- 1ICAR-Directorate of Rapeseed-Mustard Research, Bharatpur, India
- 2Genetics Division, ICAR-Indian Agricultural Research Institute, New Delhi, India
Brassica juncea is a major oilseed crop in tropical and subtropical countries, especially in south-east Asia like India, China, Bangladesh, and Pakistan. The widespread cultivation of genetically similar varieties tends to attract fungal pathogens which cause heavy yield losses in the absence of resistant sources. The conventional disease management techniques are often expensive, have limited efficacy, and cause additional harm to the environment. A substantial approach is to identify and use of resistance sources within the Brassica hosts and other non-hosts to ensure sustainable oilseed crop production. In the present review, we discuss six major fungal pathogens of B. juncea: Sclerotinia stem rot (Sclerotinia sclerotiorum), Alternaria blight (Alternaria brassicae), White rust (Albugo candida), Downy mildew (Hyaloperonospora parasitica), Powdery mildew (Erysiphe cruciferarum), and Blackleg (Leptoshaeria maculans). From discussing studies on pathogen prevalence in B. juncea, the review then focuses on highlighting the resistance sources and quantitative trait loci/gene identified so far from Brassicaceae and non-filial sources against these fungal pathogens. The problems in the identification of resistance sources for B. juncea concerning genome complexity in host subpopulation and pathotypes were addressed. Emphasis has been laid on more elaborate and coordinated research to identify and deploy R genes, robust techniques, and research materials. Examples of fully characterized genes conferring resistance have been discussed that can be transformed into B. juncea using advanced genomics tools. Lastly, effective strategies for B. juncea improvement through introgression of novel R genes, development of pre-breeding resistant lines, characterization of pathotypes, and defense-related secondary metabolites have been provided suggesting the plan for the development of resistant B. juncea.
Introduction
The Brassicaceae family has about 3709 species and 338 genera, displaying enormous diversity and used as a source of oil, vegetables, mustard condiments, and fodder (Warwick et al., 2006). Brassica juncea (L.) Czern & Coss is a natural amphidiploid (AABB, 2n = 36) of Brassica rapa (AA, 2n = 20) and Brassica nigra (BB, 2n = 16) belonging to this family and cultivated worldwide for its edible oil. The estimated area, production, and yield of rapeseed-mustard in the world were 36.59 Mha, 72.37 Mt, and 1980 kg/ha, respectively, during 2018–2019. India contributed 19.8 and 9.8% to global acreage and production, respectively (USDA, 2020). It is presumed that this species originated approximately 0.039-0.055 million years ago (Yang et al., 2016) and the present B. juncea species evolved through chromosomal triplications and other re-arrangements (Lysak et al., 2005). The linkage map of B. juncea showed that both parental genomes were conserved and have remained unchanged since hybridization (Axelsson et al., 2000). This evolution theory is supported by the genome assemblies of B. rapa (Wang et al., 2011; Cai et al., 2017; Zhang et al., 2018), B. nigra (Yang et al., 2016; Perumal et al., 2020), B. juncea (Yang et al., 2016; Paritosh et al., 2020), B. napus (Chalhoub et al., 2014; Bayer et al., 2017; Lee et al., 2020), and the pan-genomes of B. napus (Song et al., 2020). The A and B genomes of B. juncea belong to the two different lineages of the Brassicaceae family namely Nigra and Rapa/Oleracea, thus contains the most dissimilar genomes (Kaur et al., 2014).
The genetic uniformity between all cultivated varieties of B. juncea makes it vulnerable to pathogen attacks. The major biotic diseases and pathogens of mustard that cause a serious threat to B. juncea worldwide are Alternaria blight [Alternaria brassicae (Berk.) Sacc.; A. brassicicola (Schwein.) Wiltshire; A. raphani Groves & Skolko], Sclerotinia stem rot [Sclerotinia sclerotiorum (Lib.) deBary], White rust [Albugo candida (Pers.) Kunze], Clubroot [Plasmodiophora brassicae Woronin], Powdery mildew [Erysiphe polygoni DC.], Blackleg [Leptosphaeria maculans (Desmaz.) Ces.& De Not.], and Downy mildew [Hyaloperonospora parasitica (Pers.: Fr.) Fr.] (Williams and Saha, 1993). These diseases cause serious damage to mustard production worldwide owing to the lack of vertical and horizontal resistance against pathogens in current cultivating varieties. Disease management through chemical fungicides is not climate-resilient and also economically not suitable. However, despite cultural adaptations very limited efficiency in disease management has been demonstrated. The host’s genetic resistance is the most effective and consistent means to control diseases (Ren et al., 2016). Fortunately, the resistant gene(s) and quantitative trait locus (QTLs) for most of the aforementioned pathogens have been identified within the Brassicaceae family, with some wild members as a treasure trove of resistant genes (Table 1).

Table 1. The resistance genes/QTLs that have been mapped in Brassicaceae family in relation of major fungal pathogens of B. juncea (Sclerotinia sclerotiorum, Alternaria brassicae, Albugo candida, Hyaloperonospora parasitica, Erysiphe cruciferarum, Leptosphaeria maculans).
The plants respond to pathogens either through constitutive or inducible defense mechanisms to protect themselves from damage. Constitutive defense includes preformed physical barriers such as cell walls, cuticle, wax on the outer surface, bark, and cork layers. Alternately, plants produce toxic chemicals, cell lytic enzymes, and deliberate cell suicide when attacked by pathogens, which form the inducible defense systems. The inducible defense system of the plants activated only after a pathogen is recognized by his immune responsive system because of the production of chemicals or proteins. This leads to high energy costs and nutrient requirements in association with their production and management. A lot of plant pathogens, known as biotrophs, make a close connection with their host plant suppressing its defense system but keeping their host alive, use the plant’s nutrients for their growth and reproduction. However, some pathogens produced toxins, cell degrading enzymes, or proteins to overcome plant defense systems promoting the rapid release of nutrients called necrotrophs. The pathogens can infect more than one host plant called the host range. Similarly, plants can defend themselves against a particular, or wide range of pathogens, or races of the pathogen. This type of plant defense is called vertical and horizontal resistance, respectively. The vertical resistance of the host plant can protect from a particular type of pathogen or race with high intensity but does not last long against the pathogen. The vertical resistance of the host plants is due to one or two major genes and is highly specific. However, the horizontal resistance is durable and it can protect host plants against a wide range of pathogens and races. In this, a group of genes simultaneously take part in host defense and present within the QTL. The genes that take part in the host defense systems are called R genes. The plant pathogenic fungi and bacteria have Avr (avirulence) genes to identify their host plants and infect them. The Avr genes produce small secreted protein (SSP) effectors and secondary metabolites (SM), the key elements of their pathogenesis that regularize innate immunity of the host plant and facilitate infection. However, the host resistance can be established only when the R gene of the host recognizes the effectors or secondary metabolites. This recognition usually triggers its defense responses including the hypersensitive response (HR) and results in resistance from the plant to the pathogen (Freeman and Beattie, 2008).
Apart from resistance genes, plants also resist pathogenic attacks via hormone signaling pathways such as Jasmonates (JAs), Salicylates (SAs), Brassinosteroids (BR), Ethylene (ET), Abscisic acid (ABA), and Phosphoglycerolipids (PGL). Out of all the above signaling pathways, JAs and SAs are reported most effective against pathogenic diseases. The polyunsaturated fatty acids (PUFA), a group of plant oxylipins produced Jasmonates (JAs) through oxidation by using one of the seven different branches of the lipoxygenase (LOX) pathway. However, the key component of JA biosynthesis is allene oxide synthase (AOS) that utilizes 13-hydroperoxide from α-linolenic acid (18:3, α-LeA). PUFA is released from the chloroplast membrane and converted into 9 or 13-hydroperoxides through the LOX pathway, which further enters into the AOS pathway to produce free JA. There are a lot of JA derivatives present in the host plants such as free JA, methyl-jasmonate (Me-JA), cis-jasmone, Jasmonyl isoleucine (JA-Ile), and Jasmonoyl ACC (JA-ACC). The role of free JA and Me-JA is well established under various biotic and abiotic stress conditions (Sirhindi et al., 2017). Salicylic acid (SA) is a phenolic plant hormone that actively participates in defense mechanisms under biotic and abiotic stresses to protect the host plant. For instance, the basal resistance for the fungal pathogen B. cinerea is regulated by SA in tomato but by jasmonic acid and ethylene in tobacco (Loon et al., 2006). The key enzymes of the phosphoglycerolipid (PGL) pathways are phospholipase C (PLC) and phospholipase D (PLD) capable of producing phosphatidic acid (PA). Phosphatidic acid acts as a signaling molecule by binding to target proteins altering their subcellular localization and enzymatic activity. This signaling pathway is activated during plant exposure to both biotic and abiotic stresses (Pokotylo et al., 2017). The hormone signaling pathways functions systemically in host plants, thus labeled as the systemic acquired resistance (SAR) effective against a wide range of pathogens. These plant defense mechanisms regulated through resistance genes and hormone signaling pathways under strict control (Larkan et al., 2013; Ma and Borhan, 2015) exhibited highly complex and specific immune responses (Stotz et al., 2014). Very few R genes have been characterized and cloned as yet, thus resistance breeding programs are slow as they depend on chemical management. However, the next-generation sequencing projects have developed highly specific thorough genomic data of diploid and polyploidy plants, providing an opportunity to identify resistance genes.
The present study aims to review the work done on identifying the sources of these resistance genes/QTLs across the Brassicaceae family. This knowledge may help improve the resistance of B. juncea by focusing on major pathogens and corresponding diseases such as Sclerotinia stem rot, Alternaria blight, White rust, Downy mildew, and Powdery mildew (Figure 1). We shall confirm the sources of resistance genes for these pathogens in Brassicaceae to support their potential use in resistance breeding of B. juncea. Also, the challenges in identification and the use of resistance genes, and effective strategies to use resistance genes in the development of resistant B. juncea lines for such major pathogens will be discussed.

Figure 1. Major fungal diseases of B. juncea. (A) Sclerotinia stem rot; (B) Alternaria leaf blight; (C) Staghead formation in flower buds of B. juncea by A. candida; (D,E) Downy mildew symptoms at lower surface of cotyledon and mature leaf, respectively (Photo courtesy: A. K. Tewari); (F) Powdery mildew of B. juncea.
Resistance Sources for Major Pathogens
Sclerotinia Stem Rot
The Sclerotinia sclerotiorum (Lib.) deBary is a necrotrophic fungal pathogen causing stem rot disease in B. juncea worldwide (Ghasolia et al., 2004; Singh et al., 2008). Severe outbreaks of stem rot disease under cool and moist weather conditions (Purdy, 1979; Saharan and Mehta, 2008) cause heavy yield losses ranging between 5 to 100% (Ghasolia et al., 2004; McDonald and Boland, 2004; Shukla, 2005). Sclerotinia stem rot (SSR) is also now recognized as a major yield-limiting disease of B. napus in Australia, Canada, China, Europe, and India (Rana et al., 2017). However, the management of stem rot by traditional and/or biochemical practices has been a cumbersome process, owing to its wide host range (about 408 plant species of 75 families) and the potential to survive in harsh environmental conditions by forming sclerotia (Boland and Hall, 1994; Peltier et al., 2012). Genetic diversity studies of S. sclerotiorum have been conducted since last three decades in different parts of the world including Australia, Brazil, Canada, India, Iran, United Kingdom, and the United States (Kohn et al., 1991; Sexton et al., 2006; Hemmati et al., 2009; Litholdo et al., 2011; Barari et al., 2012; Clarkson et al., 2012; Mandal and Dubey, 2012; Attanayake et al., 2014). Therefore, instead of using traditional practices and chemicals, the cultivation of resistant varieties is the most economical and climate-resilient approach to manage SSR in the mustard crop. Resistant cultivars can efficiently minimize yield losses due to the disease (Westman et al., 1999; Taylor et al., 2002). Thus, the resistance sources were investigated within the Brassicaceae and out of the family to identify potential R genes for B. juncea resistance breedings and introgressions. The resistance screening experiments were mainly focused on leaf and stem at cotyledonary and adult plant stages under in vitro and/or field conditions (Garg et al., 2008; Li et al., 2008; Barbetti et al., 2014; Uloth et al., 2014; Ge et al., 2015; Naher et al., 2018; Kumari and Singh, 2019).
The species displaying a promising resistance against the SSR disease are Brassica fruticulosa, wild Brassica oleracea, Brassica napus, Camelina sativa, Diplotaxis tenuisiliqua, Erucastrum abyssinicum, E. cardaminoides, and Sinapis alba (Morrall and Dueck, 1982; Kolte, 1985; Garg et al., 2010; Mei et al., 2013; Wu et al., 2013; Rana et al., 2017, 2019; Purnamasari et al., 2019). These species were used to introgress resistance in the cultivated oilseed Brassicas. The introgression lines were developed through somatic hybridization or embryo rescue, followed by subsequent backcrossing and/or selfing. The wild crucifers viz. Erucastrum cardaminoides, Diplotaxis tenuisiliqua, and E. abyssinicum were used for resistance introgressions in B. juncea and B. napus and then evaluated for a higher level of resistance as compared to 54 Brassica germplasms of Australia, China, and India (Garg et al., 2010). These introgression lines were then used to locate the candidate gene or QTL for S. sclerotiorum resistance and physically mapped through advanced molecular tools. Rana et al. (2017) produced fertile B. juncea–B. fruticulosa introgression lines with segmental introgressions in B-genome chromosomes at terminal positions. All these lines were resistant and genotyped with 262 microsatellite markers. They have identified a total of ten significant marker-trait associations through association mapping. In another study, Rana et al. (2019) developed a set of 96 B. juncea–Erucastrum cardaminoid introgression lines for SSR resistance. All these introgression lines were genotyped by transferable microsatellite markers, followed by genotyping by sequencing to establish marker-trait associations. They have detected a total of six marker loci in both A and B genomes associated with resistance through SSR markers. However, the genome-wide association analysis (GWAS) identified a large number of single nucleotide polymorphisms (SNPs) linked to resistance in A03, A06, and B03 chromosomes. The annotation studies identified an array of resistance mechanisms, i.e., signal transduction pathways, hypersensitive responses, and production of anti-fungal proteins and metabolites. A total of five SNPs on the A03 chromosome were found to be associated with LRR-RLK genes that encoded LRR-protein kinase family proteins. They also predicted genetic factors associated with pathogen-associated molecular patterns (PAMPs) and effector-triggered immunity (ETI) on the A03 chromosome. These belonged to three R-Genes encoding TIR-NBS-LRR proteins. Hitherto, no resistant gene has been cloned for this disease (Wu et al., 2016, 2019).
The population derived from the resistant wild B. oleracea (B. incana) with the susceptible B. oleracea var. alboglabra was used to identify QTL for SSR. The F2 population exhibited a total of 12 and 6 QTLs for leaves and stem resistance, respectively. The candidate R genes were identified for Sclerotinia resistance on C09 of B. oleracea by blasting the sequence of adjacent markers to the B. rapa reference genome. The 12.8-cM genetic region on C09 contains two major QTLs for both leaf and stem resistance, while the corresponding 2.7-Mb genomic region on chromosome A09 of B. rapa harbors about 30 genes putatively encoding resistance-related and defense associated proteins. The putative genes present on the A09 chromosome belonged to the CC-NBS-LRR class (Mei et al., 2013).
Furthermore, the B. napus has been evaluated by many to identify putative QTLs for SSR resistance. The GWAS for SSR resistance in B. napus had identified three putative QTLs on C04, C06, and C08 (Wu et al., 2016), A08, C06, and C09 chromosomes (Gyawali et al., 2016). The chromosome A02, A03, C02, and C06 of B. napus were identified to have QTLs for SSR resistance through SNP-array genotyping. However, it was recorded that these genomic regions also harbored QTLs for flowering time (Wu et al., 2019). Mei et al. (2020) developed a BC1F8 population of B. napus var. Zhongshuang 9 and B. incana for pyramiding of three resistant QTLs for SSR present on C01, C09-1, and C09-2 chromosomes. They concluded that all QTLs worked cumulatively for disease and the lines having all three QTLs showed the highest resistant level. Qasim et al. (2020) identified three major QTLs through comparative transcriptomic studies harboring 36 putative candidate genes from resistant B. napus lines that might actively involved in SSR resistance. Wu et al. (2013) identified 10 and 3 QTLs for stem and leaf resistance in B. napus, respectively. Out of these, a major QTL present on the C06 chromosome was associated with the BnaC.IGMT5, a candidate resistant-gene for SSR. Theoretically, there could be a strong correlation between leaf and stem resistance as noticed in various studies of Brassica species (Mei et al., 2012; Wu et al., 2013; You et al., 2016). Yet, this observation could not be correlated practically when some genotypes of Brassica were evaluated for SSR resistance (Uloth et al., 2013; You et al., 2016). Nevertheless, some other genotypes share common genomic regions for leaf and stem resistance (Li et al., 2015). Such contradictory findings indicate that the resistance against SSR in leaves and stem is specific at subpopulation levels. The inheritance of common resistance genes on both genomes (A and C) indicated a common ancestry of chromosomes (Wei et al., 2014).
