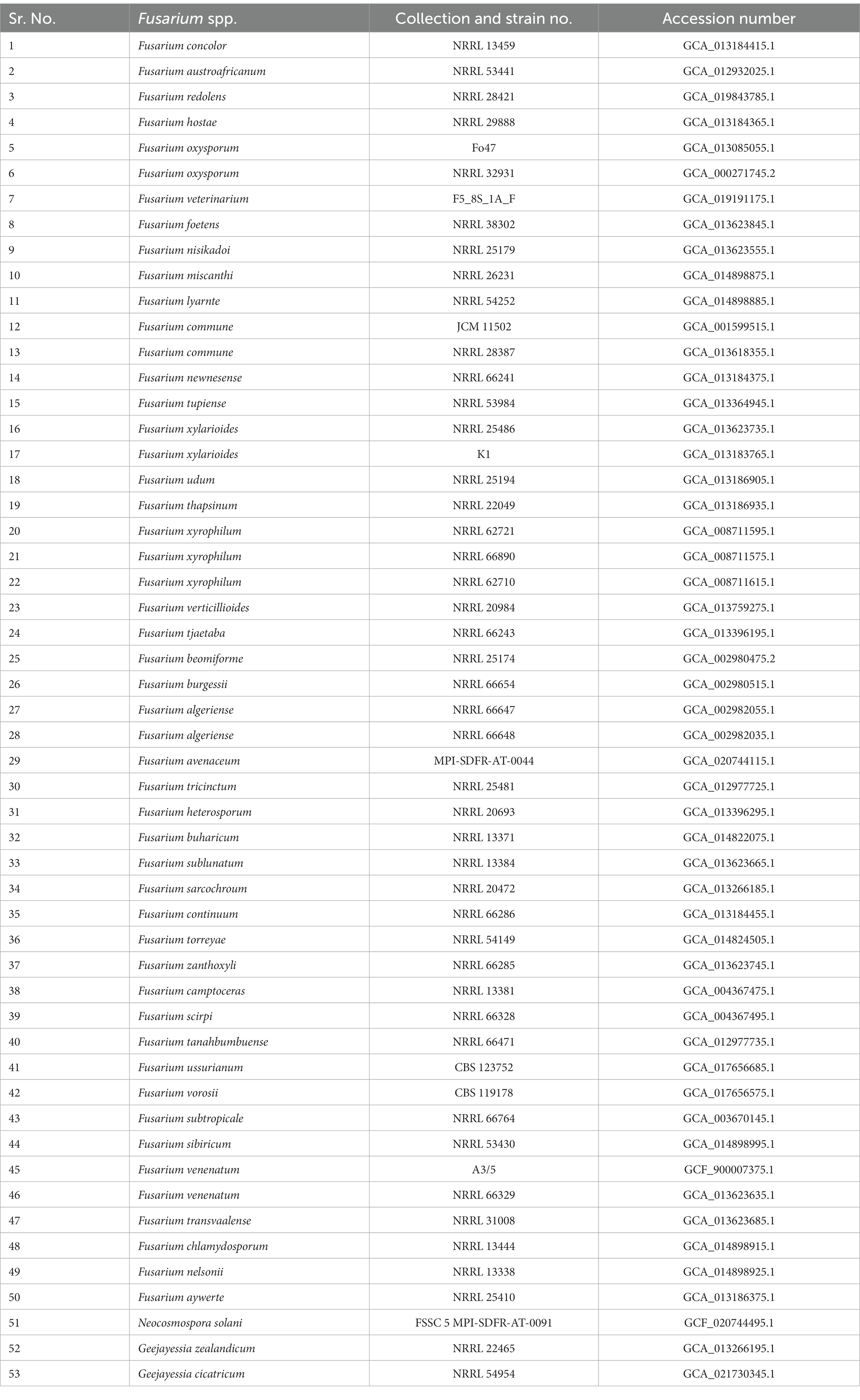
Table 1. Fusarium species used for the comparison of orthologous proteins with Fusarium indicum NFCCI 5145 and its phylogeny.
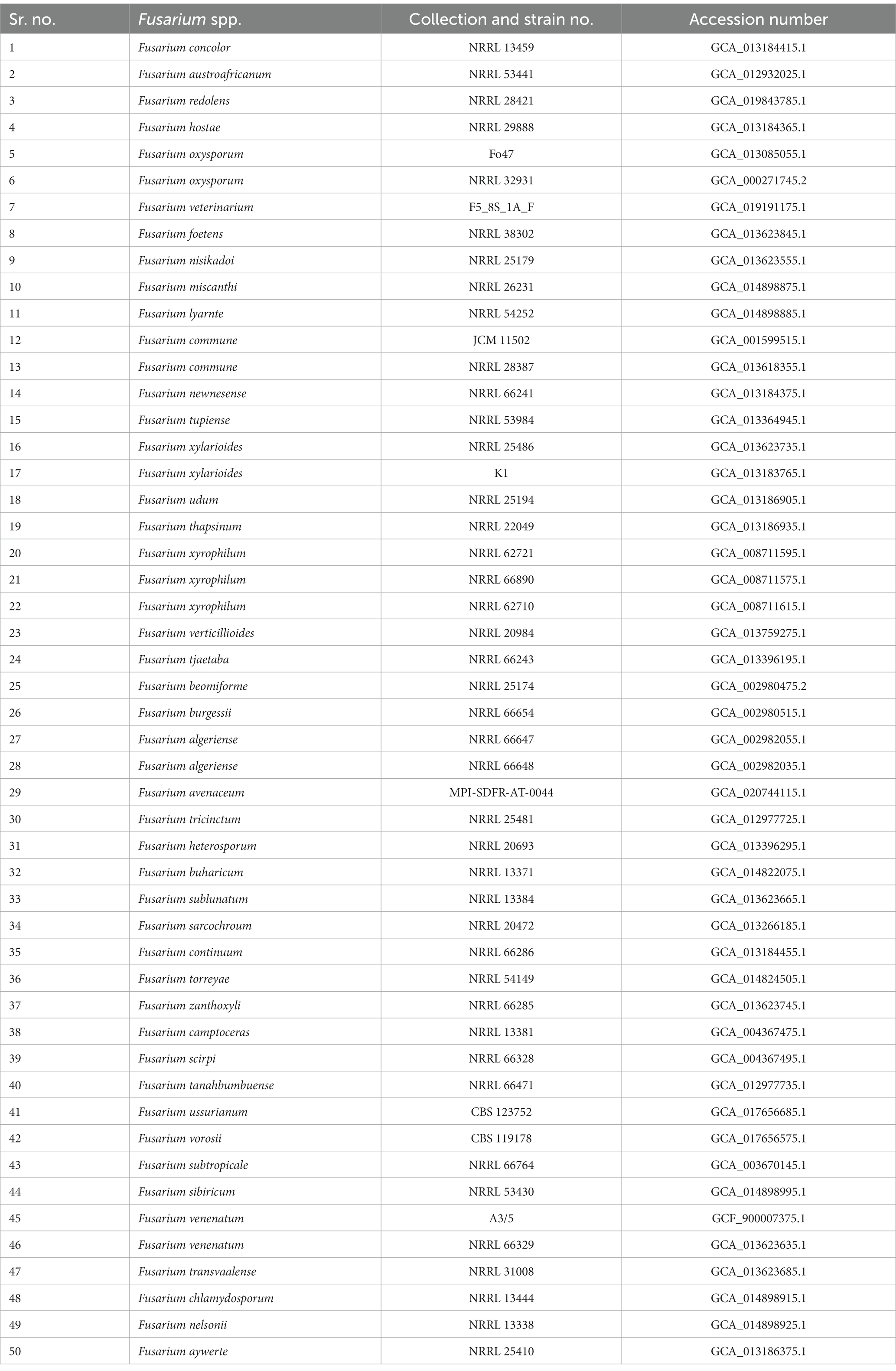
Table 2. Details of Fusarium species used for the calculation of genome-scale average nucleotide identity with Fusarium indicum NFCCI 5145.
3 Results
3.1 Taxonomy of Fusarium indicum
Fusarium indicum S. Rana and S.K. Singh, sp. nov. Figures 1A–K, 2.
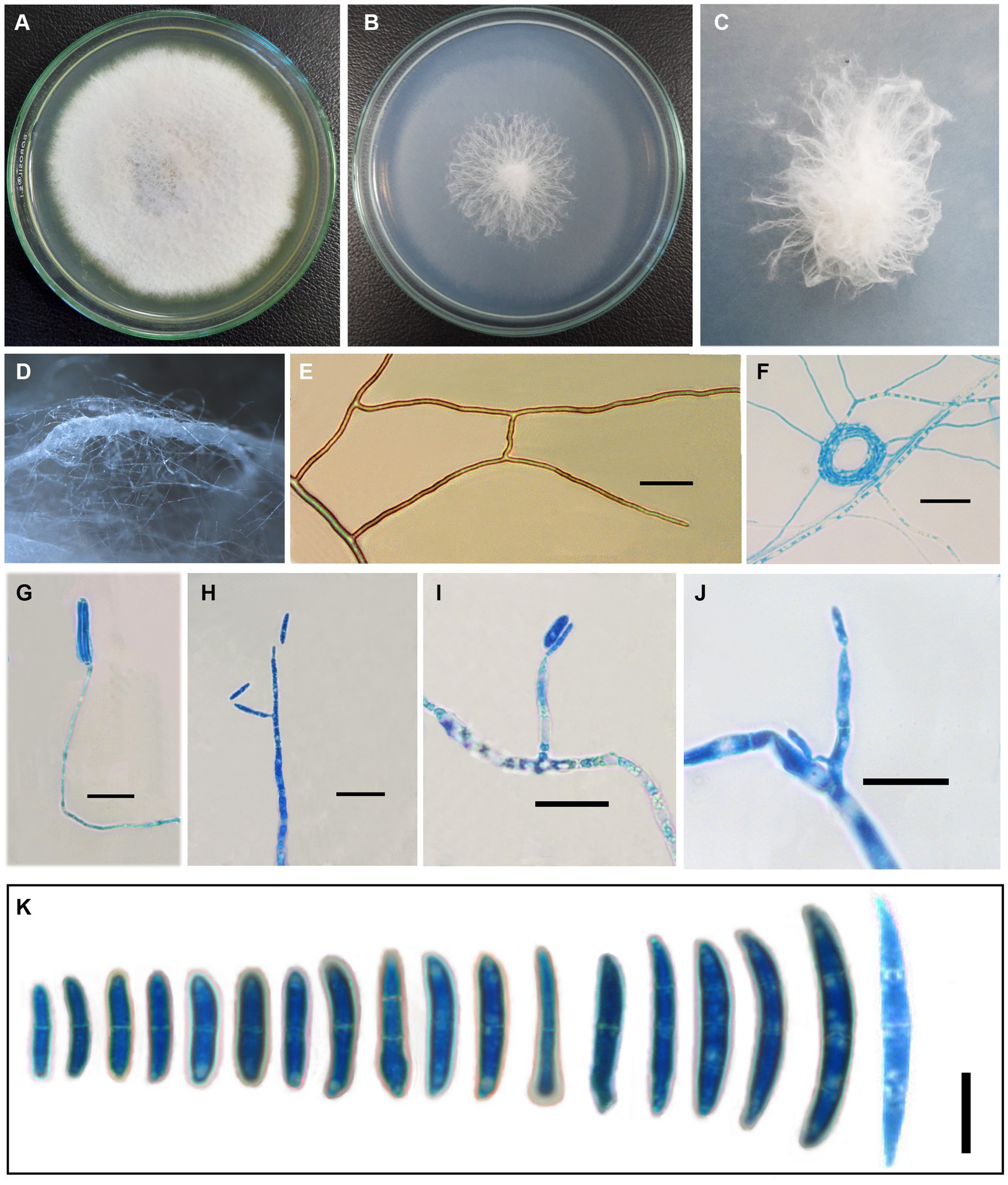
Figure 1. Fusarium indicum (NFCCI 5145); colonies of Fusarium indicum on (A) PDA; (B) SNA; (C) Stereoscopic view of colony growing on SNA; (D) Stereoscopic view of bundles of hyphae growing on SNA; (E) Hyphae showing anastomoses; (F) Coiled hyphae; (G–J) Phialides bearing microconidia; (K) Micro and macroconidia. —Scale bars: E–J = 20 μm, K = 10 μm.