Alternaria Blight
Alternaria is a key pathogenic genera of the family Brassicaceae, causing severe damage to host plants and one of the major yield-limiting factors. The Alternaria genus was first identified by Nees Von Esenbeck (1817) and Berkeley (1836) as a causal pathogen of leaf blight on Brassicaceae plants as Macrosporium brassicae Berk., which was later renamed as Alternaria brassicae (Berk.) (Saccardo, 1886). The blight disease of B. juncea is responsible for heavy yield losses ranging upto 47% in India (Kolte, 1985) and 32–57% in Nepal (Shrestha et al., 2005). The pathogen has a wide host range in cruciferous crops and is reported from all parts of the world including Canada (Berkenkamp and Kirkham, 1989; Conn and Tewari, 1990), India (Kadian and Saharan, 1983), Iran (Nourani et al., 2008), Italy (Tosi and Zezzerini, 1985), Poland (Nowicki et al., 2012), and United States, United Kingdom, Europe (Gladders, 1987). This pathogen attacks leaves, stem, and siliquae at all growth stages producing characteristic disease symptoms. However, the blighted spots produced by A. brassicae are gray as compared to the black sooty velvety spots of A. brassicicola, another leaf blight pathogen of mustard (Meena et al., 2010; Kumari and Singh, 2019). The infection spot leads to the formation of a necrotic zone which looks like a target board due to the interrupted growth of the pathogen. The pathogen resides at the center of the lesion surrounded by an uninvaded chlorotic halo created by the diffusion of pathogen toxins (Tewari, 1983; Agarwal et al., 1997). The Indian isolates of A. brassicae were grouped under three categories according to their virulence level as A (highly virulent), C (moderately virulent), and D (avirulent) collected from all over India (Vishwanath and Kolte, 1997). A total of 32 isolates of A. brassicae were examined by Random Amplified Polymorphic DNA (RAPD) and Inter Simple Sequence Repeat (ISSR) markers for genetic variability. The results of the study revealed that there was no clear grouping of isolates but little variations were recorded which may be due to ecological or survival-based adaptations (Sharma et al., 2013).
The pathogen has a very high survival rate on Brassica host debris and other alternative hosts (Anagallis arvensis, Convolvulus arvensis) carry the inoculum from one crop season to another (Tripathi and Kaushik, 1984; Verma and Saharan, 1994; Mehta et al., 2002). Thus, the air-borne spores of A. brassicae are the prime source of the inoculum of this polycyclic disease (Kolte, 1985). The pathogen can infect host plants through epidermis penetration, stomata, insects- or wounds. However, A. brassicae preferred stomatal penetration while A. brassicicola penetrated directly through the epidermis (Nowicki et al., 2012). The A. brassicae produced host-specific toxins (HST) during infection characterized as ABR toxin protein (Parada et al., 2008; Wight et al., 2009). This compound plays a vital role in the pathogenesis, also determining the virulence and pathogenicity level of the pathogen (Nishimura and Kohmoto, 1983). However, some additional toxins like destruxin B (Bains and Tewari, 1987) and it’s derivatives (homodestruxin B, desmethyldestruxin B, and destruxin B2) are also produced. All these toxins are responsible for the typical tissue necrosis and chlorosis. These toxins stimulate the phytoalexins, brassilexin, and sinalbin A (Sodelade et al., 2012). The A. brassicae produces abscisic acid causing premature leaf aging and defoliation, dropping of flowers, and premature siliquae breaking (Tewari, 1991b), while cytokines are responsible for green discolorations within the blighted spots (Tylkowska et al., 2004).
It was recorded that when Brassica rapa seedlings were inoculated with the fungal pathogen A. brassicae, 3-butenyl, and 4-pentenyl isothiocyanates were released together with dimethyl disulfide, dimethyl trisulphide, 4-oxoisophorone, and several sesquiterpenes (Kevin et al., 1996). The release of isothiocyanates is evidence for the catabolism of glucosinolates during infection, which is a prerequisite for their involvement in resistance (Doughty et al., 1996). The host resistance is the most economical and feasible mode to control yield losses. This approach allows us to decrease pesticide usage and also lowers the chemical pollution. The resistance breeding programs for Brassica improvement are hindered due to bottlenecks in the resistance introgression from the wild species into the commercial crops. The cultivated vegetable and oilseed Brassicas are devoid of genetic resistance against Alternaria blight pathogens. However, among the Brassica crops, the Ethiopian mustard has displayed the highest level of resistance against A. brassicae. The wild members of the Brassicaceae family such as S. alba (Kolte, 1985; Brun et al., 1987; Ripley et al., 1992; Sharma and Singh, 1992; Hansen and Earle, 1995, 1997; Kumari et al., 2018, 2020b), Camelina sativa, Capsella bursa-pastoris, Eruca sativa, Neslia paniculata, Alliaria petiolata, Barbarea vulgaris, Brassica elongate, B. desnottessi, B. fruticulosa, B. maurorum, B. nigra, B. souliei, B. spinescens, Camelina sativa, Capsella bursa-pastoris, Coincya spp., Diplotaxis catholica, D. berthautii, D. creacea, D. erucoides, D. tenuifolia, Erucastrum gallicum, Eruca vesicaria subsp. sativa, Hemicrambe fruticulosa, H. matronalis, Neslia paniculata, Raphanus sativus, and S. arvensis (Conn and Tewari, 1986; Brun et al., 1987; Conn et al., 1988; Tewari, 1991a; Zhu and Spanier, 1991; Tewari and Conn, 1993; Sharma et al., 2002; Warwick, 2011) reportedly possess the highest level of resistance against A. brassicae.
Camelina sativa produces phytoalexin on the landing of conidia on leaves and accumulates near and under conidial drops. This phytoalexin accumulation is a rapid response initiated just after conidial inoculation solely responsible for the inhibition of pathogen growth on the leaf surface (Jejelowo et al., 1991). The C. sativa produces two types of phytoalexins in leaves after elicitation by the pathogen, namely camalexin (C11N8N2S) and 6-methoxycamalexin (C H N SO) (Browne et al., 1991). The pathogenic attack elicits the production of camalexin, 6-methyloxycamalexin, and N-methylcamalexin (C H N S) in Capsella bursa-pastoris (Jimenez et al., 1997). The mutant Arabidopsis (phytoalexin deficient) is devoid of camalexin biosynthesis displaying higher susceptibility to A. brassicicola, establishing the contribution of camalexin in host resistance (Thomma et al., 1999b). However, it appears that the defense against A. brassicicola in Arabidopsis depends on multiple components, as Jasmonate insensitive Arabidopsis mutant coi1 was found vulnerable to this pathogen. This indicates that both components contribute to host resistance against A. brassicicola pathogen separately but in parallel pathways (Thomma et al., 1998, 1999a, 2000, 2001). The pathogen initiates signaling cascade through MAPK3 inducing the expression of LOX which further regulates the expression of MAPK3. Interaction of both the genes initiates the Jasmonate acid biosynthesis which induces the defense mechanism in B. juncea (Taj et al., 2011). The S. alba produces two types of phytoalexins (sinalexin) sinalbins A and B in the leaf and stem tissues for resistance against the A. brassicae (Pedras and Smith, 1997; Pedras and Zaharia, 2000). The variation in the elicitation level of phytoalexin is believed to be responsible for resistance status in plants (Conn et al., 1988).
Nayanakantha et al. (2016) examined five defense-related genes viz., PR-1, PR-2, PR-3, non-expresser of PR-1 (NPR-1), and plant defensin (PDF1.2) after inoculation of the seedlings of B. juncea and S. alba. The transcripts of all five defense-related genes accumulated locally as well as systemically at a greater level and earlier in S. alba than in B. juncea upon challenge inoculation with A. brassicae. The PDF1.2 was induced by SA and PR-1 by JA in both B. juncea and S. alba resulting in both JA and SA responsive genes conferring resistance against A. brassicae in S. alba. Similarly, upon challenging with A. brassicicola, the S. alba plants showed higher accumulation of abscisic acid and JA than B. juncea (Mazumder et al., 2013). The molecular basis of resistance could not be uncovered in S. alba due to the lack of molecular markers. However, the recently developed de novo genome assembly and a large number of microsatellites may assist to identify the potential genomic regions of resistance (Kumari et al., 2020a). The transgenic B. juncea cv. RLM-198 for hevein (a chitin binding lectin protein) was developed and evaluated against A. brassicae infection under glasshouse conditions. It was found that transgenics showed higher resistance levels for diseases such as longer incubation and latent period, smaller necrotic lesion size, lower disease intensity, and delayed senescence (Kanrar et al., 2002). Chhikara et al. (2012) developed transgenic B. juncea plants with barley antifungal genes class II chitinase (AAA56786) and type I ribosome-inactivating protein (RIP; AAA32951) to coexpress. These transgenics were evaluated in vitro and greenhouse conditions for resistance against A. brassicae. The study revealed a 44% reduction in hyphal growth, delayed onset of the disease, and a limited number of lesions as compared to wild resistant plants (Mondal et al., 2003). The transgenic B. juncea for tomato glucanase conferred higher resistance against A. brassicae under in vitro and polyhouse conditions (Mondal et al., 2007). Rajarammohan et al. (2017) developed three bi-parental mapping populations from three resistant (CIBC-5, Ei-2, and Cvi-0) and two susceptible (Gre-0 and Zdr-1) accessions. A total of five population-specific and one non-specific QTLs were identified governing resistance against A. brassicae. The presence of population-specific and non-specific QTLs indicated the quantitative nature of resistance to A. brassicae. The RtAbeCvG2-1 QTL consisted of 55 probable candidate genes with known functions in disease resistance pathways. A total of 27 candidate genes were identified at RtAbeCZ5-1 QTL on chromosome 5, out of which 24 belong to TIR-NBS-LRR and three genes to the CC-NBS-LRR class. The other genes present in this region involved in defense signaling pathways include BIR1 (BAK1-interacting receptor-like kinase), MYC3 (JAZ-interacting transcription factor), ERFs (Ethylene response factors), BRG (Botrytis-induced Related Gene), RBOHD (Respiratory Burst Oxidative Homolog D), NPR3 (Non-expressor of PR3) including 14 defensin and defensin-like genes known for antimicrobial/antifungal activities. The exogenous application of β-Aminobutyric Acid to B. juncea prevented plants from A. brassicae infection independent of SA and JA signaling pathways (Kamble and Bhargava, 2007). The cysteine-rich antimicrobial peptide (PmAMP1) from Pinus monticola provides resistance to B. napus against multiple fungal plant pathogens including A. brassicae, Leptosphaeria maculans, and S. sclerotiorum (Verma et al., 2012). Vishwanath et al. (1999) induced resistance in susceptible B. juncea cv. PR-15 against highly virulent isolate-A of A. brassicae by using avirulent isolate-D.
White Rust
Albugo candida (Pers. Ex. Lev.) Kuntze. is an obligate parasite of the oilseed Brassicas causing white rust and staghead disease worldwide including Brazil (Viegas and Teixeira, 1943), Canada (Petrie, 1973), China (Zhang et al., 1984), Germany (Klemm, 1938), India (Chowdhary, 1944), Japan (Hirata, 1954), Korea (Choi et al., 2011), New Zealand (Hammett, 1969), Pakistan (Perwaiz et al., 1969), Palestine (Rayss, 1938), Romania (Savulescu, 1946), Turkey (Bremer et al., 1947), United Kingdom (Berkeley, 1848), and United States (Walker, 1957). The disease can be identified as localized white to pale cream-colored spots on leaves and inflorescence (hypertrophied flowers). The formation of staghead is accounted for complete loss of seed formation which causes upto 90% yield loss. The yield loss is dependent on the disease severity and has been reported upto 60% in Brassica rapa L. in Canada (Petrie, 1973; Petrie and Vanterpool, 1974; Harper and Pittman, 1974), 23–89.8% of B. juncea in India (Bains and Jhooty, 1979; Lakra and Saharan, 1989), and about 5–10% in Australia (Barbetti, 1981; Barbetti and Carter, 1986). The A. candida has reportedly infected more than 63 genera and 241 species of the Brassicaceae family from all over the world (Farr et al., 1989; Saharan and Verma, 1992; Choi et al., 2007). The fungal pathogen can also infect plant species outside of the Brassicaceae family (Choi et al., 2006, 2007, 2008, 2009, 2011). The infection of pathogen leads to many biochemical changes in the host for a successful establishment of the pathogen and causes disease. The chlorophyll, sugars, and total phenol content was found in higher concentrations in resistant cultivars than the susceptible ones during all growth stages. However, the number of total proteins and free amino acids were increased in the susceptible cultivars in their life cycle (Singh, 2000). Gupta et al. (1997) identified a positive role of chlorophyll in A. candida resistance in B. juncea. A higher concentration of auxins (indole-3-acetonitrile [IAN], indole-3-acetic acid [IAA]) is produced in A. candida infected plants which lead to hyperplasia and hypertrophy of leaf, stem, and floral parts (Kiermayer, 1958). The abundance of nineteen proteins was found variable between the susceptible and resistant varieties of B. juncea after the pathogen invasion and out of these nineteen proteins, five were present in the resistant variety (Kaur et al., 2011a).
Cheung et al. (1998) identified Restriction Fragment Length Polymorphism (RFLP) markers linked to the white rust resistant gene (Acr) in B. juncea. The genotype non-specific intron polymorphic (IP) markers At5g41560 and At2g36360 derived from A. thaliana were successfully validated to have a close link with white rust resistant loci AcB1-A4.1 and AcB1-A5.1 of B. juncea, respectively (Singh et al., 2015). The mapping (RAC1, RAC2, and RAC3) and cloning of white rust-resistant genes (RAC1) were reported from A. thaliana (Accession: Ksk-1 and Ksk-2) for resistance against A. candida. The RAC genes belong to the Drosophila toll/interleukin-1 receptor (TIR) nucleotide-binding site leucine-rich repeat (NB-LRR) class of plant resistant gene (Borhan et al., 2001). Another white rust resistant gene, WRR4, was cloned from A. thaliana (Accession: Col) (Borhan et al., 2008) which encodes for the same TIR-NB-LRR protein and was found to be resistant against three races of A. thaliana (race 2,4,7, and 9). The expression of WRR4 in susceptible lines of B. juncea and B. napus provided complete resistance against white rust pathogen belonging to the races 2 and 7, respectively (Borhan et al., 2010).
A large number of resistant genes were identified in different Brassica species such as in B. juncea (Acr – Cheung et al., 1998; AC-21 – Prabhu et al., 1998; AC-2 – Varshney et al., 2004; ACB1-A4.1 and ACB1-a5.1 – Massand et al., 2010), B. rapa (ACA1 - Kole et al., 1996), B. napus (ACA1 – Ferreira et al., 1994; AC 2V1 – Somers et al., 2002), and A. thaliana (RAC-1, RAC-2, RAC-3, and RAC-4 – Borhan et al., 2001, 2008) conferring resistance against one or more races of A. candida. The Raphanus sativus cv. caudatus possess a dominant resistant gene for A. candida race 1 designated as AC-1 (Humaydan and Williams, 1976). However, some other Brassica species demonstrated monogenic control of the white rust pathogen race 2 such as B. nigra, B. rapa, B. carinata, and B. juncea (Delwiche and Williams, 1974; Ebrahimi et al., 1976; Thukral and Singh, 1986; Tiwari et al., 1988). The resistance to A. candida race 2 (Ac2V) can be explained in A. thaliana accessions by at least one of four genes (WRR4A, WRR4B, WRR8, and WRR9) encoding nucleotide-binding, leucine-rich repeat (NLR) immune receptors. However, the WRR12 gene identified in A. thaliana confers resistance to A. candida race 9 that infects B. oleracea. Thus, the effector-triggered immunity conferred by the distinct NLR-encoding genes in multiple A. thaliana accessions provides species-wide resistance to different races of A. candida (Volkan et al., 2019). The B. juncea—Donskaja-IV from the east European gene pool line was mapped for conferring resistance locus (AcB1-A5.1) against white rust and was found to be completely resistant to six different Indian isolates. The locus was identified to possess a single CC-NB-LRR protein-coding R gene (BjuWRR1) which provided resistance to genetically transformed susceptible Indian mustard variety Varuna from all the isolates (Arora et al., 2019). The F1DH mapping population of B. juncea var. Tumida (resistant) × B. juncea var. Varuna (susceptible) was identified with a new white rust resistance-conferring locus on LG A6 which was most likely to BjuA046215 candidate gene, a CC-NBS-LRR type R gene, and closely related to BjuWRR1 (Bhayana et al., 2020).