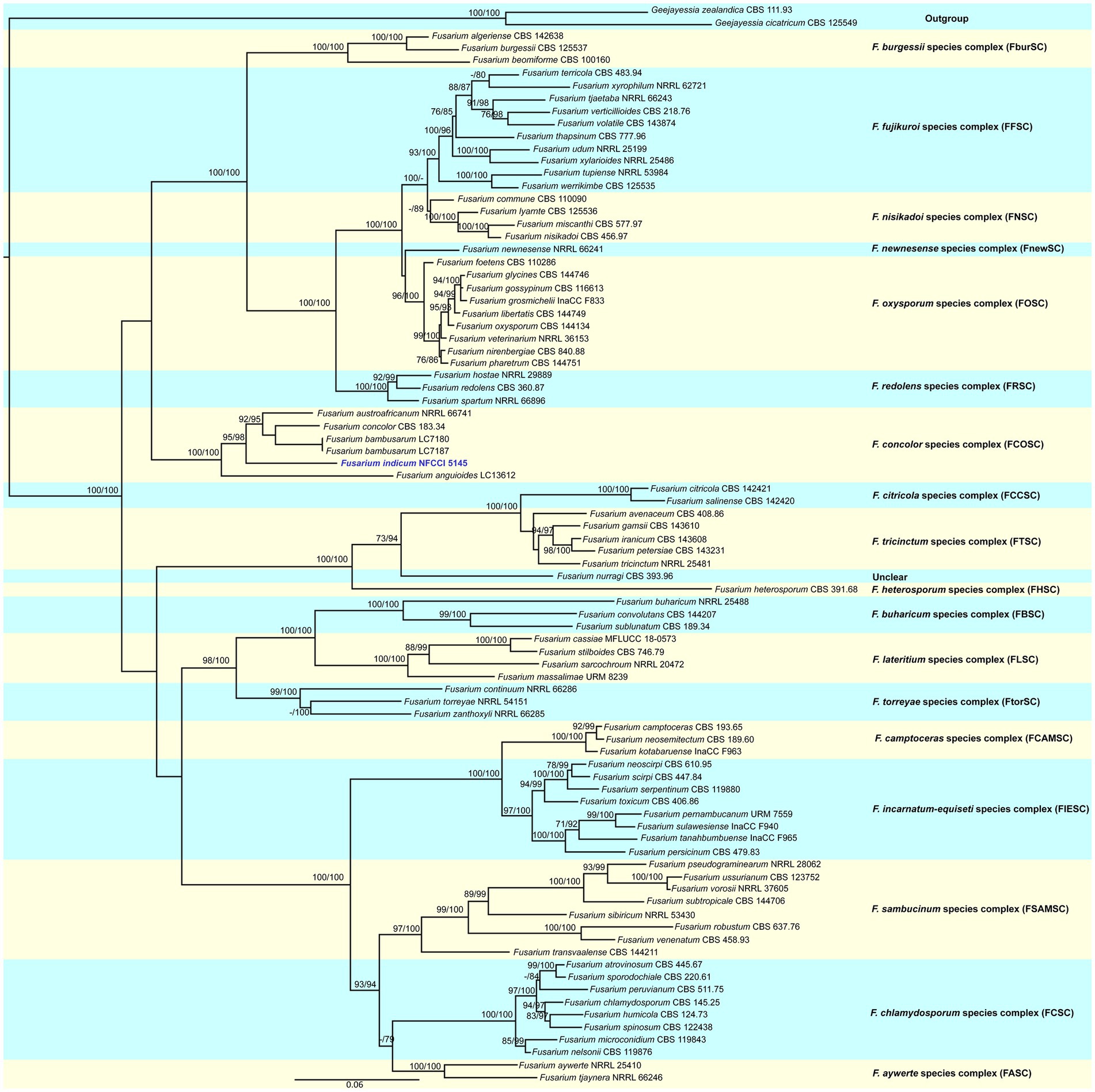
Figure 2. Molecular phylogenetic analysis of new species of Fusarium indicum (NFCCI 5145) based on maximum-likelihood (ML) method using both tef-1α and rpb2 sequence data. New species Fusarium indicum (NFCCI 5145) is shown in blue. Statistical support values are shown next to each node, UFBS values and SH-aLRT obtained from 1,000 replicates using IQ-TREE and the TIM2e + I + G4 model.
MycoBank Number: MB 847911
Holotype: AMH 10381
Etymology: Named after the country of isolation “India.”
Host/distribution: As endophyte from Bambusa sp. collected from Panchrukhi, Himachal Pradesh, India.
Original description: Asexual morph Mycelium appeared in parallel bundles of hyphae, simple to branched, septate, smooth-walled, hyaline, thin to thick, showing anastomoses, 1.0–6.5 μm wide (x̄ = 3.58 μm, n = 10). Conidiophores produced from superficial lateral hyphae, simple to branched, gradually decreasing to the length, septate, sometimes rudimentary protruding from superficial hyphae 24.67–118 × 1.35–4.20 μm (x̄ = 59.97 × 2.9 μm, n = 10). Conidiogenous cells are phialidic. Phialides produced directly from superficial hyphae, cylindrical gradually decreasing toward the length, ampuliform, monophialides to polyphialides, sometimes reduced to conidiogenous cell 4.5–43.70 × 2.25–4.90 μm (x̄ = 24.75 × 3.51 μm, n = 10). Microconidia rarely produced, hyaline, smooth-walled, fusoid to cylindrical, straight to curved, 0–1 septate, 9.5–19 × 2.15–4.5 μm (x̄ = 14.18 × 3.15 μm, n = 30). Macroconidia straight to curved, guttulate, short-beaked, 0–3 septate, foot cell prominent, 12.85–46.5 × 2.57–7.5 μm (x̄ = 23.04 × 3.73 μm, n = 30). Chlamydospores are rare, terminal to intercalary, sub-globose to globose, or solitary or in groups of 2–3.
Cultural characteristics: —Colonies growing on PDA reached 39 mm after 4 days at 25°C; yellowish white (1A2) to white (1A1), reverse pale yellow (3A3) after 1 week. In the colony, aerial mycelium is produced, colonies lacking distinct odor, colonies slightly raised, cottony, margin smooth and entire. Colonies growing on SNA reached 33 mm after 4 days of incubation at 25°C; grayish white (1B1) to white (1A1) after 1 week. Colony cottony, raised from the center, and flat toward the periphery with entire smooth margins.
Sexual morph: Not observed.
Known distribution: Panchrukhi, Himachal Pradesh, India.
Material examined: INDIA, Himachal Pradesh, Panchrukhi (32°03′01.1″N 76°34′01.8″E), from leaves of Bambusa sp., S. rana, 4 October 2021, AMH 10381(holotype), deposited in Ajrekar Mycological Herbarium (AMH) of India, ex-type culture is deposited in National Fungal Culture Collection of India (NFCCI 5145).
GenBank numbers: OM032811 (tef-1α), OM032812 (rpb2), and OM025235 (LSU).
Notes: The present species is somewhat similar to F. concolor morphologically (Reinking, 1934; Booth, 1971) in having pale pigmentation and marked heel on the foot cell of macroconidia. It is different from other taxa in the complex. The colonies of F. anguioides are carmine to ochre aerial mycelia, having red to brick tinges of pigmentation; Fusarium austroafricanum produces white to reddish white (7A2) colonies on PDA and reverse is light orange (6A4), while F. indicum produces white to yellowish white colonies on PDA. F. concolor differs from F. indicum in having smaller phialides compared with that of F. indicum and sometimes reduced to conidiogenous cell; aerial phialides monophialidic in F. bambusarum and polyphialidic in F. concolor vs. monophialidic to polyphialidic F. indicum. Macroconidia in F. concolor are usually 3–5 septate and occasionally 6–7 septate, F. austroafricanum macroconidia are 0–11 septate but mostly 3–8-septate, F. anguioides macroconidia 4–7 septate; F. bambusarum macroconidia 3–6 septate; however, F. indicum macroconidia are 0–3 septate (Booth, 1971; Jacobs-Venter et al., 2018; Yli-Mattila et al., 2018; Wang et al., 2022).
Fusarium concolor species are reported predominantly from soil; F. anguioides species have been reported from bamboo, Cordyline stricta and Alocasia odora; Fusarium austroafricanum was isolated as endophyte from Pennisetum clandestinum (kikuyu grass); Fusarium bambusarum species have been reported from bamboo (Booth, 1971; Nelson et al., 1995; Jacobs-Venter et al., 2018; Wang et al., 2022). Though F. anguioides, F. bambusarum, and F. indicum share the same host, they are quite distinct morphologically as well as based on the molecular data.
On the basis of MegaBLAST algorithm search on NCBI blastn5 for Fusarium indicum NFCCI 5145, the closest hit using tef-1α gene sequence was found to be Fusarium concolor (NRRL 13994, type) showing 92.59% (638 out of 689 bp) identity and having 15 gaps (2.17%) and with Fusarium austroafricanum (NRRL 66741, type) showing 93.48% (617 out of 660 bp) identity and having 4 gaps (0.6%); based on rpb2, closest hits were Fusarium concolor (NRRL 13994, type) showing 94.21% (766 out of 813 bp) identity and having 2 gaps (0.2%) and with Fusarium austroafricanum (NRRL 53441) showing 93.74% (644 out of 687 bp) identity and having 4 gaps (0.58%).