Downy Mildew
The disease is incited by a biotrophic fungal pathogen Hyaloperonospora parasitica (Pers.) Constant. Syn. H. brassicae which causes heavy yield loss to cruciferous crops and is distributed worldwide. In India, the disease is responsible for upto 66% yield loss in B. juncea making up a yearly loss of about 683.1 million INR, depending on the disease severity (Meena et al., 2014). The pathogen can infect all aerial parts (leaf, stem, and flowers) of the plant. However, plant resistance depends on the plant age and environmental variations (Coelho et al., 2009). Mohammed et al. (2018) screened about 154 members of the Brassicaceae family comprising Brassica napus, B. carinata, B. juncea, R. sativus, Rapistrum rugosum, B. incana, Crambe abyssinica, B. fruticulosa, Hirschfeldia incana, B. insularis, B. oleracea, and Sinapis arvensis for resistance against seven isolates of H. parasitica. They found most of the resistant lines from R. sativus, B. carinata, and B. juncea genotypes. However, they did not conduct any molecular study to identify a downy mildew resistant gene. The downy mildew resistant and white rust susceptible B. juncea genotype were inoculated with A. candida followed by H. parasitica that lead to asymptomatic systemic colonization of downy mildew pathogen and a more severe infection of white rust about four days earlier (Kaur et al., 2011b). The field resistance of B. oleracea var. tronchuda can not be predicted through cotyledon resistance as both are governed by different genetic systems (Monteiro et al., 2005). Nashaat and Awasthi (1995) screened 31 B. juncea accessions against four isolates of H. parasitica at the cotyledon stage and almost all expressed high-level resistance against the pathogen. These accessions were grouped into five categories showing differential disease responses.
The homozygous resistant and susceptible varieties (USVL012 and USVL047, respectively) of B. oleracea were used for the development of the self and backcross population. The F2 population showed segregation for resistant character and backcross population developed with resistant plant remained resistant and with susceptible plant remained susceptible. Thus, they concluded that the resistant character must be governed by two complementary dominant genes (Wang et al., 2001). In similar studies on 200 Raphanus sativus accessions, it was confirmed that the resistance was governed by a single dominant gene (Coelho and Monteiro, 2018). Dang et al. (2000) used 36 genotypes of different varieties of Brassicaceae for resistance screening against leaf pathogens during three consecutive years and found Brassica alba (S. alba) highly resistant to downy mildew disease in all 3 years. The F2 population developed from the resistant and susceptible genotypes of broccoli (B. oleracea L. Italica Group) and cauliflower (B. oleracea L. var. botrytis) showed segregation in a 3:1 (resistant:susceptible) ratio which indicated that the resistance is governed by a single dominant gene and also confirmed by test-cross (Farnham et al., 2002; Vicente et al., 2012; Verma and Singh, 2018). It was also evident from different studies that the resistance against downy mildew pathogen in B. napus and B. juncea was also governed by a single dominant gene (Nashaat et al., 1997, 2004).
Coelho et al. (1998) identified the dominant and monogenic (Pp523) inheritance of plant resistance expressed at the adult stage in broccoli located on another genetic map of RAPD and AFLP markers assigned to chromosome C8 of B. oleracea (Carlier et al., 2011), with the syntenic region at the top arm of A. thaliana’s chromosome 1 (Farinhó et al., 2007). Similarly, Singh et al. (2012) mapped a single dominant R gene, Ppa3 in B. oleracea through molecular markers. The map-based cloning of resistant genes to H. parasitica from A. thaliana was done to encode for TIR- and CC-NBS-LRR categories viz. as RPP5 (Parker et al., 1997), RPP8 (McDowell et al., 1998), RPP1 (Botella et al., 1998), RPP13 (Bittner-Eddy et al., 2000), RPP4 (Van Der Biezen et al., 2002), and RPP2A/RPP2B (Sinapidou et al., 2004). The white rust-resistant genes RAC1 and RAC3 were found closely associated with downy mildew resistant genes RPP9 and RPP8 in A. thaliana, respectively (Borhan et al., 2001). A major gene RPP31 for the adult stage resistance against downy mildew pathogen was genetically mapped in A. thaliana on chromosome 5 (McDowell et al., 2005). The mutation on the RPP-non-specific locus called EDS1 that is required for the proper functioning of RPP genes revealed enhanced susceptibility in A. thaliana (Parker et al., 1996). A large number of R genes have been mapped or cloned in A. thaliana and the orthologous genes can be searched in other Brassicaceae members through genome sequence comparisons (Yu et al., 2014) and pan-genome analysis (Golicz et al., 2016). The cDNA clones (Bcchi and BcAF) were isolated and characterized for their role in plant defense mechanisms from B. rapa ssp. chinensis L. cv. Suzhouquing against downy mildew pathogen. These clones were translated into protein products and found to be homologous to plant chitinases and defensins. The study reveals their involvement in plant resistance upon infection by a fungal pathogen (Chen et al., 2008). Yu et al. (2011) identified two SSRs (kbrb006c05-2 and kbrb058m10-1) and one Sequence Characterized Amplified Region-SCAR (SCK14-825) marker closely linked to downy mildew resistant QTL (BrDW) on chromosome A8 and used them for MAS in Brassica rapa ssp. pekinensis. A further extension to this map, a total of four major QTLs (sBrDM8, yBrDM8, rBrDM8, and hBrDM8) identical to BraDM were mapped to A08 through three SNP markers (A08-709, A08-028, and A08-018) for resistance at the seedling, young plant, rosette, and heading stages (Yu et al., 2016). Lucas et al. (1988) have identified a single dominant gene conferred resistance for downy mildew pathogen in B. napus. However, the genetic background and environment could influence the phenotypic expression of resistance. The DMR6 gene is responsible for susceptibility in A. thaliana plants on infection by downy mildew pathogen. However, the mutants for this gene have lost the susceptibility for H. parasitica. The susceptibility of DMR6 mutant A. thaliana can be restored by two closely related gene DLO1 and DLO2 (Zeilmaker et al., 2015).
Powdery Mildew
The powdery mildew of Brassicaceae is caused by biotrophic fungal pathogen Erysiphe cruciferarum Opiz ex L. Junell. The pathogen was reported from several parts of the world including Australia, China, Europe, the Former Soviet Union, India, Japan, Korea, South Africa (Kaur et al., 2008; Farr and Rossman, 2013; Kim et al., 2013) in dry-warm weather conditions with low relative humidity favoring the pathogen growth. Besides B. juncea, the powdery mildew pathogen can also infect wild members of Brassicaceae such as Camelina sativa and Sinapis arvensis (Vellios et al., 2017). The pathogen can infect all above-ground plant parts which get covered with dense cottony growth leading to premature leaf fall. The pathogen produces dark-colored cleistothecia on the plant surface, spreading secondary infection, i.e., polycyclic disease. The powdery mildew pathogen can cause upto 17% yield losses in India at harvest (Dange et al., 2002). Tonguc and Griffiths (2004) developed interspecific hybrids and backcross progeny from B. carinata and B. oleracea cultivars through embryo rescue to transfer resistance against powdery mildew pathogen. The interspecific hybrids tested with powdery mildew under greenhouse conditions were found to be completely resistant. However, only 38% of the first backcross progeny showed a resistant response against the disease. Frye and Innes (1998) have identified an Arabidopsis mutant that displays enhanced disease resistance for powdery mildew caused by Erysiphe cichoracearum. The edr1 mutant does not constitutively express the pathogenesis-related genes PR-1, BGL2, or PR-5. The edr1 mutation is recessive and maps to chromosome 1 between molecular markers ATEAT1 and NCC1. It was speculated that the edr1 mutation derepresses multiple defense responses, making them more easily induced by virulent pathogens. However, Vogel and Somerville (2000) identified 20 recessive mutants of A. thaliana that inhibit the normal growth of powdery mildew pathogen (E. cichoracearum). They concluded that resistance for powdery mildew is not simply due to constitutive activation of the salicylic acid or ethylene and jasmonic acid-dependent defense pathways, but because the mutants did not constitutively accumulate elevated levels of PR1 or PDF1.2 mRNA.
Li et al. (2016) studied multiple evolutionary events involved in maintaining homologous resistance genes in B. napus conferring broad-spectrum resistance for powdery mildew pathogen. One and three genes homologous to RPW8 were present in B. rapa and B. oleracea, respectively. There should be seven homologs of RPW8 in B. napus. They found that the copy of B. oleracea resistant genes was highly conserved, while the B. rapa homolog was variable in the B. napus genome possibly due to gene loss, point mutation, insertion, deletion, and intragenic recombination. Nanjundan et al. (2020) evaluated 1020 B. juncea accessions against E. cruciferarum under natural hot spot conditions. They have identified only one accession (RDV29) consistently resistant for 5 years. This line was used for hybridization with a highly susceptible line (RSEJ775) to obtain filial and backcross populations. The study revealed that resistance was governed by two different semi-dominant genes. Adam and Somerville (1996) used six A. thaliana accessions to identify resistant loci for powdery mildew pathogen (E. cichoracearum). Their study revealed that out of six, five accessions conferred resistance through a single locus and all were independent and the another one accession have resistance from two unlinked loci. Xiao et al. (1997) identified two independent segregating dominant loci (RPW6 and RPW7) on chromosome 5 and 3 in A. thaliana for resistance to powder mildew. The majority of R genes characterized in A. thaliana for powdery mildew disease belong to the nucleotide-binding site and C-terminal leucine-rich-repeats (NB-LRRs) and a less prevalent N-terminal transmembrane domain and a coiled-coil motif superfamily. These genes were identified as NDR1, EDS1, PAD4, NPR1, EDS5, RAR1, SGT1b, RPW8, PBS3, COI1, EIN2, and EDR1 (Xiao et al., 2005). However, the transgenic form of Nicotiana tabacum and N. benthamiana developed with RPW8.1 and RPW8.2 genes confers higher resistance against E. orontii, O. lycopersici, and E. cichoracearum but failed to provide resistance to Lycopersicum esculentum. Thus, these genes could be used to develop transgenics in other families to provide resistance against powdery mildew disease (Xiao et al., 2003). Alkooranee et al. (2015) investigated the mechanism of systemic resistance induced by T. harzianum (TH12) or its cell-free culture filtrate in B. napus and R. alboglabra to powdery mildew pathogen. The results of the study revealed that the pathogen failed to develop colonies on R. alboglabra leaves even after 10 days of inoculation while B. napus leaves have fungal colonies after 6 days of inoculation. The expression of PR-1 and PR-2 levels increased in E. cruciferarum infected leaves but decreased in the TH12 treated leaves. However, the expression of PR-3 and PDF1.2 is decreased in E. cruciferarum infected plants whereas it was increased when treated with TH12 suggesting that TH12 can be used for improving resistance to powdery mildew in hosts.
Blackleg
The blackleg, also known as phoma stem canker, is one of the most devastating diseases of Brassicaceae family that limits oilseed production worldwide such as in Australia, Canada, and Europe (Zhang and Fernando, 2018). The disease is causing by two coexisting fungal pathogens Leptosphaeria maculans (Desm.) Ces et de Not. [anamorph: Phoma lingam (Tode: Fr.) Desm.] and L. biglobosa sp. nov., Shoemaker & Brun which belongs order Ascomycota (Shoemaker and Brun, 2001). However, the former pathogen causes severe damage to crops than the latter (Petrie, 1978; Rouxel and Balesdent, 2005; Vincenot et al., 2008; Dilmaghani et al., 2009). The strains of L. maculans have been categorized into two groups according to pathogenesis, i.e., the strains that causes stem cankers on canola named aggressive, virulent, or “A” group and that do not cause cankers on canola named non-aggressive, avirulent, or “B” group (Howlett et al., 2001). The ascospores are the primary source of disease formed in pseudothecia, produced on host plant residues of the former crop season, and survive up to 5 years on residues (Petrie, 1986, 1995; West et al., 2001; Aubertot et al., 2006). The successful infection leads to the formation of small light green to pale color lesions without margins in the case of L. maculans and L. biglobosa formed dark brown or gray lesions with distinct dark margin. The asexual fruiting bodies (pycnidia) of the pathogen are formed as small dark spots on leaves and stem which contains pycnidiospores that transmitted through water splashes (Kaczmarek and Jêdryczka, 2011). The pathogens causes severe damage to host plants by girdling and lodging stems (Khangura and Barbetti, 2001). However, the pathogens can infect other parts of host plants such as roots and siliquae.
The commercial varieties of B. napus introgressed by resistant genes for blackleg from B. rapa subsp. sylvestris lost their ability within three years possibly due to the rapid increase in the frequency of L. maculans isolates (Sprague et al., 2006b). Sjodin and Glimelius (1988) screened 96 accessions of the Brassicaceae family at cotyledon, seedling, and adult plant stages against seventeen isolates of L. maculans to find out the resistant source for blackleg disease. Out of all tested accessions, only five accessions of B. juncea, two of B. nigra, and two of B. carinata were found resistant to the disease and all contain B genome (Barret et al., 1998). In another study, Dixelius and Wahlberg (1999) analyzed asymmetric progeny of B. napus with three B genome donors (B. nigra, B. juncea, and B. carinata) for the presence of resistance to blackleg disease. They have identified a total of four co-segregating RFLP markers for cotyledon and adult-leaf resistance which was associated with six loci present on linkage groups 2, 5, and 8. A triplicate region in the B- genome had preserved the resistant loci in all three species. Fredua-Agyeman et al. (2014) provided evidence for the B3 chromosome of the B genome to carry resistant genes and confirmed that the entire chromosome was associated with blackleg resistance. Thus, B. napus suffer more severely from the blackleg disease than B. juncea (Marcroft et al., 2002). However, a large number of attempts were made to transfer blackleg resistance into B. napus from members of the Brassicaceae family (Chèvre et al., 2003; Winter et al., 2003; Gaebelein et al., 2019) and stabilized (Chevre et al., 1997; Barret et al., 1998; Chèvre et al., 2008). The B. nigra monosomic lines for chromosome 4 in B. napus have the same level of resistance as in B. nigra (Chevre et al., 1996). The resistance to blackleg disease is governed by a polygenic system and present in a clustered manner. However, the resistance provided by the B genome remains constant throughout the life cycle of the host plant (Rimmer and Van-den Berg, 1992; Chèvre et al., 2003). The F2 and backcross population of two different cultivars of B. juncea (resistant- AC Vulcan and susceptible- UM3132) was segregated for blackleg resistance which suggested that the resistance was controlled by two independent dominant and recessive genes positioned on linkage group J13 and J18, respectively (Christianson et al., 2006). The NILs developed from the asymmetric somatic hybrids of B. napus with B. juncea and B. nigra showed that one single dominant allele and two independent loci govern adult plant stage resistance to blackleg in B. napus–B. juncea and B. napus–B. nigra lines, respectively (Dixelius, 1999). The B- genome introgression lines of B. napus confirmed the inheritance of recessive resistant gene, rjlm2 from B. juncea for the blackleg disease at the cotyledon stage (Saal et al., 2004).
In contrast to A- or B- genome crops where a large number of resistant genes have been identified, few resistant genes were also identified on C- genome crops (Hossain et al., 2020). During a study, Rlm1, Rlm2, Rlm3, Rlm4, Rlm9, RlmS, LepR1, and LepR2 genes were reported to present in Canadian canola varieties (Zhang et al., 2016) and also a large number of avirulence (Avr) genes (∼14) in corresponding L. maculans pathogen and cloned few of them (Liban et al., 2016). The mapping and cloning of an avirulence gene, AvrLmJ1, from L. maculans confers avirulence to B. juncea cultivars (Van-de Wouw et al., 2014). To date, at least sixteen resistant genes for blackleg were mapped in B. napus and allied Brassica species. Out of these resistant genes, many were present in an association of others or clustered together (Delourme et al., 2004). The inheritance of resistance against blackleg in B. juncea was breached in the recent past by newly evolving strains of L. maculans in Australia (Elliott et al., 2015) but the breakdown of Rlm3 resistance was not reported until recently in Canadian canola varieties (Zhang and Fernando, 2018). However, some other reports indicated that Rlm1, Rlm3, Rlm6, Rlm7, LepR1, and LepR3 lost their effectiveness in the field against the blackleg pathogen (Rouxel et al., 2003; Sprague et al., 2006a,b; Brun et al., 2010; Winter and Koopmann, 2016; Zhang et al., 2016; Van-de Wouw et al., 2017). In a study, the blackleg resistant gene Rlm2 was located on chromosome A10 of the B. napus cv. Glacier with the help of tightly linked microsatellites (sR1448, sN8502, sN1982, and sN8474). The gene was localized to a 5.8 cM interval corresponding to approximately 873 kb of the B. napus chromosome A10 (Larkan et al., 2014).
The Rlm4 quantitative trait locus was characterized for harboring 18 candidate resistant genes (BLR1–BLR18) for blackleg in B. napus through NGS (Tollenaere et al., 2012). The LmR1 locus controls seedling resistance for blackleg in B. napus cv. Shiralee was positioned on linkage group N7, find out after fine mapping of 2500 backcross lines. A total of three microsatellites were found associated with resistance and co-segregated with this gene. However, an additional seedling resistance gene, ClmR1, was identified in the same region of LmR1 (Mayerhofer et al., 2005). Yu et al. (2005) have identified and mapped two resistant alleles, LepR1 and LepR2, for blackleg resistance in B. napus lines introgressed from B. rapa subsp. sylvestris that were located on linkage group N2 and N10, respectively. In addition to both these, they have mapped a third resistant gene LepR3 by microsatellite markers on linkage group N10 at below the LepR2 gene in B. napus “Surpass 400” (Yu et al., 2008). Raman et al. (2018) assessed quantitative resistance (QR) for blackleg in DH lines of Darmor-bzh/Yudal (DYDH) population in three successive years and identified a total of 27 significant QTLs on 12 different chromosomes. Out of which, seven were repeatedly present on chromosomes A02, A07, A09, A10, C01, and C09 in all experiments. They have identified eight stable QTLs for blackleg in three diverse growing conditions of Australia, France, and the United Kingdom. In another study, genome-wide association analysis found extensive variations in resistance to blackleg at the adult plant stage in B. napus. A total of 59 statistically significant SNPs were identified on seventeen chromosomes of B. napus genome that were responsible for variations to blackleg resistance (Raman et al., 2020). Huang et al. (2019) identified a total of five QTLs on linkage groups A02, A03, A10, C01, and C09 for resistance to L. maculans growth on oilseed rape DY (“Darmor-bzh” × “Yudal”). Mapping population contributed about 35 percent phenotypic variations.
Bottlenecks in Identification of Resistance Sources
Genome Complexity in B. juncea
The diploid ancestral species (B. rapa and B. nigra) of B. juncea have evolved from a common ancestor followed by ancient genome triplication along with structural and numerical changes about 7.9–14.6 million years ago (Lysak et al., 2005). B. juncea developed through the spontaneous hybridization of these two ancestral Brassica species by combining the genome to give rise to an allotetraploid species. This event was followed by a natural chromosome doubling. It is suggested that B. rapa was a cytoplasmic donor in B. juncea development process (Banga et al., 1983; Warwick and Black, 1991; Pradhan et al., 1992; Prakash et al., 2009). The crop was diversified into vegetable and oil-producing subvarieties and cultivation was started about 6000–7000 years ago in China (Yang et al., 2018) and 2300 BC in India (Prakash and Hinata, 1980). However, it was assumed that B. juncea formed about 0.039–0.055 million years ago (mya) while B. napus formed about 0.038–0.051 mya, slightly after the formation of B. juncea. B. juncea was estimated by flow cytometry to possess a 922 Mb genome size. The B. juncea genome has 316.1 Mb of repetitive sequence, out of which 131.2 Mb are from B. rapa and 216.5 Mb from B. nigra genomes. During the speciation process, it was estimated that a total of 562 and 545 genes from B. rapa and B. nigra subgenomes of B. juncea were lost, respectively, as compared to their common ancestral genomes. This number is higher than the gene loss estimated in B. napus subgenomes (BnaA and BnaC) as compared to their common ancestral genomes. However, the number of genes lost in B. juncea and B. napus was consistent since their formation. The A subgenome of B. juncea and B. napus had divergent origins. It was discovered that A subgenome of B. juncea might be derived from Asian B. rapa ssp. tricolaris, while the subgenome A of B. napus might be derived from European B. rapa ssp. rapa. Thus, both A subgenomes of allotetraploids had independent geographical origins. It was also discovered that the chromosomal regions of B. juncea had gone through various rearrangements and formed the current species contributing to gene duplications and losses. The polyploidy nature of B. juncea played an essential role in genome speciation and plasticity (Yang et al., 2016). The A subgenome of B. juncea remained intact while the B subgenome has changed considerably. However, the B subgenome in B. carinata is unchanged during evaluation (Prakash et al., 2009). B. rapa possesses a rich genetic diversity with various desirable agronomic traits, i.e., rapid growth and tolerance to nutrient-deficient soil and low temperatures (Franks, 2011; Waalen et al., 2011; Ahmed et al., 2012).
The cultivated B. juncea varieties have very lower genetic diversity due to the unidirectional selection force for yield characteristics. This is also evident from recently developed genome assemblies of B. Juncea var. tumida (Yang et al., 2016), BjVaruna (Paritosh et al., 2020), and it’s parent species B. rapa (Wang et al., 2011; Cai et al., 2017), and B. nigra (Yang et al., 2016). However, the complete genome sequences were not developed during the assembling process which leads to deletion (∼10–20%) of sequences containing potential R genes. Contrastingly, the artificial B. juncea was synthesized by combining the Asian oil crop B. rapa (ArAr) and the Bc subgenome from the African oil crop B. carinata (BcBcCcCc) and synthesized allohexaploid (ArArBcBcCcCc), crossed with traditional B. juncea to generate pentaploid F1 hybrids (ArAjBcBjCc), with subsequent self-pollination to obtain newly synthesized B. juncea (Ar/jAr/jBc/jBc/j). The genetically stable new type of B. juncea population was obtained at the F6 generation retaining good fertility and rich genetic diversity while being distinct from the traditional B. juncea. This newly developed B. juncea had more than half a modified genome due to exotic introgressions and novel variations in gene copies, numbers, and sequences (Wei et al., 2016). This innovatively developed B. juncea can be utilized for the identification of novel R genes and the improvement of genetic base.
The R genes controlling vertical resistance and function of the host against a specific race or strain of the pathogen can be altered easily for resistance-breeding, compared to polygenic host resistance which involves the collective action of more than one gene. However, the identification of R genes in B. juncea is challenging due to complex genomic structures. The B. juncea genome was identified to contain a total of 289 NBS-LRR type resistance genes, more than its diploid progenitors (B. rapa and B. nigra). A total of 4 and 7 QTLs for the white rust and blackleg resistance in B. juncea respectively, were identified. The white rust resistant locus AcB1-A04.1 was present on the linkage group A04 and B01, while another locus AcB1-A05.1 was present on the same linkage group on an overlapping position at A05 with a similar copy at B06. Similarly, blackleg-resistant quantitative loci-PhR2 were identified on the linkage groups A03 and B03, and another QTL, RLM6, identified on three linkage groups (A07, A09, B01). However, PhR2 and LMJR1 resistant loci for blackleg were present on linkage group B03. This study concluded that the B. juncea genome has duplicated sequences (Inturrisi, 2018). The different resistant genes or QTLs were clustered together in the B. juncea genome. The clustering effect of resistant gene/QTL for white rust and blackleg diseases directs to the complexity of the B. juncea genome. The gene clustering is commonly found in plant genomes to adopt a quick response against pathogens through recombination incidents (Hulbert et al., 2001; Meyers et al., 2003). The information regarding resistant QTLs/genes for major diseases is useful to develop durable resistant cultivars effective against a wide range of pathogen strains or races. The overlapping, clustering, or allele copies of R genes can cause problems in the identification of candidate resistant genes as many such regions may not be a part of sequenced reference genome assemblies. The dependency on a single reference genome assembly to identify resistant genes can obstruct efforts in the identification and use of such R genes. All possible future endeavors should add innovative technologies for the sequencing of target chromosomes or regions.
Genetic Variations in Pathogens
The genome complexity of B. juncea in terms of resistant genes and resistance levels is also variable in different hosts for the major disease-causing pathogens, as discussed before. It has been noticed frequently that variable disease symptoms were produced when infected by different isolates/strains obtained from a different source or geographical region of the same source. These variable symptoms could be due to genetic variations within the host genotypes and/or within the pathogen. The leaf blight or black spot pathogen of B. juncea has very high genetic diversity all over the world. A. brassicae has three races viz. RM-1, RM-2, and V-3 in India which are virulent on the rapeseed-mustard group of crops (Saharan and Kadian, 1983). However, in different investigations on the diversity of A. brassicae, Awasthi and Kolte (1989), Kolte et al. (1991), Kumar et al. (2003), and Sangwan and Mehta (2007) identified various races from Indian regions. In recent studies on the identification of genetic variability of A. brassicae in 32 isolates collected from Himachal Pradesh (India), a total of seven pathogenic races were identified designated as Abr1 to Abr7 (Kumar et al., 2014). The races/pathotypes identified by various studies presented a highly virulent to non-virulent response on screening with different Brassica crops. Sharma and Tewari (1998) collected isolates of A. brassicae from different geographical regions of the world (India, Canada, France, Costa-Rica, England, and Poland) and analyzed them with RAPD and RFLP markers. Having tested a total of 20 decamer primers of arbitrary nucleotide sequences for PCR amplification of A. brassicae genomic DNA, only five primers with amplified genomic DNA from 20 isolates of A. brassicae were selected and classified into four groups. However, the variations between isolates collected from intra-geographical regions were very less apparent. Kaur et al. (2007) evaluated 322 isolations of A. brassicae collected from a wide geographic area of north-west India. Out of these, 114 were identified as pathogenic and all were categorized under seven broad groups based on morphological characters showing a wide range of diversity. Goyal et al. (2013) characterized 13 isolates of A. brassicae collected from different parts of India through 100 RAPD decamer primers and found genetic variability among the isolates at the genomic level, but not in the highly conserved regions of the genome of A. brassicae. Aneja et al. (2014) studied genetic diversity through RAPD markers in 32 isolates of A. brassicae collected from eight north Indian states and found a total of 57–78% genetic diversity within all isolates with no correlation established. They determined the genetic relationship among 32 A. brassicae isolates by UPGMA revealing the clustering into four major classes that were further subdivided into nine subgroups. Rajarammohan et al. (2019a) identified 460 A. brassicae specific genes which included many secreted proteins and effectors. They also identified gene clusters responsible for the production of pathogenicity specific Destruxin-B (a cyclic depsipeptide). However, in the recent past, a chromosome level about a complete genome assembly of A. brassicae was developed that could be useful to uncover the B. juncea–A. brassicae pathosystem related genes (Rajarammohan et al., 2019b).
The races of white rust (A. candida) isolated from four Brassicaceae members behaved as four independent races in Saskatchewan (Canada). These races were evaluated for their virulence in various Brassica members collected from different parts of the world. The results showed that certain crop species were susceptible to specific race but not to all races. However, some accessions of Brassica show resistance to some of these races, but not all (Petrie, 1988). Jouet et al. (2019) analyzed 85 isolates of A. candida for genetic diversity collected from many European countries infecting various Brassicaceae hosts.
Impact of Environmental Conditions on Disease Development
Environmental factors play a key role in pathogen inoculum distribution, host–pathogen interaction, disease development, and its severity. These factors included soil pH, moisture, and temperature, available soil nutrients, environmental temperature, relative humidity (RH), and inoculums dispersal conditions. All these environmental factors directly influence the host-resistance status by affecting pathogen growth and reproduction potential. The sclerotia developed by S. sclerotiorum at the end of each successful disease cycle can be germinated between 10 and 15°C temperatures, however above this temperature the germination is affected severely (Jones and Gray, 1973). The carpogenic germination of sclerotia was affected by variable soil moistures and relative humidity (Hao et al., 2003; Matheron and Porchas, 2005). The development of the stem rot lesion is favored by moist conditions and a temperature range of 20–25°C. However, the lesion growth will interrupt under dry and warm conditions but may resume when favorable conditions appeared again (Canola Council of Canada, 2020). The sporulation in A. brassicae on naturally infected leaves of oilseed rape needs a relative humidity of 91.5% or above with temperature ranging between 18 and 24°C. However, the sporulation is inhibited just above the 24°C temperatures (Jones and Phelps, 1989). For successful infection, a minimum wetness period of 4 h is required by A. brassicae at 18°C and disease severity increases with increasing wetness period up to 12 h. The disease severity also increases with increasing inoculum concentration from 80 to 660 spores ml–1 with increasing leaf age from 4 days (Hong and Fitt, 1995). The severity of white rust on B. juncea leaves was favored by >40% afternoon (minimum) RH, >97 mornings (maximum) RH with 16–24°C daily temperatures. However, the staghead formation was influenced positively between temperature ranges of 20–29°C and >97% morning RH (Chattopadhyay et al., 2011). In contrast, the powdery mildew pathogen of B. juncea needs warmer (24–30°C) environmental conditions and low relative humidity (∼65 RH) for successful disease establishment and conidial dispersion (Desai et al., 2004).
Re-emergence of Virulent Pathogen Races and Host–Pathogen Interactions
The hybridization between two races/isolates of the pathogen is recognized as a major force in the evolution of new races or adaptation of exiting plant pathogenic race (Brasier, 2000, 2001). The global agriculture commodity trade moves pathogens and hosts around the world and has played a key role in the emergence of new races or diseases (Brown and Hovmoller, 2002). The hybrid race or pathogen can be formed by sexual hybridization between parental species or fusion between hyphae or vegetative cells. The fusion of vegetative cells can be followed by parasexual reproduction where mitotic crossing over generates recombinant cells or new races (Schardl and Craven, 2003). The cultivation of resistant varieties containing the R gene for vertical resistance against a particular pathogen race or isolate exerts selection pressure on the pathogen that is better adapted to breach the host defense system. The resistant crops cultivated continuously in a particular field should be avoided due to the direct exposure to virulent strains of pathogens that can overcome the host resistance. Thus, the vertical host resistance may effectively work against a particular pathogen race for a short period and lose its effectiveness after 3–5 crop seasons. A new race of A. candida was identified on Lepidium latifolium (pepperweed) in California which was unable to infect fifteen host differentials of the Brassicaceae family. Thus, the host range information of any particular pathogen is important to identify new races (Koike et al., 2011).
The host resistance mechanism is dependent on a particular host pathosystem as a single major gene or a group of genes involved in the defense mechanism. In the B. juncea–S. sclerotiorum pathosystem, a highly resistant response was recorded for stem rot in cotyledon and stem assays. The expression of differential resistance response in introgression lines showed quantitative trait inheritance governed by more than one gene which works cumulatively against S. sclerotiorum. In this pathosystem, the resistance at seedling or adult plant stage and in cotyledon, leaf, or stem were expressed independently. The resistance shown by a particular resistant host is also dependent on the pathogen aggressiveness or virulence. A. thaliana–A. brassicae pathosystem has a quantitative inheritance for resistance by major independent loci. There were common and population-specific QTLs and also a chance of different genes governing resistance to the pathogen. Out of these, one QTL expressed about 50% variation in disease resistance with the genes present within probably contributing to resistance even in heterogeneous conditions (Rajarammohan et al., 2017). In contrast, the genetic mechanism of host resistance in B. juncea is different for the white rust pathogen. The host resistance was reported to be governed by a single locus on LG A4, LG A5, and LG A6 in Heera, Donskaja-IV, and Tumida lines of B. juncea, respectively. However, the resistant genes conferring resistance in these loci were found closely related to R gene BjWRR1 (Bhayana et al., 2020). It will be beneficial for molecular studies to identify the hosts conferring resistance against a broadspectrum of pathogens i.e. hosts showing resistance to all races or isolates of the pathogen.
The use of different disease scoring parameters has also created troubles in the identification of true resistant lines resulting in misidentification of R genes. Atleast threescoring systems are used in Alternaria blight disease screening (Conn et al., 1990; Vishwanath et al., 1999; Meena et al., 2016; Ayuke et al., 2017). Similarly in S. sclerotiorum-B. juncea pathosystems, there are two scoring systems to identify resistant crop plants, and different plant parts and host-age used in the evaluation of resistance (Kumari and Singh, 2019; Gupta et al., 2020). Thus, it is necessary to develop a robust screening method for each pathosystem to identify the same or different races/pathotypes, as well as host resistance status.