Additionally, the tef-1α gene sequence of Fusarium indicum NFCCI 5145 showed 93.54% (623 out of 666 bp) identity and had 6 gaps when compared with Fusarium bambusarum LC7180, and with Fusarium anguioides LC13612 showed 89.90% (595 out of 667 bp) identity and 10 gaps (1%; 10 out of 667 bp). Additionally, the rpb2 gene sequence of Fusarium indicum NFCCI 5145 showed 93.63% (735 out of 785 bp) identity when compared with Fusarium bambusarum LC7180 and with Fusarium anguioides LC13612 showed 91.84% (721 out of 785 bp) identity.
3.2 Phylogenetic analysis
The sequence alignments of both tef-1α and rpb2 were used to confirm the identity of this isolate. The concatenated file had sequence data of 86 taxa (Supplementary Table 1). Alignment contained 1,671 columns, 777 parsimony-informative, 1,061 distinct patterns, 113 singleton sites, and 781 constant sites. TIM2e + I + G4 was considered to be the best model and was selected based on the Bayesian Information Criterion (BIC). The phylogenetic tree was generated using the maximum likelihood method based on the above-mentioned model. The log-likelihood of the consensus tree was −24373.628. Rate parameters were A–C: 1.95361, A–G: 4.81458, A–T: 1.95361, C–G: 1.00000, C–T: 9.32964, and G–T: 1.00000; base frequencies were A: 0.250, C: 0.250, G: 0.250, and T: 0.250, and the proportion of invariable sites was 0.387 and gamma shape alpha parameter was 1.138. Combined phylogenetic analysis using tef-1α and rpb2 nested this isolate, Fusarium indicum, in a distinct and unique clade in the Fusarium concolor species complex. The clade was well supported with strong SH-like aLRT and ultrafast bootstrap (UFBoot; Figure 2).
3.3 Genome sequencing and assembly of Fusarium indicum NFCCI 5145
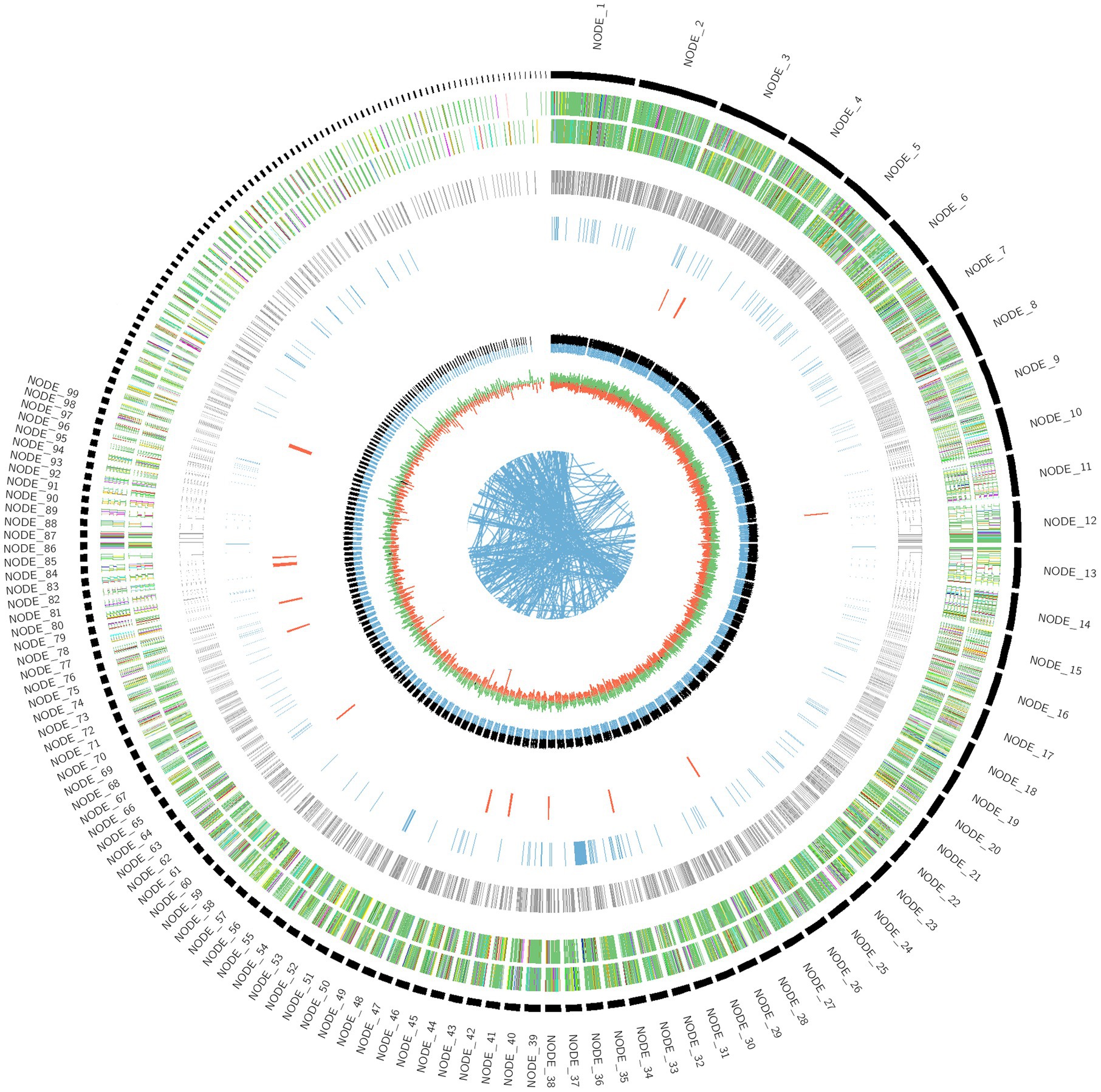
The genome sequence of Fusarium indicum NFCCI 5145 was assembled and deposited in the NCBI GenBank database (SRA accession No. PRJNA993417; BioProject PRJNA993417; BioSample SAMN36633317; accession number JAWDHB000000000). The genome diagram of Fusarium indicum NFCCI 5145 shows that there are nine circles in the circle diagram (Figure 3), which are as follows from inside to outside: the first blue line shows in-paralog regions; the second circle shows the GC skew, with the green part showing a positive GC SKEW and the orange part showing a negative GC SKEW; the third circle shows the GC content (%; negative gene; positive gene); the fourth circle shows secondary metabolites; the fifth circle shows ncRNA; the sixth circle shows repeats; the seventh circle and the eighth circle display CDS annotation information. The seventh circle indicates that CDS is in a positive chain, and the eighth circle indicates that CDS is in a negative chain. The outer rim shows the contigs.
The whole genome size of Fusarium indicum NFCCI 5145 was 40.2 Mb. This consisted of 513 scaffolds with an N50 of 0.46 Mb and 48.39% of GC content. A total of 7,884,552 raw reads and 7,847,766 clean and high-quality reads (99.53%) were generated in the Illumina sequencing. Before filtering, total reads were 7.884552 M, total bases were 1.182683 G, Q20 bases were 1.162804 G (98.319170%), Q30 bases were 1.121368 G (94.815622%), and GC content was 48.005470%. After filtering, total reads were 7.847766 M, total bases were 1.176427 G, Q20 bases were 1.158196 G (98.450277%), Q30 bases were 1.117366 G (94.979678%), and GC content was 48.000585%. Reads that passed the filters were 7.847766 M (99.533442%), reads with low quality were 33.424000 K (0.423918%), reads with too many N were 2.328000 K (0.029526%), and reads which were too short were 1.034000 K (0.013114%). The total number of scaffolds was 513, the total number of bases (bp) were 40,226,774 (~40.2 Mb), the minimum scaffold length (bp) was 300, the maximum scaffold length (bp) was 1,668,425, scaffolds ≥ 200 kbp were 513, scaffolds ≥ 500 kbp were 347, scaffolds ≥ 1 kbp were 246, scaffolds ≥ 10 kbp were 169, scaffolds ≥ 1 Mbp were 6, and N50 value was 461,173. The Augustus prediction method was used to predict the encoding gene. In total, 12,724 protein-coding genes were predicted using highly annotated databases such as Uniprot. Gene total length (bp) was 6,336,400, gene average length (bp) was 498, maximum gene length (bp) was 8,599, and minimum gene length (bp) was 24. Simultaneously, the Tandem Repeats Finder Program (trf) was found to predict the number of repeats, which were found to be 3,232.
3.4 Genome sequence annotation of Fusarium indicum NFCCI 5145 using KEGG, COG, and GO
For the prediction of the protein sequences, 12,963 non-redundant genes of Fusarium indicum NFCCI 5145 were subjected to a similarity search on the basis of various public databases. Many genes were mapped using the Uniprot database (12,638 genes/97.49%), Clusters of Orthologous Groups (COG; 6,509 genes/50.21%), and Kyoto Encyclopedia of Genes and Genomes (KEGG; 9,155 genes/70.62%).
As per COG database, “General function prediction only” was related with many genes (1436), followed by “Carbohydrate transport and metabolism (624),” “Amino acid transport and metabolism (489),” “Function unknown (422),” “Post Translational modification, protein turnover, chaperones (405),” and “Translation, ribosomal structure and biogenesis (401)” (Figure 4; Natale et al., 2000). These results depict that Fusarium indicum NFCCI 5145 possesses a varied and enriched array of functions for carbohydrates and amino acid metabolism that may result in better energy conversion efficiency.