Strategies for Brassica Improvement
Identification and Introgression of Novel R Genes
The Brassica coenospecies or wild members of the Brassicaceae family have a high degree of resistance against S. sclerotiorum such as B. fruticulosa, S. alba, B. incana, C. sativa, D. tenuisiliqua, E. abyssinicum, and E. cardminoides. These species were used to introgress resistance into B. juncea and B. napus through somatic hybrid production through PEG mediated protoplast fusion, followed by backcrossing with the cultivated parent. The introgression lines possessing segmental or chromosomal introgression demonstrated promising resistance against stem rot disease (Figure 2A). However, the screening experiments should be performed with virulent races/isolates and introgression lines through robust screening assays to identify true resistant breeding lines. Next-generation sequencing will help in developing a near-complete genome assembly of these lines serving as a good source in the identification of R genes through comparative genome analysis. This approach will help broaden the genetic base by introgression of novel R genes into Brassica lines.
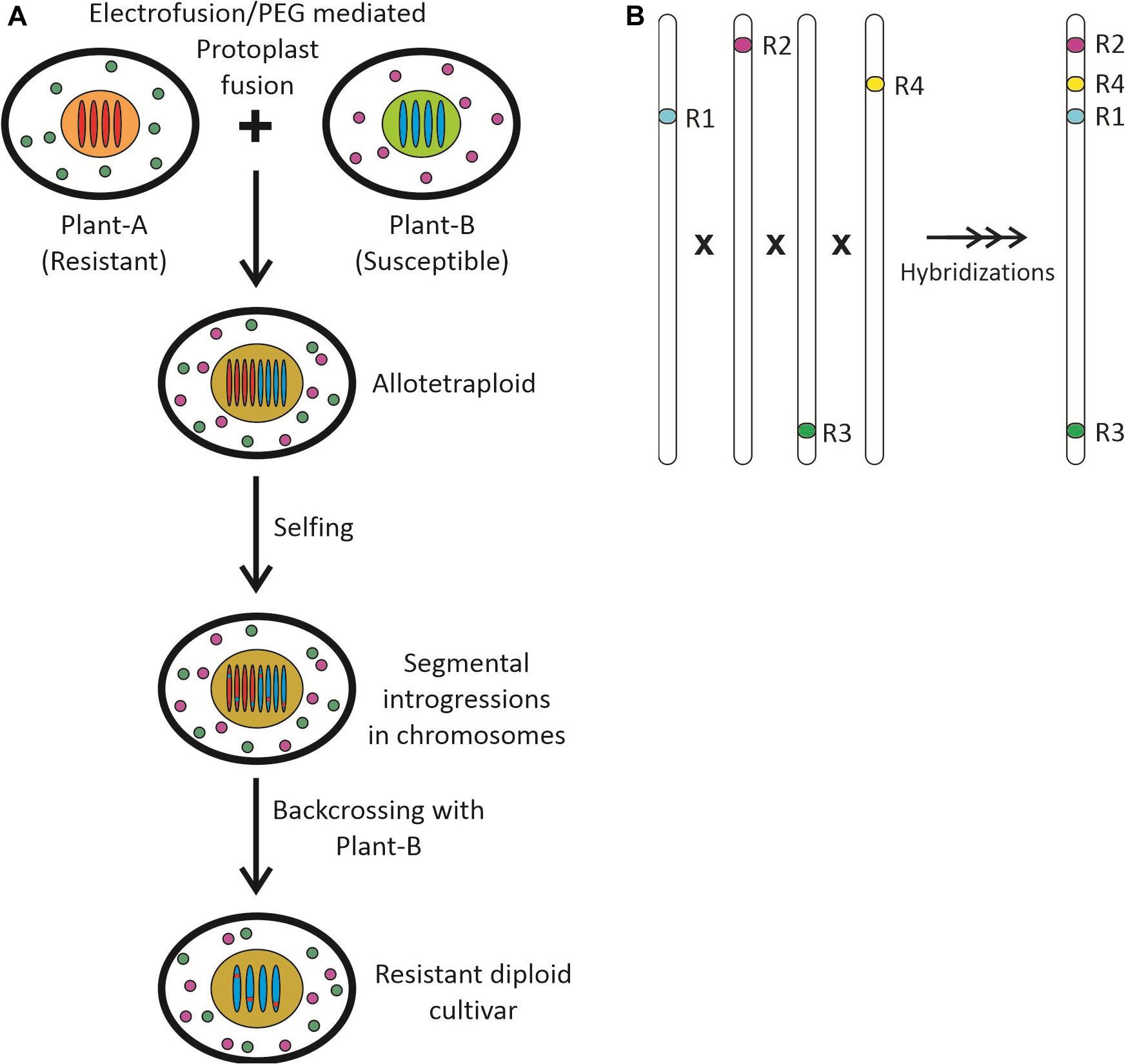
Figure 2. (A) The development of resistant segmental introgression lines from susceptible cultivars through PEG mediated protoplast fusion and subsequent backcrossing; (B) The gene pyramiding approach by wide hybridization and selection.
In recent years, a large number of candidate R genes have been identified for various diseases of the mustard plant (Table 1). The information regarding these gene sequences will enhance the cloning capacity and also be transferred into cultivated species to develop resistant transgenic plants. However, there has been very little progress in the cloning of R genes. The use of completely characterized antifungal genes to develop transgenic Brassica lines will be beneficial to confer resistance against a range of pathogens. The hevein producing gene was used to develop transgenic Brassica which displayed enhanced resistance against leaf blight pathogen (Kanrar et al., 2002). There are some more examples for use of antifungal genes to develop transgenic Brassica for improving resistance such as barley antifungal genes class II chitinase (AAA56786) and type I ribosome-inactivating protein (RIP; AAA32951), and tomato glucanase. All these provide higher resistance to Brassica lines against A. brassicae (Mondal et al., 2003, 2007). The B. juncea transgenic lines developed with antimicrobial gene msrA1 have shown upto 85 and 71.5% disease reduction caused by A. brassicae and S. sclerotiorum, respectively (Rustagi et al., 2014). Moreover, BjNPR1 transgenic lines of B. juncea showed improved resistance to A. brassicae and E. cruciferarum as there was an impediment in symptom development and reduced disease severity than non-transgenic plants (Ali et al., 2017). The chickpea lectin transgenic of B. juncea conferred resistance against A. brassicae showing upto 60% disease reduction with additional resistance against salinity and drought stresses (Kumar et al., 2015). The transfer of resistant genes from non-host plants conferring resistance to biotic diseases into the high-yielding susceptible host is a promising strategy toward the development of resistant Brassica varieties. The use of non-host genes into host plants has opened a new way of crop improvement and protect susceptible lines that lack resistance sources.
Resistance Breeding and Development of Pre-breeding Resistant Lines
Incessant efforts are being put in to introgress resistance into cultivated B. juncea lines by breeding with resistant allied wild genera or species. B. incana was identified to possess a high degree resistance against stem rot disease which can be transferred into cultivated susceptible B. oleracea var. alboglabra. The candidate R genes for SSR were identified on the C09 chromosome for leaf and stem rot which belongs to CC-NBS-LRR class (Mei et al., 2013). The B. napus lines evaluated for SSR resistance showed many putative QTLs on C02, C04, C06, C08, C09, A02, A03, and A08 chromosomes (Wu et al., 2013, 2016, 2019; Gyawali et al., 2016). The SSR resistant loci working cumulatively, thus pyramiding of resistant loci, could efficiently protect against a wide range of S. sclerotiorum races/isolates (Figure 2B). The B. juncea introgression lines carrying chromosomal segments of wild B. fruticulosa showed a resistant response against SSR and marker-trait association (Rana et al., 2017).
The related wild species of B. juncea was reported to possess genes conferring resistance against Alternaria blight disease that can be used for resistance breeding programs (Fatima et al., 2019). The advanced biotechnological approaches such as tissue culture and genome transformation have also been used for developing resistant Brassica lines. An in vitro ovary and ovule culture was attempted to transfer resistance against A. brassicae from S. alba to B. napus (Chevre et al., 1994) and E. cardaminoides to B. oleracea var. alboglabra (Mohanty et al., 2009). Agnihotri et al. (1991) obtained interspecific hybrids of B. campestris (B. rapa) and B. spinescens through ovary, ovule, and embryo rescue conferring resistance for Alternaria blight. Moreover, the somatic hybridization (protoplast fusion) technique has been used for transferring resistance from S. alba to B. napus (Primard et al., 1988; Wang et al., 2005), B. juncea (Gaikwad et al., 1996; Kumari et al., 2018, 2020c; Kumari and Bhat, 2019), B. oleracea (Hansen and Earle, 1997), B. carinata to B. juncea (Sharma and Singh, 1992), B. nigra to B. oleracea (Jourdan and Salazar, 1993), and B. spinescens to B. juncea (Kirti et al., 1991). There were some attempts made to introduce somaclonal variations to incorporate disease tolerance for Alternaria blight through mutagenesis (Verma and Rai, 1980; Agnihotri et al., 2009). The introgression lines developed for Alternaria blight resistance were used in the resistant breeding programs.
The B. juncea genome confers a good resistance source to white rust disease caused by A. candida. The marker-trait association was established in RAPD markers and white rust resistance that can be used for marker-assisted resistant breeding programs (Prabhu et al., 1998). The potential resistance genes against white rust come from the B genome of B. nigra and B. juncea (Delwiche and Williams, 1974; Massand et al., 2010). Moreover, some other lines of B. rapa, B. napus, B. carinata, and A. thaliana also possess genetic resistance to the white rust pathogen. However, the monogenic resistance was found effective against a particular race of A. candida. Thus, the resistant lines can be selected for resistant breeding programs according to the pathogenic race that is prevalent in a particular region or for resistant gene pyramiding program to develop lines governing resistance for more than one pathogenic race. In the breeding programs, the selection of parents who carry the R genes is important since their function may be masked in the newly evolved genome of offsprings due to an epistatic interaction (Werner et al., 2007). However, this problem can be managed successfully through the genome editing technique, the CRISPR-Cas (Pickar-Oliver and Gersbach, 2019). To select parent genotypes for resistant breeding, the field analysis of pathogen isolates for their virulent/avirulent genes should be considered. To select parent genotypes for resistant breeding, the field analysis of pathogen isolates for its virulence/avirulence genes should be considered that are adapted for host and identify the genetic variations in the pathogen races that can overcome the host resistance. Furthermore, the consideration of the host R gene is also important as pathogen pressure can affect the performance of R genes (Neik et al., 2017). B. napus cv. Surpass 400 was released in the year 2000 as the most resistant cultivar to L. maculans carrying a single dominant resistant gene from B. rapa subsp. sylvestris. However, in 2001 during an experiment, the cv. Surpass 400 and the susceptible cv. Westar were inoculated with pycnidiospore suspension of 18 isolates of L. maculans. The inoculated cotyledons of cv. Surpass 400 showed characteristic disease symptoms while susceptible cv. Westar was collapsed. The calculation of disease severity revealed that a total of 54% of cv. Surpass 400 and 100% plants of cv. Westar were susceptible to the disease. This study confirmed the ability of L. maculans isolates to overcome the resistance governed by a single dominant gene present in cv. Surpass 400. The information regarding the co-evolution of resistant genes and pathotypes is also important before the selection of a resistant host to develop durable resistant lines (McDonald and Linde, 2002).
Co-inoculation to Improve the Resistance of Susceptible Brassica Lines
In the L. maculans–B. napus pathosystem, the B. napus cv. Madrigal seedlings were pre-treated with ascospores of L. biglobosa, foliar sprays of ASM, or MSB and showed improved resistance against L. maculans (Liu et al., 2007). Pseudomonas syringae pv. tomato, the causal pathogen of bacterial speck disease in tomato and Arabidopsis induces resistance on pre-inoculation in Chinese cabbage to Erwinia carotovora subsp. carotovora that causes soft rot disease. It was found that Pst activates both salicylate-dependent and salicylate-independent defense responses in Chinese cabbage (Park et al., 2005). The pre-inoculation of the virulent strain of H. parasitica enhanced resistance against A. candida in white rust susceptible B. juncea host (Singh et al., 2002). The susceptible B. juncea cv. PR-15 showed resistance against virulent isolates A and C of A. brassicae when pre-treated with avirulent isolate D of A. brassicae. However, A. alternata failed to induce resistance for A. brassicae isolates A and C, rather it induced susceptibility to them (Vishwanath et al., 1999). The pre-inoculation of B. oleracea var. italica (Accession: Milady) seedlings with avirulent isolates of P. parasitica induced systemic resistance against virulent isolates and reduced disease incidence upto 70% (Monot et al., 2002). In contrast, it was recorded that the successful infection by a pathogen of a susceptible host could enhance the susceptibility of the host to secondary infection even for avirulent pathogens. The host defense system was suppressed by oomycete pathogen A. candida in A. thaliana and B. juncea. A pre-infection of B. juncea by A. candida can suppress immunity in cotyledons to downy mildew pathogen (H. parasitica), however, B. juncea still possesses resistance to mildew pathogen. The pre-infection with A. candida can successfully suppress a broadspectrum resistance conferred by RPW8 to the two morphotypes of Erysiphe spp. (a powdery mildew pathogen) in A. thaliana (Cooper et al., 2008). In the H. parasitica–B. juncea pathosystems, the pre-inoculation of A. candida to B. juncea followed by inoculation of avirulent H. parasitica increased susceptibility for white rust pathogen. However, the host plant that was resistant to downy mildew was also found to be infected systemically by H. parasitica. This work suggested that the host resistance could be determined by pathogen strain, infection sequence, or virulence status of the pathogenic strain (Kaur et al., 2011b). However, the molecular basis of enhanced resistance or susceptibility performance of host plants is still unknown when co-infected by pathogens.
Defense-Related Secondary Metabolites and Proteins
The plants have developed some alternative defense systems, such as secondary metabolites (Glucosinolate-myrosinase, phytoalexins, and phytoanticipins) and defense-related proteins (chitinases, glucanases, thionins, chitin-binding lectins, ribosome-inactivating proteins) to protect them from pathogens or stressful conditions other than occupying resistant genes (Terras et al., 1993; Kumar, 2017). The production of a large number of secondary metabolites from primary metabolites is induced when plants face biotic or abiotic stresses and they get accumulated in plant cells (Rejeb et al., 2014; Caretto et al., 2015; Narayani and Srivastava, 2018). These secondary metabolites usually belong to one of the three large chemical groups terpenoids, phenolics, and alkaloids. The gossypol, a member of the terpenoid class is produced by Gossypium hirsutum and has strong antibacterial and antifungal properties. Additionally, saponins (glycosylated triterpenoids) that are present in the cell membranes have detergent-like properties and disintegrate cell membranes of fungal pathogens. However, some fungal pathogens have saponin-degrading capacity causing diseases like Botrytis cinerea, Fusarium oxysporum, and Septoria lycopersici. The phytoalexins (camalexin, brassinin, and rapalexin A) have antibacterial and antifungal properties that are produced on the pathogenic attack. These are toxic molecules that disturb the metabolism or cellular structure of pathogens but are often toxic to a specific pathogen (Freeman and Beattie, 2008). The glucosinolates are sulfur-containing secondary metabolites synthesized by members of the Brassicaceae family. These glucosinolates are hydrolyzed by myrosinase enzyme secreted as a consequence of cell wall lysis (Osbourn, 1996). The reaction by-products are predominantly isothiocyanates (ITCs) having biocidal distinctiveness against a wide range of insects, nematodes, fungal pathogens, bacteria, and weeds (Brown and Morra, 1997; Bednarek, 2012).
The defensins are small cysteine-rich proteins that show broad-spectrum anti-microbial activity and were first isolated from the endosperm of Hordeum vulgare and Triticum aestivum. They are predominantly characterized in the seeds but can be present in all types of plant tissues (leaves, pods, tubers, fruit, roots, bark, and flowers). The specific mechanisms employed by defensins to inhibit fungal growth are still being characterized, but they appear to act upon molecular targets in the plasma membrane of pathogens. The chitinases and glucanases are enzymes that catalyze the degradation of chitin and glucans, respectively, polymers with a backbone similar to cellulose that is present in the cell walls of true fungi. The in vitro analysis has confirmed the anti-fungal activities of these enzymes, and transgenic plants expressing a high concentration of these enzymes exhibit increased resistance to a wide range of pathogens (Freeman and Beattie, 2008). Another class of cysteine-rich anti-fungal proteins is 2S albumin, AFP1, and AFP2 commonly present in members of the Brassicaceae family displaying broad antifungal properties (Terras et al., 1993). The receptor-like protein (RLP) encoded by a Ve1 gene from tomato has fungicidal properties and is transformed successfully into A. thaliana. However, very little is known about the RLP signaling in pathogen resistance (Fradin et al., 2011). Thionins are cysteine-rich proteins of low molecular weight (5 kDa) with anti-fungal activities and mainly accumulated in seeds but may be present in stems, roots, or leaves (Bohlmann, 1994; Bohlmann et al., 1998). These are toxic to a broad range of fungal pathogens, presumably attacking the cell membrane to increase their permeability and consequently leading to cell death (Cammue et al., 1992; Terras et al., 1995; Epple et al., 1997; Holtorf et al., 1998; Chan et al., 2005). The transgenics overexpressing thionin genes showed enhanced resistance against phytopathogens (Kanzaki et al., 2002; Khan et al., 2006; Almasia et al., 2008; Hao et al., 2016). A large number of thionin producing genes have been identified in members of the Brassicaceae family such as Arabidopsis, Eutrema, and Raphanus (Hoshikawa et al., 2012). The Kelch domain protein of Trichoderma harzianum T34 encoded by the Thkel1 gene was found to confer salt tolerance and also improve resistance against phytopathogens by induction of Jasmonic acid-mediated systemic resistance and myrosinase activity when overexpressed in transformed B. napus (Poveda et al., 2019). It was shown that a pair of A. thaliana TIR-NLR proteins, RRS1 and RPS4, function together in disease resistance against different phytopathogen isolates. These dual R proteins also confer resistance to Ralstonia solanacearum wilt in transgenic Brassica crops (Narusaka et al., 2014). The in vitro bioassays demonstrated that ARACIN1 and ARACIN2 peptides have antifungal properties against necrotrophic fungi such as B. cinerea, A. brassicicola, Fusarium graminearum, and S. sclerotiorum, and yeast (Saccharomyces cerevisiae). Moreover, the transgenic A. thaliana plants expressing ARACIN1 were well protected from infections of B. cinerea and A. brassicicola (Neukermans et al., 2015).
Characterization of Pathotypes and Pathogen Effectors/Elicitors
The identification of pathogen race or strain is important for the successful management of disease outbreaks in a particular region/country as some resistant genes were identified to work against a particular race or strain. This goal can be achieved by the characterization of pathotypes through genetic markers such as race/strain/isolate specific molecular markers, proteins, and genes. The strains of A. alternata can be identified by the comparative analysis of ITS, GAPDH, and RPB2 genetic markers (Zheng et al., 2017). Rajarammohan et al. (2019a) have identified a total of 460 genes specific to A. brassicae which were absent in all other Alternaria species. These genes have included 35 secreted protein-coding genes and out which 11 were predicted to be effectors. A large number of these proteins were uncharacterized proteins with no known function. These A. brassicae specific genes and effectors could be a potential source for categorization and identification of A. brassicae races.
The advanced sequencing technology provides near-complete genome assemblies that serve better in gene predictions and will also provide improved results for structures and functions of effector proteins. The characterization of effector genes/proteins can be a potential identification source for pathotypes (Gibriel et al., 2016). However, the effector candidates characterized functionally to date were below 300 amino acids with ∼4 cysteine residues (Stergiopoulos and de Wit, 2009). These characteristics can not be used as identification criteria because well-characterized effectors lack these properties (Lo Presti et al., 2015; Sperschneider et al., 2015). Therefore, the universal characters of effector candidates such as their secretion and differential expression in crops should be considered (Sperschneider et al., 2015). The effector candidates identified by comparative genome-based studies can be used in R gene identification (Vleeshouwers et al., 2008). The monitoring of the effector candidate diversity of pathotypes and deployment of plants carrying related R gene would contribute to disease management (Gibriel et al., 2016).
Sclerotinia sclerotiorum carried SCFE1 elicitor which evokes MTI responses and is sensed by RLP30, which confirms the association of MTI in resistance to S. sclerotiorum (Zhang et al., 2013). Some other elicitors, such as HRE (A heat-released elicitor) (Bertinetti and Ugalde, 1996), SsCut (cutinase) (Zhang et al., 2014), and SsSm1 (a Cerato-platanin family protein) (Pan et al., 2018), have also been identified from S. sclerotiorum. On being challenged by Sclerotinia, these elicitors can activate the plant immune system against the invading pathogen (Wang et al., 2019). A large number of chemical elicitors such as salicylic acid, benzothiadiazole (Thakur et al., 2014), Chitosan, b-aminobutyric acid, 2, 6-dichloro-isonicotinic acid (Mamgain et al., 2019), methyl jasmonate, glucose (Augustine and Bisht, 2015), arachidonic acid (Neerja and Sohal, 2012) were applied in the management of the Alternaria blight disease. Thus, the molecular studies should focus on the characterization of pathogen-specific genes and effectors which contribute to virulence and susceptibility of the host, further influencing the improvement in our understanding of the R gene-mediated resistance.
Conclusion
The identification and introgression of resistant gene sources into B. juncea against major fungal pathogens is a difficult procedure owing to the close association of R genes with other agronomic traits, variations in host resistance-response against pathogenic races, the polygenic inheritance of resistance, and gene interactions. Also, the identification of R genes is cumbersome because of factors like genetic variations in host subpopulations and pathotypes, the emergence of new pathotypes, genomic tools used, environmental factors, resistance evaluation techniques, scoring criteria, etc. However, continuous efforts are being made to identify R genes or quantitative trait loci for major fungal diseases of Brassica crops. A stable and long-lasting resistance source is a prerequisite for resistance breeding programs. The cloning of previously characterized R genes in Brassica will help in the identification of plant defense mechanisms that support the search of novel R gene sources. The studies on host–pathogen interactions are important to have an insight into the pathogenesis and response of the host against pathogen infection. The genome assembly database played a vital role in identifying novel R genes as the advanced sequencing technologies provide near-complete genome assemblies of diploids or polyploids. This may allow us to identify race-specific or non-specific R genes.
There is a need for universally accepted robust screening and scoring methods for each host-pathogen systems so that true R gene sources could be identified. To increase host resistance for wider pathogen races, gene pyramiding provides durable resistance and sustainable establishment of crop plants in disease-prone areas. Improving knowledge about elicitors and effectors is also important to develop successful disease management strategies. The development of novel pre-breeding material from close or distant resistant wild relatives and marker-assisted selection approach may help to efficiently facilitate Brassica-improvement programs. This review presents a comprehensive analysis of the R gene sources and their utilization in B. juncea for improvement through advanced molecular techniques.
Author Contributions
KS and PK conceptualized and drafted the manuscript, prepared the tables and figures. PK and PR edited and finalized the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
PK acknowledges financial support under WOS-A scheme (File No. SR/WOS-A/LS-373/2018) from the Department of Science and Technology (DST), Ministry of Science and Technology, Govt. of India, New Delhi. KS acknowledges for fellowship support [File No. 19/1247(0001)/19-EMR-I] from HRDG-Council of Scientific & Industrial Research, New Delhi.
References
Adam, L., and Somerville, S. C. (1996). Genetic characterization of five powdery mildew disease resistant loci in Arabidopsis thaliana. Plant J. 9, 341–356. doi: 10.1046/j.1365-313x.1996.09030341.x
Agarwal, R. A. K., Kumar, A., and Thakur, H. L. (1997). Effect of Sclerotinia rot on oil quality in low erucic acid cultivars of rapeseed. Crucif. Newslett. 19, 103–104.
Agnihotri, A., Gupta, K., Prem, D., Sarkar, G., Mehra, V. S., and Zargar, S. M. (2009). “Genetic enhancement in rapeseed- mustard for quality and disease resistance through in vitro techniques,” in Proceedings of 16th Australian Research Assembly on Brassicas, Ballarat.
Agnihotri, A., Lakshmikumaran, M. S., Jagannathan, V., and Shivanna, K. R. (1991). Wide hybridization for improvement in cultivated Brassicas. Acta Hortic. 289, 213–214. doi: 10.17660/actahortic.1991.289.49
Ahmed, N. U., Park, J. I., Jung, H. J., Seo, M. S., Kumar, T. S., Lee, I. H., et al. (2012). Identification and characterization of stress resistance related genes of Brassica rapa. Biotechnol. Lett. 34, 979–987. doi: 10.1007/s10529-012-0860-4
Ali, S., Mir, Z. A., Tyagi, A., Mehari, H., Meena, R. P., Bhat, J. A., et al. (2017). Overexpression of NPR1 in Brassica juncea confers broad spectrum resistance to fungal pathogens. Front. Plant Sci. 8:1693. doi: 10.3389/fpls.2017.01693
Alkooranee, J. T., Yin, Y., Aledan, T. R., Jiang, Y., Lu, G., Wu, J., et al. (2015). Systemic resistance to powdery mildew in Brassica napus (AACC) and Raphanus alboglabra (RRCC) by Trichoderma harzianum TH12. PLoS One 10:e0142177. doi: 10.1371/journal.pone.0142177
Almasia, N. I., Bazzini, A. A., Hopp, H. E., and Vazquez-Rovere, C. (2008). Overexpression of snakin-1 gene enhances resistance to Rhizoctonia solani and Erwinia carotovora in transgenic potato plants. Mol. Plant Pathol. 9, 329–338. doi: 10.1111/j.1364-3703.2008.00469.x
Aneja, J. K., Agarwal, A., and Agnihotri, A. (2014). Inter and intra-specific diversity in Alternaria species infecting oilseed Brassicas in India. J. Oilseed Brass. 5, 102–117.
Arora, H., Padmaja, K. L., Paritosh, K., Mukhi, N., Tewari, A. K., Mukhopadhyay, A., et al. (2019). BjuWRR1, a CC-NB-LRR gene identified in Brassica juncea, confers resistance to white rust caused by Albugo candida. Theoret. Appl. Genet. 132, 2223–2236. doi: 10.1007/s00122-019-03350-z
Attanayake, R. N., Tennekoon, V., Johnson, D. A., Porter, L. D., del Río-Mendoza, L., Jiang, D., et al. (2014). Inferring outcrossing in the homothallic fungus Sclerotinia sclerotiorum using linkage disequilibrium decay. Heredity 113, 353–363. doi: 10.1038/hdy.2014.37
Aubertot, J. N., Sohbi, Y., Brun, H., Penaud, A., and Nutter, F. W. (2006). Phomadidacte: a computer aided training program for the severity assessment of phoma stem canker of oilseed rape. IOBC Bull. 29, 247–254.
Augustine, R., and Bisht, N. C. (2015). Biotic elicitors and mechanical damage modulate glucosinolate accumulation by co-ordinated interplay of glucosinolate biosynthesis regulators in polyploid Brassica juncea. Phytochemistry 117, 43–50. doi: 10.1016/j.phytochem.2015.05.015
Awasthi, R. P., and Kolte, S. J. (1989). Variability in Alternaria brassicae affecting rapeseed and mustard. Indian Phytopath. 42:275.
Axelsson, T., Bowman, C. M., Sharpe, A. G., Lydiate, D. J., and Lagercrantz, U. (2000). Amphidiploid Brassica juncea contains conserved progenitor genomes. Genome 43, 679–688. doi: 10.1139/gen-43-4-679
Ayuke, F. O., Lagerlöf, J., Jorge, G., Söderlund, S., Muturi, J. J., Sarosh, B. R., et al. (2017). Effects of biocontrol bacteria and earthworms on the severity of Alternaria brassicae disease and the growth of oilseed rape plants (Brassica napus). Appl. Soil Ecol. 117–118, 63–69. doi: 10.1016/j.apsoil.2017.04.019
Bains, P. S., and Tewari, J. P. (1987). Purification, chemical characterization and host-specificity of the toxin produced by Alternaria brassicae. Physiol. Mol. Plant Pathol. 30, 259–271. doi: 10.1016/0885-5765(87)90039-7
Bains, S. S., and Jhooty, J. S. (1979). Mixed infections by Albugo candida and Peronospora parasitica on Brassica juncea inflorescence and their control. Indian Phytopathol. 32, 268–271.
Banga, S. S., Banga, S. K., and Labana, K. S. (1983). “Nucleo-cytoplasmic interactions in Brassica,” in Proceedings of 6th International Rapeseed Congress, 602-606, Paris, France. Groupe Consultatif International de Recherche sur la Colza [International Consultative Group for Research on Rapeseed], Paris.
Barari, H., Alavi, V., and Badalyan, S. M. (2012). Genetic and morphological differences among populations of Sclerotinina sclerotiorum by microsatellite markers, mycelial compatibility groups (MCGs) and aggressiveness in North Iran. Roman. Agric. Res. 29, 323–331.
Barbetti, M. J. (1981). Effects of sowing date and oospore seed contamination upon subsequent crop incidence of white rust (Albugo candida) in rapeseed. Aus. J. Plant Pathol. 10, 44–46. doi: 10.1071/app9810044
Barbetti, M. J., Banga, S. K., Fu, T. D., Li, Y. C., Singh, D., Liu, S. Y., et al. (2014). Comparative genotype reactions to Sclerotinia sclerotiorum within breeding populations of Brassica napus and B. juncea from India and China. Euphytica 197, 47–59. doi: 10.1007/s10681-013-1049-1
Barbetti, M. J., and Carter, E. C. (1986). “Diseases of rapeseed,” in Rapeseed in Western Australia, ed. J. A. Lawson (South Perth, WA: Western Australian Department of Agriculture), 14–19. Bulletin No. 4105.
Barret, P., Guerif, J., Reynoird, J. P., Delourme, R., Eber, F., Renard, M., et al. (1998). Selection of stable Brassica napus-Brassica juncea recombinant lines resistant to blackleg (Leptosphaeria maculans). 2. A ‘to and fro’ strategy to localize and characterize inter-specific introgressions on the B. napus genome. Theor. Appl. Genet. 96, 1097–1103. doi: 10.1007/s001220050844
Bayer, P. E., Hurgobin, B., Golicz, A. A., Chan, C. K. K., Yuan, Y., Lee, H. T., et al. (2017). Assembly and comparison of two closely related Brassica napus genomes. Plant Biotechnol. J. 15, 1–9. doi: 10.1111/pbi.12742
Bednarek, P. (2012). Sulfur-containing secondary metabolites from Arabidopsis thaliana and other Brassicaceae with function in plant immunity. Chembiochem 13, 1846–1859. doi: 10.1002/cbic.201200086
Berkenkamp, B., and Kirkham, C. (1989). Canola disease survey in N.E. Saskatchewan, 1988. Can. Plant Dis. Sur. 69:62.
Bertinetti, C., and Ugalde, R. A. (1996). Studies on the response of carrot cells to a Sclerotinia sclerotiorum elicitor: induction of the expression of an extracellular glycoprotein mRNA. Mol. Plant Microb. Interact. 9, 658–663. doi: 10.1094/MPMI-9-0658
Bhayana, L., Paritosh, K., Arora, H., Yadava, S. K., Singh, P., Nandan, D., et al. (2020). A mapped locus on LG A6 of Brassica juncea line Tumida conferring resistance to white rust contains a CNL type R gene. Front. Plant Sci. 10:1690. doi: 10.3389/fpls.2019.01690
Bittner-Eddy, P. D., Crute, I. R., Holub, E. B., and Beynon, J. L. (2000). RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 21, 177–188. doi: 10.1046/j.1365-313x.2000.00664.x
Bohlmann, H. (1994). The role of thionins in plant protection. Crit. Rev. Plant Sci. 13, 1–16. doi: 10.1080/713608052
Bohlmann, H., Vignutelli, A., Hilpert, B., Miersch, O., Wasternack, C., and Apel, K. (1998). Wounding and chemicals induce expression of the Arabidopsis thaliana gene Thi2.1, encoding a fungal defense thionin, via the octadecanoid pathway. FEBS Lett. 437, 281–286. doi: 10.1016/s0014-5793(98)01251-4
Boland, G. J., and Hall, R. (1994). Index of plant hosts of Sclerotinia. Can. J. Plant Pathol. 16, 93–108. doi: 10.1080/07060669409500766
Borhan, M. H., Brose, E., Beynon, J. L., and Holub, E. B. (2001). White rust (Albugo candida) resistance loci on three Arabidopsis chromosomes are closely linked to downy mildew (Peronospora parasitica) resistance loci. Mol. Plant Pathol. 2, 87–95. doi: 10.1046/j.1364-3703.2001.00056.x
Borhan, M. H., Gunn, N., Cooper, A., Gulden, S., Tor, M., Rimmer, S. R., et al. (2008). WRR4 encodes a TIR-NB-LRR protein that confers broad-spectrum white rust resistance in Arabidopsis thaliana to four physiological races of Albugo candida. MPMI 21, 757–768. doi: 10.1094/mpmi-21-6-0757
Borhan, M. H., Holub, E. B., Kindrachuk, C., Omidi, M., Ozorgmanesh-Frad, G., and Rimmer, S. R. (2010). WRR4, a broad-spectrum TIR-NB-LRR gene from Arabidopsis thaliana that confers white rust resistance in transgenic oilseed Brassica crops. Mol. Plant Pathol. 11, 283–291.