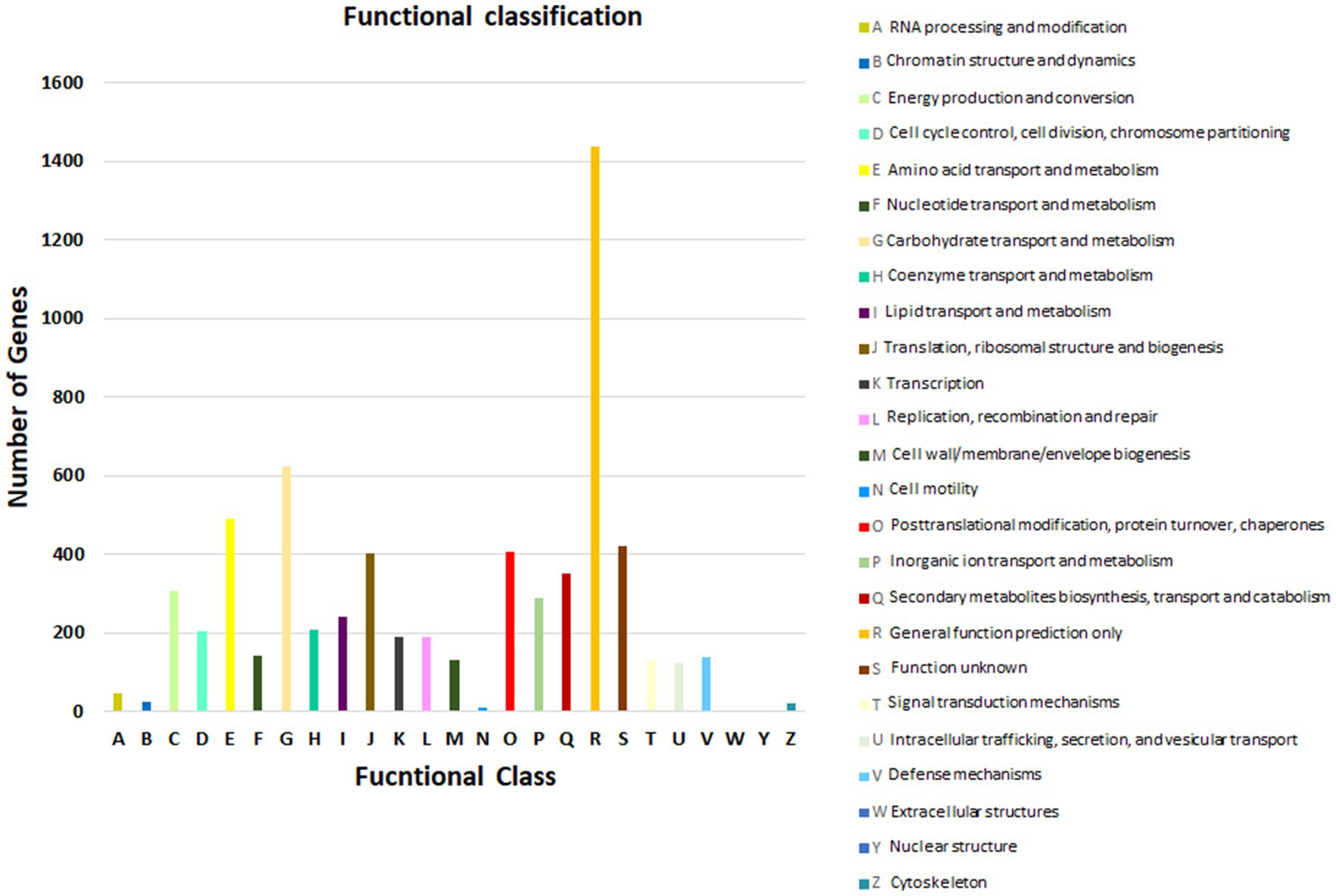
Figure 4. Functional annotation of Fusarium indicum NFCCI 5145 genes encoding for proteins using Clusters of Orthologous Genes (COG) database.
The findings from KEGG functional classification suggest that the predicted proteins fell under various categories, such as amino acid metabolism (2451), metabolism of cofactors and vitamins (1682), global map (1526), and carbohydrate metabolism (1525; Figure 5; Yamada et al., 2021). The results indicate that there is a varied and enriched array of various metabolic functions present that probably will provide higher secondary metabolism efficacy.
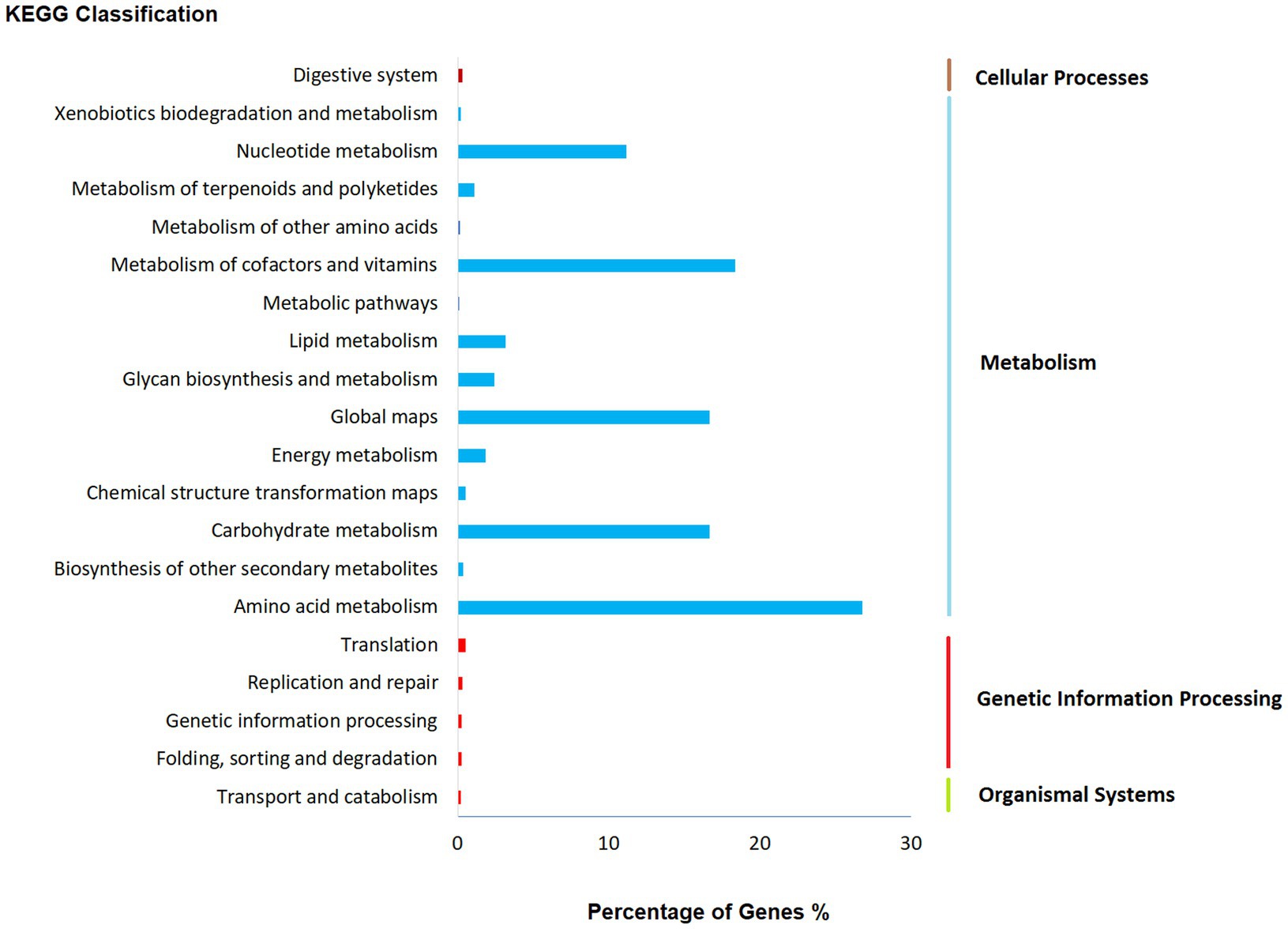
Figure 5. Functional annotation of Fusarium indicum NFCCI 5145 genes encoding for proteins using Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis.
GO annotation depicts varied genes possessed by Fusarium indicum, which may be involved in biological processes, cellular components, and molecular functions (Figure 6; Huntley et al., 2014).
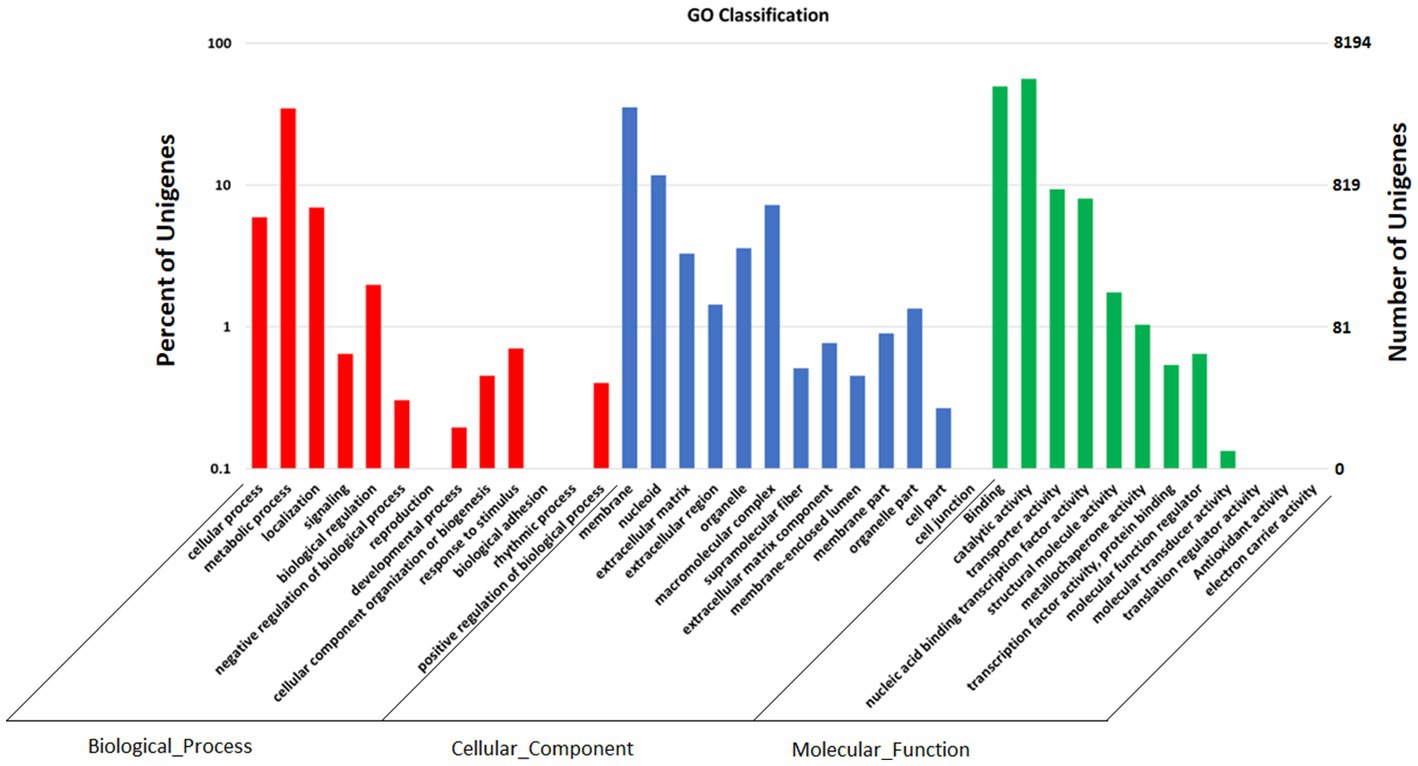
Figure 6. Functional annotation of Fusarium indicum NFCCI 5145 genes encoding for proteins using Gene Ontology (GO) analysis.