Botella, M. A., Parker, J. E., Frost, L. N., Bittner-Eddy, O. D., Beynon, J. L., Daniels, J. B., et al. (1998). Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10, 1847–1860. doi: 10.2307/3870908
Brasier, C. M. (2000). Plant pathology: the rise of the hybrid fungi. Nature 405, 134–135. doi: 10.1038/35012193
Brasier, C. M. (2001). Rapid evolution of introduced plant pathogens via intespecific hybridization is leading to rapid evolution of Dutch elm disease and other fungal plant pathogens. Bioscience 51, 123–133. doi: 10.1641/0006-3568(2001)051[0123:reoipp]2.0.co;2
Bremer, H., Lsmen, H., Karel, G., Ozkan, H., and Ozkan, M. (1947). Contribution to knowledge of the parasitic fungi of Turkey. Rev. Facul. Sci. Univ. Istanbul Ser. B 13, 122–172.
Brown, J. K. M., and Hovmoller, M. S. (2002). Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297, 537–541. doi: 10.1126/science.1072678
Brown, P. D., and Morra, M. J. (1997). Control of soil-borne plant pests using glucosinolate-containing plants. Adv. Agron. 61, 167–231. doi: 10.1016/s0065-2113(08)60664-1
Browne, L. M., Conn, K. L., Ayer, W. A., and Tewari, J. P. (1991). The Camalexins: new phytoalexins produced in the leaves of Camelina sativa (Cruciferae). Etrahedron 47, 3909–3914. doi: 10.1016/s0040-4020(01)86431-0
Brun, H., Chèvre, A. M., Fitt, B. D., Powers, S., Besnard, A. L., Ermel, M., et al. (2010). Quantitative resistance increases the durability of qualitative resistance to Leptosphaeria maculans in Brassica napus. New Phytol. 185, 285–299. doi: 10.1111/j.1469-8137.2009.03049.x
Brun, H., Plessis, J., and Renard, M. (1987). “Resistance of some crucifers to Alternaria brassicae (Berk.) Sacc,” in Proceedings of the Seventh International Rapeseed Congress, Poznan.
Cai, X., Wang, X., Liu, B., Wu, J., Liang, J., Cui, Y., et al. (2017). Brassica rapa genome 2.0: a reference upgrade through sequence re-assembly and gene re-annotation. Mol. Plant 10, 649–651. doi: 10.1016/j.molp.2016.11.008
Cammue, B. P. A., De Bolle, M. F. C., Terras, F. R. G., Proost, P., Van Damme, J., Rees, S. B., et al. (1992). Isolation and characterization of a novel class of plant antimicrobial peptides form Mirabilis jalapa L. seeds. J. Biol. Chem. 267, 2228–2233. doi: 10.1016/s0021-9258(18)45866-8
Canola Council of Canada (2020). Sclerotinia Stem Rot. Available online at: https://www.canolacouncil.org/canola-encyclopedia/diseases/sclerotinia-stem-rot/ (accessed June 28, 2020).
Caretto, S., Linsalata, V., Colella, G., Mita, G., and Lattanzio, V. (2015). Carbon fluxes between primary metabolism and phenolic pathway in plant tissues under stress. Int. J. Mol. Sci. 16, 26378–26394. doi: 10.3390/ijms161125967
Carlier, J. D., Alabaça, C. A., Coelho, P. S., Monteiro, A. A., and Leitão, J. M. (2011). The downy mildew resistance locus Pp523 is located on chromosome C8 of Brassica oleracea L. Plant Breed. 131, 170–175. doi: 10.1111/j.1439-0523.2011.01904.x
Carlsson, M., Bothmer, R. V., and Merker, A. (2004). Screening and evaluation of resistance to downy mildew (Peronospora parasitica) and clubroot (Plasmodiophora brassicae) in genetic resources of Brassica oleracea. Hereditas 141, 293–300. doi: 10.1111/j.1601-5223.2004.01818.x
Cevik, V., Boutrot, F., Apel, W., Robert-Seilaniantz, A., Furzer, O. J., Redkar, A., et al. (2019). Transgressive segregation reveals mechanisms of Arabidopsis immunity to Brassica-infecting races of white rust (Albugo candida). Proc. Natl. Acad. Sci. U.S.A. 116, 2767–2773. doi: 10.1073/pnas.1812911116
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chan, Y. L., Prasad, V., Sanjaya, L., Chen, K. H., Liu, P. C., Chan, M. T., et al. (2005). Transgenic tomato plants expressing an Arabidopsis thionin (Thi2.1) driven by fruit-inactive promoter battle against phytopathogenic attack. Planta 221, 386–393. doi: 10.1007/s00425-004-1459-3
Chattopadhyay, C., Agarwal, R., Kumar, A., Meena, R. L., Faujdar, K., Chakravarthy, N. V. K., et al. (2011). Epidemiology and development of forecasting models for White rust of Brassica juncea in India. Archiv. Phytopathol. Plant Protect. 44, 751–763. doi: 10.1080/03235400903458571
Chen, X., Hou, X., Zhang, J., and Zheng, J. (2008). Molecular characterization of two important antifungal proteins isolated by downy mildew infection in non-heading Chinese cabbage. Mol. Biol. Rep. 35, 621–629. doi: 10.1007/s11033-007-9132-0
Cheung, W. Y., Gugel, R. K., and Landry, B. S. (1998). Identification of RFLP markers linked to the white rust resistance gene (Acr) in mustard (Brassica juncea (L.) Czern. and Coss.). Genome 41, 626–628. doi: 10.1139/gen-41-4-626
Chevre, A. M., Barret, P., Eber, F., Dupuy, P., Brun, H., Tanguy, X., et al. (1997). Selection of stable Brassica napus-B. juncea recombinant lines resistant to blackleg (Leptosphaeria maculans). 1. Identification of molecular markers, chromosomal and genomic origin of the introgression. Theor. Appl. Genet. 95, 1104–1111. doi: 10.1007/s001220050669
Chèvre, A. M., Brun, H., Eber, F., Letanneur, J. C., Vallee, P., Ermel, M., et al. (2008). Stabilization of resistance to Leptosphaeria maculans in Brassica napus-B. juncea recombinant lines and its introgression into spring-type Brassica napus. Plant Dis. 92, 1208–1214. doi: 10.1094/pdis-92-8-1208
Chevre, A. M., Eber, F., Margale, E., and Kerlan, M. C. (1994). Comparison of somatic and sexual Brassica napus Sinapis alba hybrids and their progeny by cytogenetic studies and molecular characterization. Genome 37, 367–371. doi: 10.1139/g94-052
Chevre, A. M., Eber, F., This, P., Barret, P., Tanguy, X., Brun, H., et al. (1996). Characterization of Brassica nigra chromosomes and of blackleg resistance in B. napus-B. nigra addition lines. Plant Breed. 115, 113–118. doi: 10.1111/j.1439-0523.1996.tb00884.x
Chèvre, A. M., Leon, A. P., Jenczewski, E., Eber, F., Delourme, R., et al. (2003). “Introduction of blackleg resistance from Brassica rapa into Brassica napus,” in Proceedings of the 6th Blackleg Workshop, Copenhagen.
Chhikara, S., Chaudhury, D., Dhankher, O. P., and Jaiwal, P. K. (2012). Combined expression of a barley class II chitinase and type I ribosome inactivating protein in transgenic Brassica juncea provides protection against Alternaria brassicae. Plant Cell Tiss. Organ Cult. 108, 83–89. doi: 10.1007/s11240-011-0015-7
Choi, Y. J., Hong, S. B., and Shin, H. D. (2006). Genetic diversity within the Albugo candida complex (Peronosporales, Oomycota) inferred from phylogenetic analysis of ITS rDNA and COX2 mtDNA sequences. Mol. Phylogenet. Evol. 40, 400–409. doi: 10.1016/j.ympev.2006.03.023
Choi, Y. J., Park, M. J., Park, J. H., and Shin, H. D. (2011). White blister rust caused by Albugo candida on oilseed rape in Korea. Plant Pathol. J. 27, 192. doi: 10.5423/ppj.2011.27.2.192
Choi, Y. J., Shin, H. D., Hong, S. B., and Thines, M. (2007). Morphological and molecular discrimination among Albugo candida materials infecting Capsella bursa-pastoris world-wide. Fungal Divers. 27, 11–34.
Choi, Y. J., Shin, H. D., Hong, S. B., and Thines, M. (2009). The host range of Albugo candida extends from Brassicaceae through cleomaceae to capparaceae. Mycol. Prog. 8, 329–335. doi: 10.1007/s11557-009-0604-6
Choi, Y. J., Shin, H. D., Ploch, S., and Thines, M. (2008). Evidence for uncharted biodiversity in the Albugo candida complex, with the description of a new species. Mycol. Res. 112, 1327–1334. doi: 10.1016/j.mycres.2008.04.015
Christianson, J. A., Rimmer, S. R., Good, A. G., and Lydiate, D. J. (2006). Mapping genes for resistance to Leptosphaeria maculans in Brassica juncea. Genome 49, 30–41. doi: 10.1139/G05-085
Clarkson, J. P., Coventry, E., Kitchen, J., Carter, H. E., and Whipps, J. M. (2012). Population structure of Sclerotinia sclerotiorum in crop and wild hosts in the UK. Plant Pathol. 62, 309–324. doi: 10.1111/j.1365-3059.2012.02635.x
Coelho, P. S., Leckie, D., Bahcevandziev, K., Valério, L., Astley, D., Louise, N., et al. (1998). The relationship between cotyledon and adult plant resistance to downy mildew (Peronospora parasitica) in Brassica oleracea. Acta Hortic. 459, 335–342. doi: 10.17660/actahortic.1998.459.39
Coelho, P. S., and Monteiro, A. A. (2018). Genetic and histological characterization of downy mildew resistance at the cotyledon stage in Raphanus sativus L. Euphytica 214:208. doi: 10.1007/s10681-018-2289-x
Coelho, P. S., Valério, L., and Monteiro, A. A. (2009). Leaf position, leaf age and plant age affect the expression of downy mildew resistance in Brassica oleracea. Eur. J. Plant Pathol. 125, 179–188. doi: 10.1007/s10658-009-9469-4
Conn, K. L., and Tewari, J. P. (1986). Hypersensitive reaction induced by Alternaria brassicae in Eruca sativa, an oil-yielding crucifer. Can. J. Plant Pathol. 8:348.
Conn, K. L., and Tewari, J. P. (1990). Survey of Alternaria blackspot and Sclerotinia stem rot in central Alberta in 1989. Can. Plant Dis. Surv. 70, 66–67.
Conn, K. L., Tewari, J. P., and Awasthi, R. P. (1990). A disease assessment key for Alternaria black spot in rapeseed and mustard. Can. Plant Dis. Surv. 70:1.
Conn, K. L., Tewari, J. P., and Dahiya, J. S. (1988). Resistance to Alternaria brassicae and phytoalexins-elicitation in rapeseed and other crucifers. Plant Sci. 56, 21–25. doi: 10.1016/0168-9452(88)90180-X
Cooper, A. J., Latunde-Dada, A. O., Woods-Tor, A., Lynn, J., Lucas, J. A., Crute, I. R., et al. (2008). Basic compatibility of Albugo candida in Arabidopsis thaliana and Brassica juncea causes broad-spectrum suppression of innate immunity. Mol. Plant Microb. Interact. 21, 745–756. doi: 10.1094/MPMI-21-6-0745
Dang, J. K., Sangwan, M. S., Mehta, N., and Kaushik, C. D. (2000). Multiple disease resistance against four fungal foliar diseases of rapeseed-mustard. Indian Phytopath. 53, 455–458.
Dange, K. K., Patel, R. L., Patel, S. I., and Patel, K. K. (2002). Assessment of losses in yield due to powdery mildew disease in mustard under north Gujarat conditions. J. Mycol. Plant Pathol. 32, 249–250.
Delourme, R., Pilet-Nayel, M. L., Archipiano, M., Horvais, R., Tanguy, X., Rouxel, T., et al. (2004). A cluster of major specific resistance genes to Leptosphaeria maculans in Brassica napus. Phytopathology 94, 578–583. doi: 10.1094/phyto.2004.94.6.578
Delwiche, P. A., and Williams, P. H. (1974). Resistance to Albugo candida race 2 in Brassica sp. Proc. Amer. Phytopathol. Soc. 1:66.
Desai, A. G., Chattopadhyay, C., Agrawal, R., Kumar, A., Meena, R. L., Meena, P. D., et al. (2004). Brassica juncea powdery mildew epidemiology and weatherbased forecasting models for India - a case study. J. Plant Dis. Protect. 111, 429–438.
Dickson, M. H., and Petzoldt, R. (1993). Plant age and isolate source affect expression of downy mildew resistance in broccoli. Hortscience 28, 730–731. doi: 10.21273/hortsci.28.7.730
Dilmaghani, A., Balesdent, M., Didier, J., Wu, C., Davey, J., Barbetti, M. J., et al. (2009). The Leptosphaeria maculans-Leptosphaeria biglobosa species complex in the American continent. Plant Pathol. 58, 1044–1058.
Dixelius, C. (1999). Inheritance of the resistance to Leptosphaeria maculans of Brassica nigra and B. juncea in near isogenic lines of B. napus. Plant Breed. 118, 151–156. doi: 10.1046/j.1439-0523.1999.118002151.x
Dixelius, C., and Wahlberg, S. (1999). Resistance to Leptosphaeria maculans is conserved in a specific region of the Brassica B genome. Theor. Appl. Genet. 99, 368–372. doi: 10.1007/s001220051246
Doughty, K. J., Blight, M. M., Bock, C. H., Fieldsend, J. K., and Pickett, J. A. (1996). Release of alkenyl isothiocyanates and other volatiles from Brassica rapa seedlings during infection by Alternaria brassicae. Phytochemistry 43, 371–374. doi: 10.1016/0031-9422(96)00189-6
Ebrahimi, A. G., Delwiche, P. A., and Williams, P. H. (1976). Resistance in Brassica juncea to Peranospora parasítica and Albugo candida Race 2. Proc. Am. Phytopathol. Soc. 3:273.
Elliott, V. L., Norton, R. M., Khangura, R. K., Salisbury, P. A., and Marcroft, S. J. (2015). Incidence and severity of blackleg caused by Leptosphaeria spp. in juncea canola (Brassica juncea L.) in Australia. Austral. Plant Pathol. 44, 149–159. doi: 10.1007/s13313-014-0337-0
Epple, P., Apel, K., and Bohlmann, H. (1997). Overexpression of an endogenous thionin enhances resistance of Arabidopsis against Fusarium oxysporum. Plant Cell 9, 509–520. doi: 10.2307/3870503
Farinhó, M., Coelho, P., Monteiro, A., and Leitão, J. (2007). SCAR and CAPS markers flanking the Brassica oleracea L. Pp523 downy mildew resistance locus demarcate a genomic region syntenic to the top arm end of Arabidopsis thaliana L. chromosome 1. Euphytica 157, 215–221. doi: 10.1007/s10681-007-9414-6
Farnham, M. W., Wang, M., and Thomas, C. E. (2002). A single dominant gene for downy mildew resistance in broccoli. Euphytica 128, 405–407.
Farr, D. F., Bills, G. F., Chamuris, G. P., and Rossman, A. Y. (1989). “Ipomoea,” in Fungi on Plants and Plant Products in the United States, eds D. F. Farr, F. G. Billis, G. P. Chamuris, and A. Y. Rossman (St Paul: APS Press), 142–143.
Farr, D. F., and Rossman, A. Y. (2013). Fungal Databases,” Systematic Mycology and Microbiology Laboratory, ARS, USDA. Available online at: http://nt.Ars-grin.gov/fungaldatabases/ (accessed June 28, 2020).