3.5 Genome sequence annotation of Fusarium indicum NFCCI 5145 for carbohydrate genes
Carbohydrate-active enzymes (CAZymes) are classes of enzymes that catalyze the breakdown and assembly of glycoconjugates as well as glycans (oligosaccharides and polysaccharides; Garron and Henrissat, 2019). These enzymes play an important role in fungal metabolism as they are accountable for carbohydrate degradation and modification as well as biosynthesis (Liu et al., 2022). The CAZy is a specialized database for carbohydrate enzymes that have the capability to create, modify, and degrade glycosidic bonds (Liu et al., 2022). The analysis showed 1,012 genes that encode for carbohydrate-active enzymes (CAZy) dispersed in the genome of Fusarium indicum NFCCI 5145. These include 109 auxiliary activities (AAs), 115 carbohydrate-binding modules (CBMs), 64 carbohydrate esterases (CEs), 458 glycoside hydrolases (GHs), 238 glycosyltransferases (GTs), and 28 polysaccharide lyases (PLs; Figure 7). AAs mainly included AA1-16 16 families. CBMs distributed mainly across 23 families, including CBM1–2, CBM5–6, CBM9, CBM12–14, CBM18, CBM20–21, CBM24, CBM32, CBM35, CBM38, CBM41–43, CBM48, CBM50, CBM63, CBM67, and CBM87. Carbohydrate esterases were classified as CE1–5, CE8–9 CE12, CE14, and CE16 across 10 families. Glycoside hydrolases were distributed across 76 families, including GH1–3, GH5, GH7, GH9–13, GH15–18, GH20, GH23–33, GH35–39, GH43, GH47, GH49, GH51, GH53, GH55, GH62–65, GH67, GH71–79, GH81, GH86, GH88, GH92–95, GH103, GH105, GH114–115, GH123, GH125, GH128, GH130–132, GH135–136, GH139, GH141, GH146, GH152, GH154, and GH162. GTs contained 43 families, including GT1–4, GT8, GT10, GT11, GT15, GT17–18, GT20–24, GT28, GT30–35, GT39, GT41, GT43, GT47–48, GT50–51, GT57–59, GT61–62, GT64, GT66, GT69, GT71, GT76, GT90, GT106, GT107, and GT109. PLs are mainly distributed in eight families, including PL1, PL4, PL3, PL9, PL22, PL26, PL35 and PL42. These findings show that Fusarium indicum NFCCI 5145 can prove to show great ability to make and break complex carbohydrates making it capable of capturing more energy, thus possibly having great industrial applicability.
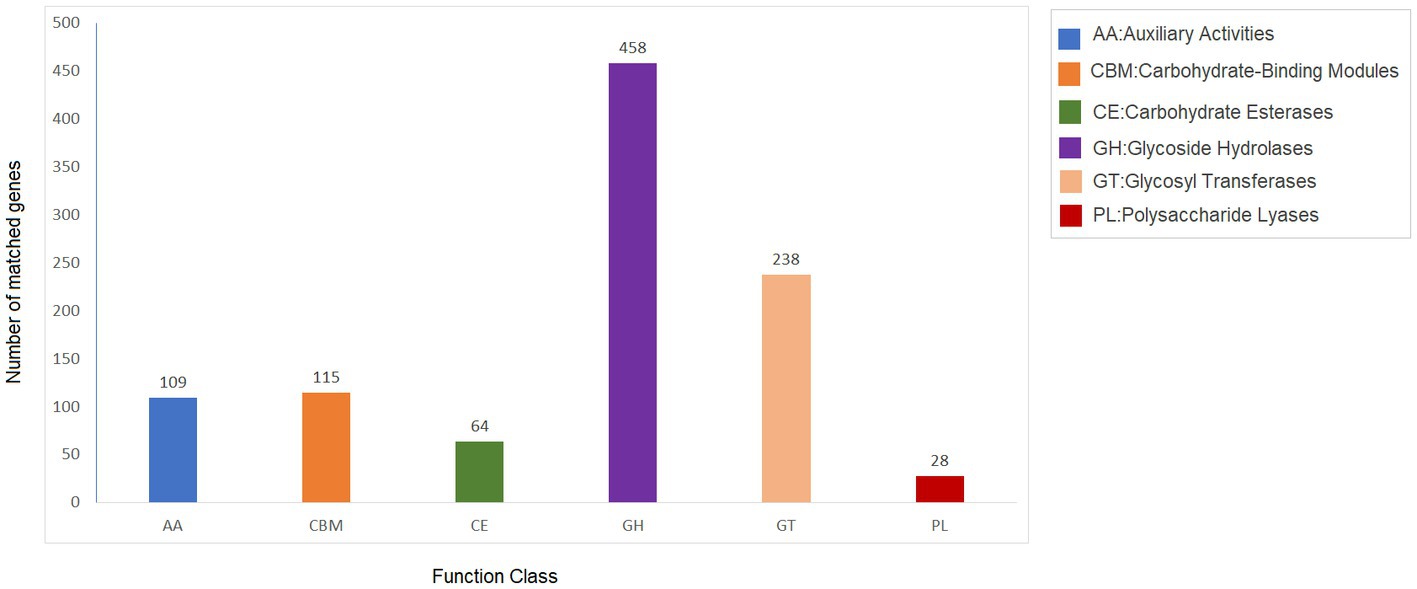
Figure 7. Carbohydrate-active enzymes (CAZy) functional classification and corresponding genes present in the genome of Fusarium indicum NFCCI 5145.
3.6 Genome sequence annotation of Fusarium indicum NFCCI 5145 for pathogen host interactions
The Pathogen Host Interactions Database (PHI-base) is a database that is manually curated by experts based on experimental evidence that consists of genes related to virulence, effector, and pathogenicity from fungi, bacteria, and oomycete that infect plants, animals, insects, and fungi (Urban et al., 2022). The amino acid sequence of Fusarium indicum NFCCI 5145 was compared with the PHI database. As shown in Figure 8, Fusarium indicum NFCCI 5145 possesses abundant PHI-base genes, including reduced virulence (853), increased virulence (hypervirulence; 66), loss of pathogenicity (154), lethal (99), unaffected pathogenicity (867), sensitivity to chemical (2), effector (plant avirulence determinant; 21), and resistance to chemical (6; Pernas, 2021). Unaffected pathogenicity and reduced virulence were the major annotation genes, indicating that Fusarium indicum NFCCI 5145 is not a pathogenic strain as expected as it was isolated as an endophyte.
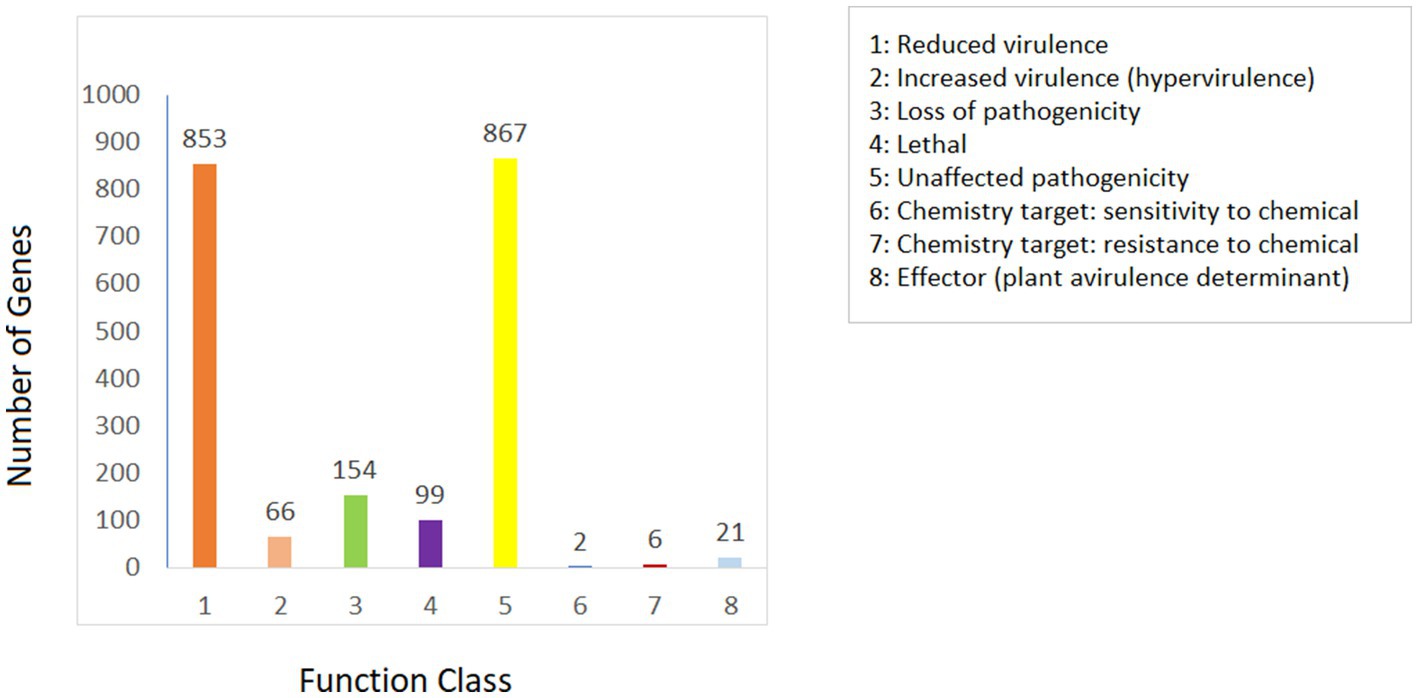
Figure 8. Distribution map of mutation types in the pathogen PHI phenotype of Fusarium indicum NFCCI 5145.
3.7 AntiSMASH analysis of Fusarium indicum
AntiSMASH results indicated that Fusarium indicum isolate NFCCI 5145 contains 45 secondary metabolite biosynthetic gene clusters (BGCs), including eight NRPS-like fragments (NRPS-like), nine nonribosomal peptide synthetases (NRPSs), six terpene synthases, 12 Type I PKS (polyketide synthase; T1PKS), one Type III PKS (polyketide synthase; T3PKS), one beta-lactone containing protease inhibitor (betalactone), three hybrid NRPS + T1PKS, two indoles, one hybrid indole + NRPS, one hybrid T1PKS and NRPS, and one hybrid NRPS, T1PKS and indole (Table 3; Figure 9A–N).
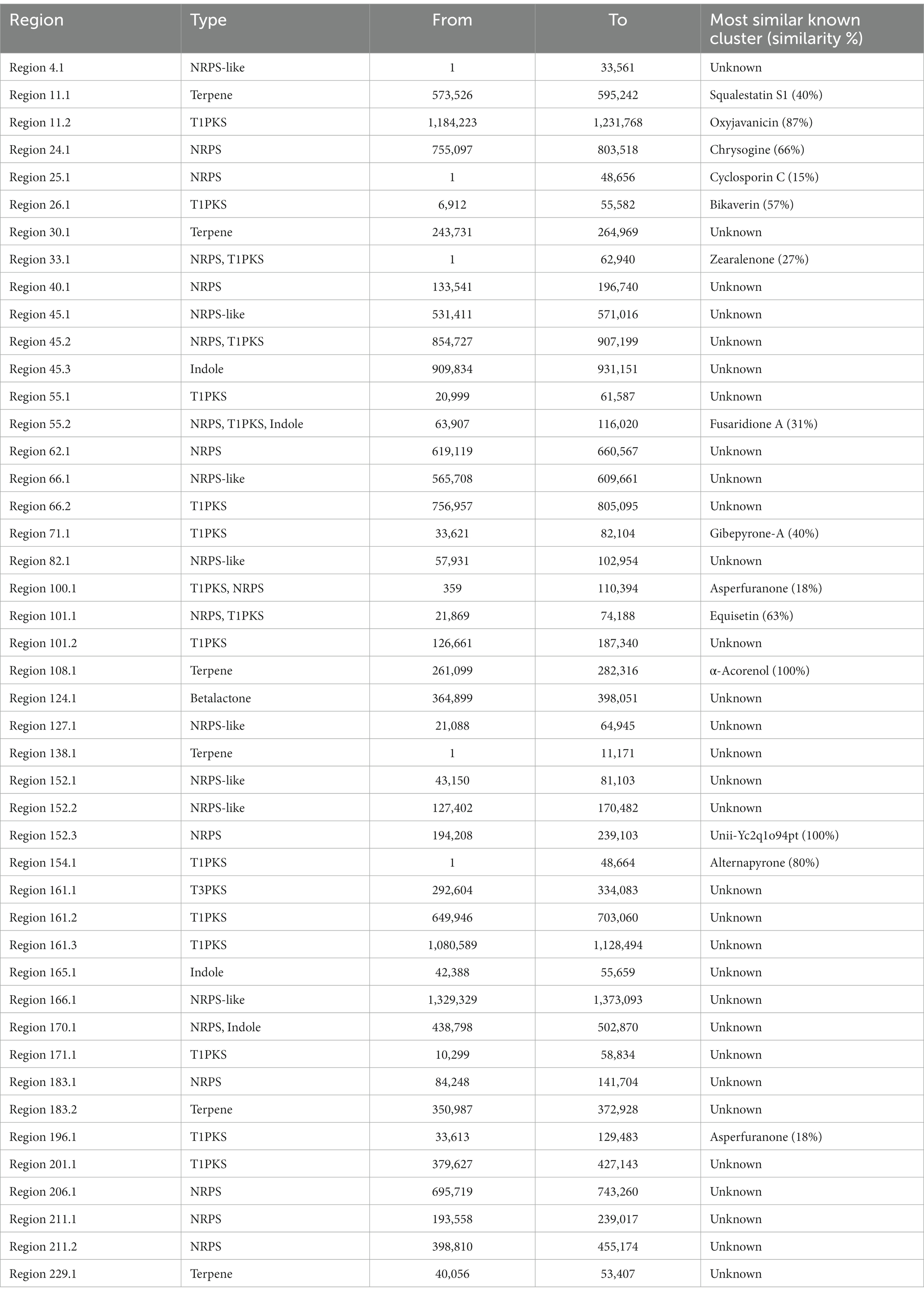
Table 3. Putative biosynthetic gene clusters (BGCs) coding for secondary metabolites in strain Fusarium indicum NFCCI 5145.
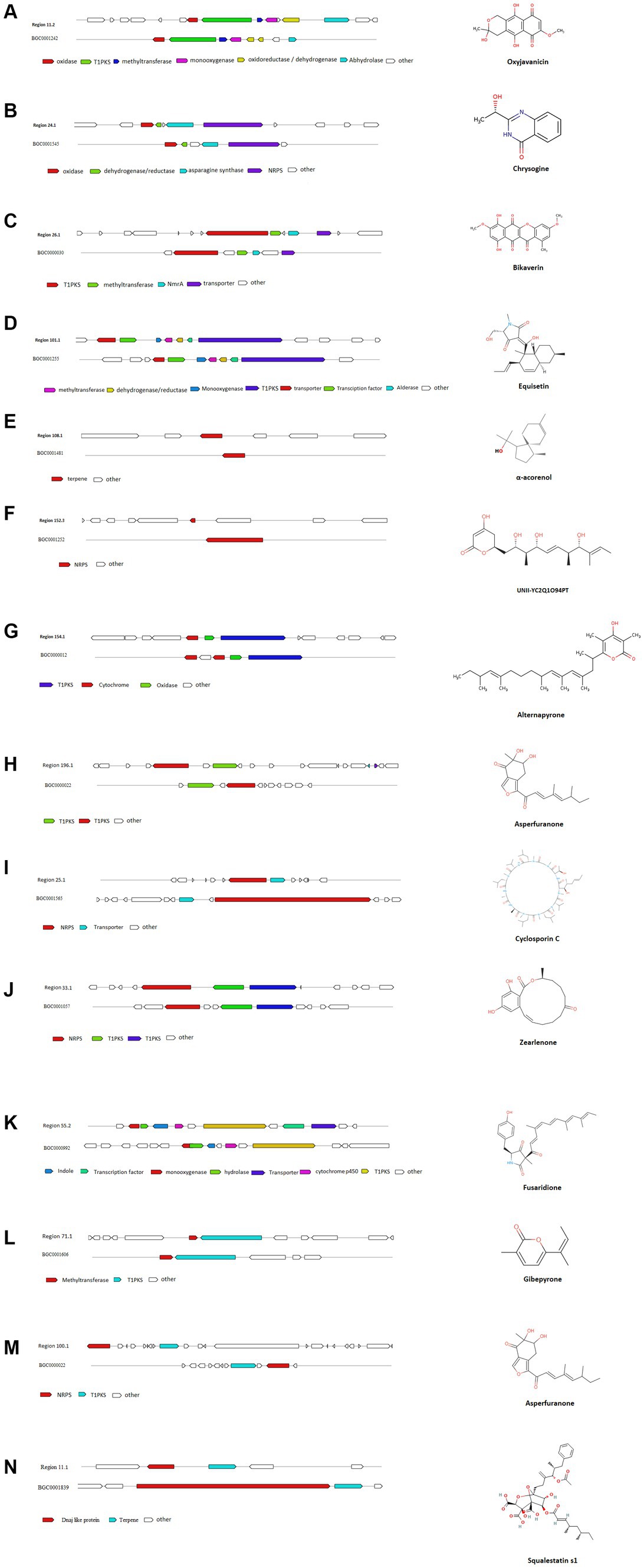
Figure 9. Comparison of biosynthetic gene cluster components in Fusarium indicum NFCCI 5145 with known biosynthetic gene clusters for biosynthesis of (A) Oxyjavanicin; (B) Chrysogine; (C) Bikaverin; (D) Equisetin; (E) α-Acorenol; (F) Unii-Yc2q1o94pt; (G) Alternapyrone; (H) Asperfuranone; (I) Cyclosporin C; (J) Zearalenone; (K) Fusaridione A; (L) Gibepyrone-A; (M) Asperfuranone; (N) Squalestatin S1.
AntiSMASH results revealed the potential of this isolate to produce interesting compounds, such as α-acorenol, oxyjavanicin, chrysogine, equisetin, bikaverin, squalestatin S1, and many other known and unknown secondary metabolites. α-Acorenol is a highly oxygenated sesquiterpene that has antioxidant activity (Elshamy et al., 2019). Oxyjavanicin belongs to the naphthoquinone class of compounds and is similar to Fusarubin (C15H14O7), a red antibiotic (Ruelius and Gauhe, 1950). Many studies have reported its antituberculosis, cytotoxic, and antimicrobial activities; it can be a beneficial drug for the treatment of asthma as it regulates cytokine balance in OVA-sensitized (Hong et al., 2022). Fusarubin is also known to inhibit proliferation and also increase apoptosis in cell lines derived from hematological cancers (Adorisio et al., 2019). Chrysogine is a yellow pigment known to lack antimicrobial or anticancer activity (Viggiano et al., 2018). Equisetin is more known for its antibiotic and cytotoxic activity; it also inhibits HIV-1 integrase. It can also potentiate antibiotic activity against multi-drug-resistant gram-negative bacteria (Zhang et al., 2021). It is known to inhibit bacterial acetyl-CoA carboxylase (ACC), which is the first step of fatty acid synthesis (Larson et al., 2020). Bikaverin belongs to the polyketide class of compounds and is a reddish pigment having many biological properties including antitumoral activity against different cancer cell lines (Limón et al., 2010). Squalestatin S1 is a potent inhibitor of squalene synthase with potential use in the control of cholesterol biosynthesis (Lebe and Cox, 2018). The functions associated with other cryptic BGCs can be characterized with the help of gene knockout and heterogeneous expression experiments, along with the help of LC-MS analysis.
3.8 Comparative phylogenetics and genomics
3.8.1 The average nucleotide identity
The average nucleotide identity (ANI) performed on the Fusarium indicum NFCCI 5145 genome provided an overall idea of the sequence identity between the Fusarium strains under comparison with the F. indicum strain NFCCI 5145 as depicted in the heatmap in Figure 10. Fusarium indicum NFCCI 5145 was grouped with F. concolor and F. austroafricanum, thus confirming the clustering of our strain Fusarium indicum NFCCI 5145 within the Fusarium concolor species complex. ANIb analysis included 51 genomes of various species of Fusarium.
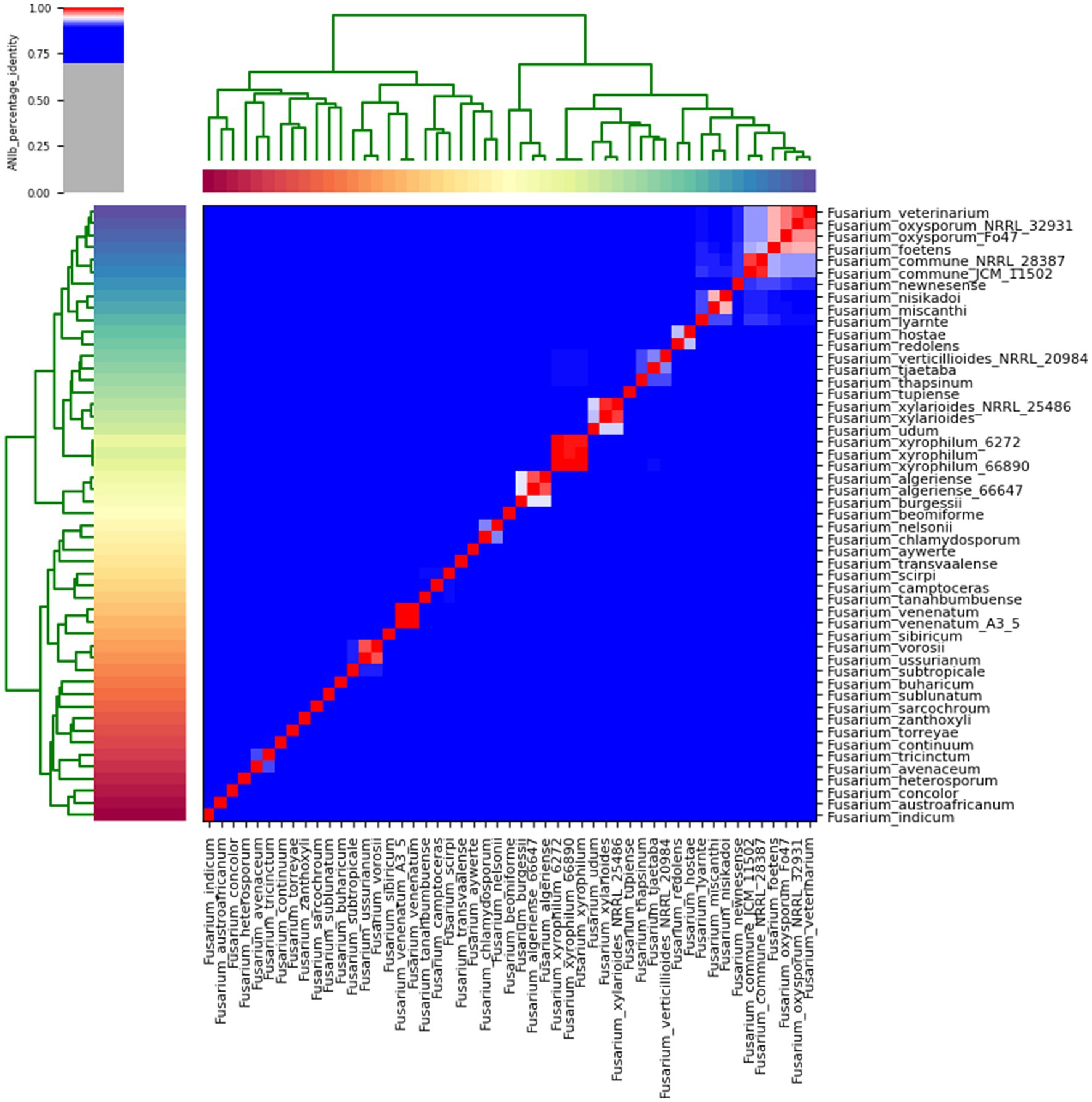
Figure 10. Heatmap of ANIb percentage identity between the Fusarium strains under comparison with the Fusarium indicum strain NFCCI 5145. ANIb was for all 51 genomes calculated based on genome sequences.
3.8.2 Phylogeny based on orthologous proteins
Fusarium indicum strain NFCCI 5145 was grouped in the Fusarium concolor species complex, which presently contains two species, F. concolor and F. austroafricanum. This was based on the phylogenetic tree constructed using the orthologous proteins of Fusarium strains, figured out using orthofinder. This confirms the clustering of the strain NFCCI 5145 in the Fusarium concolor species complex (Figure 11).
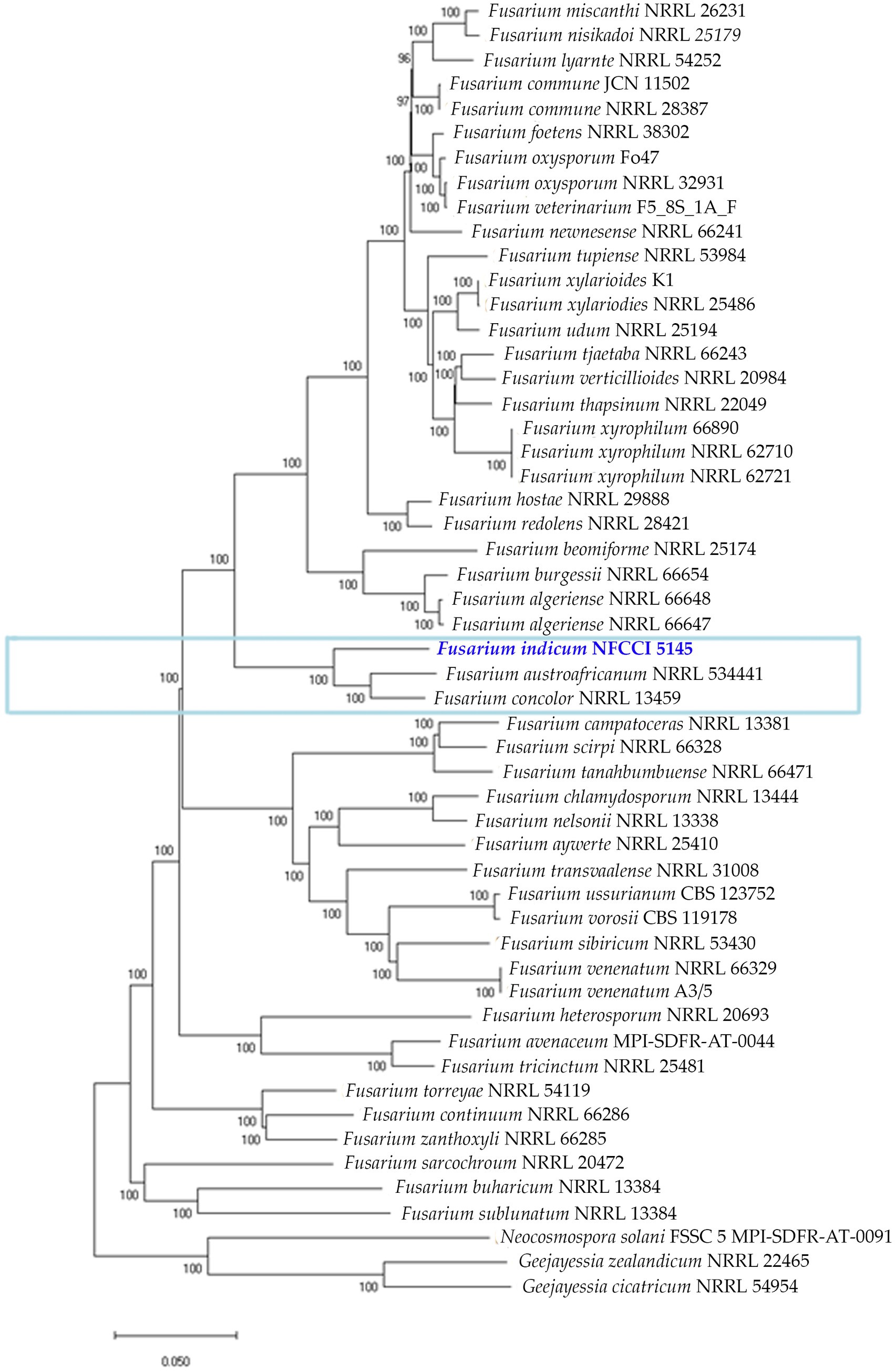
Figure 11. Phylogenetic analysis of 53 Fusarium strains based on the orthologous proteins identified using OrthoFinder. The new species is represented with blue bold and the Fusarium concolor species complex is marked with a blue rectangular box.
4 Conclusion
Species of Fusarium produce a variety of secondary metabolites. In this study, a novel endophytic, Fusarium indicum, isolated from Bambusa sp., was obtained and comprehensively examined by gene prediction and annotation. This isolate has many functional genes for energy production and conversion, amino acid and carbohydrate metabolism, secondary metabolites biosynthesis, transport, and catabolism. AntiSMASH analysis showed that it could produce secondary metabolites for drug development. According to the reports, species belonging to Fusarium concolor can produce ligninolytic enzymes, such as lignin peroxidase, laccase, and manganese peroxidase on wheat straw, thus leading to efficient delignification under solid-state fermentation conditions (Li et al., 2008). Species falling under this complex have also been known to treat poplar chemi-thermomechanical pulp to inhibit light-induced yellowing and enhance the brightness of the pulp (Daolei et al., 2020). Reports also establish the usage of an endophytic Fusarium concolor in synthesizing silver nanoparticles (Almeida et al., 2021). This newly isolated strain may be used for similar applications, such as the pretreatment of lignocellulose materials before bio-pulping or the bioconversion to fuel (Li et al., 2008). Nowadays, nanoparticles are used for the controlled release of pesticides or nanocides and the production of nanofertilizers. This isolate can be used to produce nanoparticles that may have great applicability in agriculture, biotechnology, and medicine.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
SR: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft. SS: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, and/or authorship this article. The publication of the article was supported by MACS-Agharkar Research Institute.
Acknowledgments
The authors thank Dr. Prashant Dhakephalkar, Director of MACS-Agharkar Research Institute, Pune, for providing the necessary facilities and encouragement to carry out the research work. SR acknowledges the University Grants Commission (U.G.C.), New Delhi, for granting a Senior Research Fellowship (SRF).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1266620/full#supplementary-material
Footnotes
1. ^https://github.com/OpenGene/fastp+
2. ^http://cab.spbu.ru/software/spades/
3. ^http://bioinf.uni-greifswald.de/webaugustus/
4. ^https://tandem.bu.edu/trf/trf.html
5. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn on date 18.07.2023
References
Aamir, S., Sutar, S., Singh, S. K., and Baghela, A. (2015). A rapid and efficient method of fungal genomic DNA extraction, suitable for PCR based molecular methods. Plant Pathol. Quar. J. Fungal Biolog. 5, 74–81. doi: 10.5943/ppq/5/2/6
Adorisio, S., Fierabracci, A., Muscari, I., Liberati, A. M., Cannarile, L., Thuy, T. T., et al. (2019). Fusarubin and anhydrofusarubin isolated from a Cladosporium species inhibit cell growth in human cancer cell lines. Toxins 11:503. doi: 10.3390/toxins11090503
Al-Hatmi, A. M., Hagen, F., Menken, S. B., Meis, J. F., and de Hoog, G. S. (2016). Global molecular epidemiology and genetic diversity of fusarium, a significant emerging group of human opportunists from 1958 to 2015. Emerg. Microbes Infect. 5:e124. doi: 10.1038/emi.2016.126
Almeida, A. F., Junior, A. C., and Bentes, J. L. S. (2021). Synthesis of silver nanoparticles (AgNPs) by fusarium concolor and inhibition of plant pathogens. Summa Phytopathol. 47, 9–15. doi: 10.1590/0100-5405/235097
Arney, K. L., Tiernan, R., and Judson, M. A. (1997). Primary pulmonary involvement of fusarium solani in a lung transplant recipient. Chest 112, 1128–1130. doi: 10.1378/chest.112.4.1128
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580. doi: 10.1093/nar/27.2.573
Bills, G. F., and Polishook, J. D. (1992). Recovery of endophytic fungi from Chamaecyparis thyoides. Sydowia 44, 1–12.
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., van Wezel, G. P., Medema, M. H., et al. (2021). AntiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Chambers, S. C. (1972). Fusarium species associated with wheat in Victoria. Aust. J. Exp. Agric. 12, 433–436. doi: 10.1071/EA9720433
Daolei, Z., Li, X., and Zhao, J. (2020). Fusarium concolor X4 pretreatment suppresses light-induced yellowing of high-yield pulp. Biomed. Res. Int. 2020, 1–8. doi: 10.1155/2020/9497215
Dobranic, J. K., Johnson, J. A., and Alikhan, Q. R. (1995). Isolation of endophytic fungi from eastern larch (Larix laricina) leaves from New Brunswick, Canada. Can. J. Microbiol. 41, 194–198. doi: 10.1139/m95-026
Elshamy, A. I., Abd-ElGawad, A. M., El-Amier, Y. A., El Gendy, A., and Al-Rowaily, S. L. (2019). Interspecific variation, antioxidant and allelopathic activity of the essential oil from three Launaea species growing naturally in heterogeneous habitats in Egypt. Flavour Fragr. J. 34, 316–328. doi: 10.1002/ffj.3512
Emms, D. M., and Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238–214. doi: 10.1186/S13059-019-1832-Y
Garron, M. L., and Henrissat, B. (2019). The continuing expansion of CAZymes and their families. Curr. Opin. Chem. Biol. 53, 82–87. doi: 10.1016/j.cbpa.2019.08.004
Hong, J., Wang, F., Yang, D., and Kan, J. (2022). Anti-asthmatic effect of fusarubin in an ovalbumin-induced asthmatic rat model. Trop. J. Pharm. Res. 21, 87–92. doi: 10.4314/tjpr.v21i1.14
Huntley, R. P., Harris, M. A., Alam-Faruque, Y., Blake, J. A., Carbon, S., Dietze, H., et al. (2014). A method for increasing expressivity of gene ontology annotations using a compositional approach. BMC Bioinformatics 15:155. doi: 10.1186/1471-2105-15-155
Ingle, A. P. (2017). Diversity and identity of fusarium species occurring on fruits, vegetables and food grains. Nusantara Biosci 9, 44–51. doi: 10.13057/nusbiosci/n090108
Jacobs-Venter, A., Laraba, I., Geiser, D. M., Busman, M., Vaughan, M. M., Proctor, R. H., et al. (2018). Molecular systematics of two sister clades, the fusarium concolor and F. Babinda species complexes, and the discovery of a novel microcycle macroconidium-producing species from South Africa. Mycologia 110, 1189–1204. doi: 10.1080/00275514.2018.1526619
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kornerup, A., and Wanscher, J. H. (1978). Metheun's handbook of colours, 3rd Edn. London: Metheun and Co. Ltd.
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Larson, E. C., Lim, A. L., Pond, C. D., Craft, M., Čavužić, M., Waldrop, G. L., et al. (2020). Pyrrolocin C and equisetin inhibit bacterial acetyl-CoA carboxylase. PloS One 15:e0233485. doi: 10.1371/journal.pone.0233485
Larsson, A. (2014). AliView: a fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 30, 3276–3278. doi: 10.1093/bioinformatics/btu531
Lebe, K. E., and Cox, R. J. (2018). Oxidative steps during the biosynthesis of squalestatin S1. Chem. Sci. 10, 1227–1231. doi: 10.1039/c8sc02615g
Leslie, J. F., and Summerell, B. A. (2006). The fusarium laboratory manual. Oxford, UK: Blackwell Publishing Ltd.
Li, L., Li, X. Z., Tang, W. Z., Zhao, J., and Qu, Y. B. (2008). Screening of a fungus capable of powerful and selective delignification on wheat straw. Lett. Appl. Microbiol. 47, 415–420. doi: 10.1111/j.1472-765X.2008.02447.x
Limón, M. C., Rodríguez-Ortiz, R., and Avalos, J. (2010). Bikaverin production and applications. Appl. Microbiol. Biotechnol. 87, 21–29. doi: 10.1007/s00253-010-2551-1
Link, H. F. (1809). Observationes in ordines plantarum naturales. Mag. Ges. Naturf. Freunde Berlin 3:10.
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Liu, Y., Xu, M., Tang, Y., Shao, Y., Wang, H., and Zhang, H. (2022). Genome features and AntiSMASH analysis of an endophytic strain fusarium sp. R1. Metabolites 12:521. doi: 10.3390/metabo12060521
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Murad, A., and Baiti, N. (2017). Genetic diversity of fusarium spp. isolated from fruit rot of banana in peninsular Malaysia. Masters thesis. [Malaysia]: Universiti Putra.
Natale, D. A., Shankavaram, U. T., Galperin, M. Y., Wolf, Y. I., Aravind, L., and Koonin, E. V. (2000). Towards understanding the first genome sequence of a crenarchaeon by genome annotation using clusters of orthologous groups of proteins (COGs). Genome Biol. 1:RESEARCH0009. doi: 10.1186/gb-2000-1-5-research0009
Nelson, P. E., Dignani, M. C., and Anaissie, E. J. (1994). Taxonomy, biology, and clinical aspects of fusarium species. Clin. Microbiol. Rev. 7, 479–504. doi: 10.1128/CMR.7.4.479-504.1994
Nelson, P. E., Toussoun, T. A., and Marasas, W. F. O. (1995). Neotypification and emended description of fusarium anguioides. Mycologia 87, 543–546. doi: 10.2307/3760773
Nguyen, L., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
O’Donnell, K., Kistler, H. C., Cigelnik, E., and Ploetz, R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. U. S. A. 95, 2044–2049. doi: 10.1073/pnas.95.5.2044
Pernas, L. (2021). Cellular metabolism in the defense against microbes. J. Cell Sc. 134:jcs252023. doi: 10.1242/jcs.252023
Pritchard, L., Glover, R. H., Humphris, S., Elphinstone, J. G., and Toth, I. K. (2016). Genomics and taxonomy in diagnostics for food security: softrotting enterobacterial plant pathogens. Anal. Methods 8, 12–24. doi: 10.1039/C5AY02550H
Reinking, O. A. (1934). Interesting new fusaria. Zentralblatt für Bakteriologie und Parasitenkunde Abteilung 2, 509–514.
Ruelius, H. W., and Gauhe, A. (1950). Uber Fusarubin, einen Naphthochinonfarbstoff aus Fusarien. Annalen der Chemie 569, 38–59. doi: 10.1002/jlac.19505690106
Saadabi, A. M. A. (2006). On the fungal Flora of Saudi Arabian soils. Res. J. Microbiol. 1, 280–284. doi: 10.3923/jm.2006.280.284
Torbati, M., Arzanlou, M., and da Silva Santos, A. C. (2021). Fungicolous fusarium species: ecology, diversity, isolation, and identification. Curr. Microbiol. 78, 2850–2859. doi: 10.1007/s00284-021-02584-9
Urban, M., Cuzick, A., Seager, J., Wood, V., Rutherford, K., Venkatesh, S. Y., et al. (2020). PHI-base: the pathogen-host interactions database. Nucleic Acids Res. 48, D613–D620. doi: 10.1093/nar/gkz904
Urban, M., Cuzick, A., Seager, J., Wood, V., Rutherford, K., Venkatesh, S. Y., et al. (2022). PHI-base in 2022: a multi-species phenotype database for pathogen-host interactions. Nucleic Acids Res. 50, D837–D847. doi: 10.1093/nar/gkab1037
Viggiano, A., Salo, O., Ali, H., Szymanski, W., Lankhorst, P. P., Nygård, Y., et al. (2018). Pathway for the biosynthesis of the pigment chrysogine by Penicillium chrysogenum. Appl. Environ. Microbiol. 84, e02246–e02217. doi: 10.1128/AEM.02246-17
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Vilgalys, R., and Sun, B. L. (1994). Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA-sequences. Proc. Natl. Acad. Sci. U. S. A. 91, 4599–4603. doi: 10.1073/pnas.91.10.4599
Wang, M. M., Crous, P. W., Sandoval-Denis, M., Han, S. L., Liu, F., Liang, J. M., et al. (2022). Fusarium and allied genera from China: species diversity and distribution. Persoonia 48, 1–53. doi: 10.3767/persoonia.2022.48.01
Yamada, Y., Hirakawa, H., Hori, K., Minakuchi, Y., Toyoda, A., Shitan, N., et al. (2021). Comparative analysis using the draft genome sequence of California poppy (Eschscholzia californica) for exploring the candidate genes involved in benzylisoquinoline alkaloid biosynthesis. Biosci. Biotechnol. Biochem. 85, 851–859. doi: 10.1093/bbb/zbaa091
Yli-Mattila, T., Hussien, T., Gavrilova, O., and Gagkaeva, T. (2018). Morphological and molecular variation between fusarium avenaceum, fusarium arthrosporioides and fusarium anguioides strains. Pathogens 7:94. doi: 10.3390/pathogens7040094
Young, N. A., Kwon-Chung, K. J., Kubota, T. T., Jennings, A. E., and Fisher, R. I. (1978). Disseminated infection by fusarium moniliforme during treatment for malignant lymphoma. J. Clin. Microbiol. 7, 589–594. doi: 10.1128/jcm.7.6.589-594.1978
Keywords: asexual morph, novel taxon, Fusarium, genome mining, antiSMASH, secondary metabolites
Citation: Rana S and Singh SK (2023) Insights into the genomic architecture of a newly discovered endophytic Fusarium species belonging to the Fusarium concolor complex from India. Front. Microbiol. 14:1266620. doi: 10.3389/fmicb.2023.1266620
Edited by:
Rajesh Jeewon, University of Mauritius, MauritiusReviewed by:
Bernhard Timo Werner, University of Giessen, GermanyDurgesh K. Jaiswal, Graphic Era University, India
Sunil Kumar Deshmukh, The Energy and Resources Institute (TERI), India
Copyright © 2023 Rana and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sanjay K. Singh, sksingh@aripune.org
 Shiwali Rana
Shiwali Rana Sanjay K. Singh
Sanjay K. Singh