Fatima, U., Bhorali, P., Borah, S., and Kumar, M. S. (2019). Perspectives on the utilization of resistance mechanisms from host and nonhost plants for durable protection of Brassica crops against Alternaria blight. PeerJ 7:e7486. doi: 10.7717/peerj.7486
Ferreira, M. E., Williams, P. H., and Osborn, T. C. (1994). RFLP mapping of Brassica napus using doubled haploid lines. Theor. Appl. Genet. 89, 615–621. doi: 10.1007/bf00222456
Fradin, E. F., Abd-El-Haliem, A., Masini, L., Van den Berg, G. C. M., Joosten, M. H. A. J., and Thomma, B. P. H. J. (2011). Interfamily Transfer of Tomato Ve1 Mediates Verticillium Resistance in Arabidopsis. Plant Physiol. 156, 2255–2265. doi: 10.1104/pp.111.180067
Franks, S. J. (2011). Plasticity and evolution in drought avoidance and escape in the annual plant Brassica rapa. New Phytol. 190, 249–257. doi: 10.1111/j.1469-8137.2010.03603.x
Fredua-Agyeman, R., Coriton, O., Huteau, V., Parkin, I. A. P., Chevre, A. M., and Rahman, H. (2014). Molecular cytogenetic identification of B genome chromosomes linked to blackleg disease resistance in Brassica napus × B. carinata interspecific hybrids. Theor. Appl. Genet. 127, 1305–1318. doi: 10.1007/s00122-014-2298-7
Freeman, B. C., and Beattie, G. A. (2008). An overview of plant defenses against pathogens and herbivores. Plant Health Instr. doi: 10.1094/phi-i-2008-0226-01
Frye, C. A., and Innes, R. W. (1998). An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10, 947–956. doi: 10.2307/3870681
Gaebelein, R., Alnajar, D., Koopmann, B., and Mason, A. (2019). Hybrids between Brassica napus and B. nigra show frequent pairing between the B and A/C genomes and resistance to blackleg. Chrom. Res. 27, 221–236. doi: 10.1007/s10577-019-09612-2
Gaikwad, K., Kirti, P. B., Sharma, A., Prakash, S., and Chopra, V. L. (1996). Cytogenetical and molecular investigations on somatic hybrids of Sinapis alba and Brassica juncea and their backcross progeny. Plant Breed. 115, 480–483. doi: 10.1111/j.1439-0523.1996.Tb00961.x
Garg, H., Atri, C., Sandhu, P. S., Kaur, B., Renton, M., Banga, S. K., et al. (2010). High level of resistance to Sclerotinia sclerotiorum in introgression lines derived from hybridization between wild crucifers and the crop Brassica species B. napus and B. juncea. Field Crops Res. 117, 51–58. doi: 10.1016/j.fcr.2010.01.013
Garg, H., Sivasithamparam, K., Banga, S. S., and Barbetti, M. J. (2008). Cotyledon assay as a rapid and reliable method of screening for resistance against Sclerotinia sclerotiorum in Brassica napus genotypes. Austral. Plant Pathol. 37, 106–111. doi: 10.1071/ap08002
Ge, X. T., You, M. P., and Barbetti, M. J. (2015). Virulence differences among Sclerotinia sclerotiorum isolates determines host cotyledon resistance responses in Brassicaceae genotypes. Eur. J. Plant Pathol. 143, 527–541. doi: 10.1007/s10658-015-0696-6
Ghasolia, R. P., Shivpuri, A., and Bhargava, A. K. (2004). Sclerotinia rot of Indian mustard (Brassica juncea) in Rajasthan. Indian Phytopathol. 57, 76–79.
Gibriel, H. A., Thomma, B. P., and Seidl, M. F. (2016). The age of effectors: genome-based discovery and applications. Phytopathology 106, 1206–1212. doi: 10.1094/PHYTO-02-16-0110-FI
Gladders, P. (1987). Current status of diseases and disease control in winter oilseed rape in England and Wales. Bull. SROP 10, 7–10.
Golicz, A. A., Bayer, P. E., Barker, G. C., Edger, P. P., Kim, H., Martinez, P. A., et al. (2016). The pangenome of an agronomically important crop plant Brassica oleracea. Nat. Commun. 7:13390. doi: 10.1038/ncomms13390
Goyal, P., Chattopadhyay, C., Mathur, A. P., Kumar, A., Meena, P. D., Datta, S., et al. (2013). Pathogenic and molecular variability among Brassica isolates of Alternaria brassicae from India. Ann. Plant Protect. Sci. 21, 349–359.
Gupta, M. L., Singh, G., Raheja, R. K., Ahuja, K. L., and Banga, S. K. (1997). Chlorophyll content in relation to white rust (A. candida) resistance in Indian mustard. Crucif. Newslett. 19, 105–106.
Gupta, N. C., Sharma, P., Rao, M., Rai, P. K., and Gupta, A. K. (2020). Evaluation of non-injury inoculation technique for assessing Sclerotinia stem rot (Sclerotinia sclerotiorum) in oilseed Brassica. J. Microbiol. Methods 175:105983. doi: 10.1016/j.mimet.2020.105983
Gyawali, S., Harrington, M., Durkin, J., and Hornerauthor, K. (2016). Microsatellite markers used for genome-wide association mapping of partial resistance to Sclerotinia sclerotiorum in a world collection of Brassica napus. Mol. Breed. 36:72. doi: 10.1007/s11032-016-0496-5
Hansen, L. H., and Earle, E. D. (1995). Transfer of resistance to Xanthomonas cam-pestris pv. campestris(L.) by proto-plast fusion. Theor. Appl. Genet. 91, 1293–1300. doi: 10.1007/BF00220944
Hansen, L. H., and Earle, E. D. (1997). Somatic hybrids between Brassica oleracea (L.) and Sinapis alba (L.) with re-sistance to Alternaria brassicae (Berk.) Sacc. Theor. Appl. Genet. 94, 1078–1085. doi: 10.1007/s001220050518
Hao, G., Stover, E., and Gupta, G. (2016). Overexpression of a modified plant thionin enhances disease resistance to citrus canker and huanglongbing (HLB). Front. Plant Sci. 7:1078. doi: 10.3389/fpls.2016.01078
Hao, J. J., Subbarao, K. V., and Duniway, J. M. (2003). Germination of Sclerotinia minor and S. sclerotiorum sclerotia under various soil moisture and temperature combinations. Phytopathology 93, 443–450. doi: 10.1094/phyto.2003.93.4.443
Harper, F. R., and Pittman, U. J. (1974). Yield loss by Brassica campestris and Brassica napus from systemic stem infection by Albugo curciferarum. Phytopathology 64, 408–410. doi: 10.1094/phyto-64-408
Hemmati, R., Javan-Nikkhah, M., and Linde, C. C. (2009). Population genetic structure of Sclerotinia sclerotiorum on canola in Iran. Eur. J. Plant Pathol. 125, 617–628. doi: 10.1007/s10658-009-9510-7
Hirata, S. (1954). Studies on the phytohormone in the malformed portion of the diseased plants. I. The relation between the growth rate and the amount of free auxin in the fungous galls and virusinfected plants. Ann. Phytopathol. Soc. Jpn. 19, 33–38. doi: 10.3186/jjphytopath.19.33
Holtorf, S., Ludwig-Müller, J., Apel, K., and Bohlmann, H. (1998). High-level expression of a viscotoxin in Arabidopsis thaliana gives enhanced resistance against Plasmodiophora brassicae. Plant Mol. Biol. 36, 673–680.
Hong, C. X., and Fitt, B. D. L. (1995). Effects of inoculum concentration, leaf age and wetness period on the development of dark leaf and pod spot (Alternaria brassicae) on oilseed rape (Brassica napus). Plant Dis. 127, 283–295. doi: 10.1111/j.1744-7348.1995.Tb06673.x
Hoshikawa, K., Ishihara, G., Takahashi, H., and Nakamura, I. (2012). Enhanced resistance to gray mold (Botrytis cinerea) in transgenic potato plants expressing thionin genes isolated from Brassicaceae species. Plant Biotechnol. 29, 87–93. doi: 10.5511/plantbiotechnology.12.0125a
Hossain, M. R., Ferdous, M. J., Park, J., Robin, A. H. K., Natarajan, S., Jung, H., et al. (2020). In-silico identification and differential expression of putative disease resistance-related genes within the collinear region of Brassica napus blackleg resistance locus LepR2’ in Brassica oleracea. Hortic. Environ. Biotechnol. 61, 879–890. doi: 10.1007/s13580-020-00271-5
Howlett, B. J., Idnurm, A., and Pedras, M. S. C. (2001). Leptosphaeria maculans, the causal agent of blackleg disease of Brassicas. Fungal Genet. Biol. 33, 1–14. doi: 10.1006/fgbi.2001.1274
Huang, Y. J., Jestin, C., Welham, S. J., King, G. J., Manzanares-Dauleux, M. J., Fitt, B. D. L., et al. (2016). Identification of environmentally stable QTL for resistance against Leptosphaeria maculans in oilseed rape (Brassica napus). Theor. Appl. Genet. 129, 169–180. doi: 10.1007/s00122-015-2620-z
Huang, Y. J., Paillard, S., Kumar, V., King, G. J., Fitt, B. D. L., and Delourme, R. (2019). Oilseed rape (Brassica napus) resistance to growth of Leptosphaeria maculans in leaves of young plants contributes to quantitative resistance in stems of adult plants. PLoS One 14:e0222540. doi: 10.1371/journal.pone.0222540
Hulbert, S. H., Webb, C. A., Smith, S. M., and Sun, Q. (2001). Resistance gene complexes: evolution and utilization. Ann. Rev. Phytopathol. 39, 285–312. doi: 10.1146/annurev.phyto.39.1.285
Humaydan, H. S., and Williams, P. H. (1976). Inheritance of seven characters in Raphanus sativus. Hort. Sci. 11, 146–147.
Inturrisi, F. C. (2018). Genome-Wide Analysis of NBS-LRR genes in Indian Mustard (Brassica juncea) and Prediction of Candidate Disease Resistance Genes. Ph. D. thesis, The University of Western Australia, Perth. doi: 10.26182/5b57f1254d265
Jejelowo, O. A., Conn, K. L., and Tewari, J. P. (1991). Relationship between conidial concentration, germling growth and phytoalexin production by Camelina sativa leaves inoculated with Alternaria brassicae. Mycol. Res. 95, 928–934. doi: 10.1016/s0953-7562(09)80089-0
Jensen, B. D., Værbak, S., Munk, L., and Andersen, S. B. (1999). Characterization and inheritance of partial resistance to downy mildew, Peronospora parasitica, in breeding material of broccoli, Brassica oleracea convar. botrytis var. italica. Plant Breed. 118, 549–554. doi: 10.1046/j.1439-0523.1999.00409.x
Jimenez, L. D., Ayer, W. A., and Tewari, J. P. (1997). Phytoalexins produced in leaves of Capsella bursa-pastoris (Shepherd’s Purse). Phytoprotection 78, 99–103. doi: 10.7202/706124ar
Jones, D., and Gray, E. G. (1973). Factors affecting germination of sclerotia of Sclerotinia sclerotiorum from peas. Trans. Br. Mycol. Soc. 60, 495–500. doi: 10.1016/s0007-1536(73)80033-6
Jones, F. M. H., and Phelps, K. (1989). Climatic factors influencing spore production in Alternaria brassicae and Alternaria brassicicola. Plant Pathol. 114, 449–458. doi: 10.1111/j.1744-7348.1989.Tb03360.x
Jouet, A., Saunders, D. G. O., McMullan, M., Ward, B., Furzer, O., Jupe, F., et al. (2019). Albugo candida race diversity, ploidy and host-associated microbes revealed using DNA sequence capture on diseasedplants in the field. New Phytol. 221, 1529–1543. doi: 10.1111/nph.15417
Jourdan, P., and Salazar, E. (1993). Brassica carinata resynthesized by protoplast fusion. Theor. Appl. Genet. 86, 567–572. doi: 10.1007/BF00838710
Kaczmarek, J., and Jêdryczka, M. (2011). Characterization of two coexisting pathogen populations of Leptosphaeria spp., the cause of stem canker of Brassicas. Acta Agrobot. 64, 3–14. doi: 10.5586/aa.2011.012
Kadian, A. K., and Saharan, G. S. (1983). Symptomatology, host range and assessment oflosses due to Alternaria brassicae infection inrapeseed and mustard. Indian J. Mycol. Pl. Pathol. 13, 319–323.
Kamble, A., and Bhargava, S. (2007). β-Aminobutyric acid-induced resistance in Brassica juncea against the necrotrophic pathogen Alternaria brassicae. J. Phytopathol. 155, 152–158. doi: 10.1111/j.1439-0434.2007.01209.x
Kanrar, S., Venkateswari, J. C., Kirti, P. B., and Chopra, V. L. (2002). Transgenic expression of hevein, the rubber tree lectin, in Indian mustard confers protection against Alternaria brassicae. Plant Sci. 162, 441–448. doi: 10.1016/s0168-9452(01)00588-x
Kanzaki, H., Nirasawa, S., Saitoh, H., Ito, M., Nishihara, M., Terauchi, R., et al. (2002). Overexpression of the wasabi defensin gene confers enhanced resistance to blast fungus (Magnaporthe grisea) in transgenic rice. Theor. Appl. Genet. 105, 809–814. doi: 10.1007/s00122-001-0817-9
Kaur, P., Banga, S., Kumar, N., Gupta, S., Akhatar, J., and Banga, S. S. (2014). Polyphyletic origin of Brassica juncea with B. rapa and B. nigra (Brassicaceae) participating as cytoplasm donor parents in independent hybridization events. Am. J. Bot. 101, 1157–1166. doi: 10.3732/ajb.1400232
Kaur, P., Jost, R., Sivasithamparam, K., and Barbetti, M. J. (2011a). Proteome analysis of the Albugo candida-Brassica juncea pathosystem reveals that the timing of the expression of defense-related genes is a crucial determinant of pathogenesis. J. Exper. Bot. 62, 1285–1298. doi: 10.1093/jxb/erq365
Kaur, P., Sivasithamparam, K., Li, H., and Barbetti, M. J. (2011b). Pre-inoculation with Hyaloperonospora parasitica reduces incubation period and increases severity of disease caused by Albugo candida in a Brassica juncea variety resistant to downy mildew. J. Gen. Plant Pathol. 77, 101–106. doi: 10.1007/s10327-011-0293-2
Kaur, P., Li, C. X., Barbetti, M. J., You, M. P., Li, H., and Sivasithamparam, K. (2008). First report of powdery mildew caused by Erysiphe cruciferarum on Brassica juncea in Australia. Plant Dis. 92:650. doi: 10.1094/pdis-92-4-0650c
Kaur, S., Singh, G., and Banga, S. S. (2007). “Documenting variation in Alternaria brassicae isolates based on conidial morphology, fungicidal sensitivity and molecular profile,” in Proceedings of the International Rapeseed Congress-iv on Sustainable Development in Cruciferous Oilseed Crops Production, Wuhan.
Kevin, J. D., Blight, M. M., Bock, C. H., Fieldsend, J. K., and Picket, J. A. (1996). Release of alkenyl isothiocyanates and other volatiles from brassica rapa seedlings during infection by Alternaria brassicae. Phytochemistry 43, 371–374.
Khan, R. S., Nishihara, M., Yamamura, S., Nakamura, I., and Mii, M. (2006). Transgenic potatoes expressing wasabi defensin peptide confer partial resistance to gray mold (Botrytis cinerea). Plant Biotechnol. 23, 179–183. doi: 10.5511/plantbiotechnology.23.179
Khangura, R., and Barbetti, M. J. (2001). Prevalence of blackleg (Leptosphaeria maculans) on canola (Brassica napus) in Western Australia. Austr. J. Exper. Agric. 41, 71–80. doi: 10.1071/ea00068
Kiermayer, O. (1958). Paper chromatographic studies of the growth substances of Capsella bursa-pastoris after infection by Albugo candida and Peronospora parasitica. Osterr. Bot. Z. 105, 515–528.
Kim, J. Y., Kim, B. S., Cho, S. E., and Shin, H. D. (2013). First report of powdery mildew caused by Erysiphe cruciferarum on Indian mustard (Brassica juncea) in Korea. Plant Dis. 97:1383. doi: 10.1094/PDIS-04-13-0378-PDN
Kim, S., Song, Y. H., Lee, J.-Y., Choi, S. R., Dhandapani, V., Jang, C. S., et al. (2011). Identification of the BrRHP1 locus that confers resistance to downy mildew in Chinese cabbage (Brassica rapa ssp. pekinensis) and development of linked molecular markers. Theor. Appl. Genet. 123, 1183–1192. doi: 10.1007/s00122-011-1658-9
Kirti, P. B., Prakash, S., and Chopra, V. L. (1991). Interspecific hybridization between Brassica juncea and B. spinescens through protoplast fusion. Plant Cell Rep. 9, 639–642. doi: 10.1007/BF00231806
Klemm, M. (1938). The most important diseases and pests of Colza and Rape. Dtsch. Landw. 20, 251–252.
Kohn, L. M., Stasovski, E., Carbone, I., Royer, J., and Anderson, J. B. (1991). Mycelial incompatibility and molecular markers identify genetic variability in field populations of Sclerotinia sclerotiorum. Phytopathology 81, 480–485. doi: 10.1094/Phyto-81-480
Koike, S. T., Sullivan, M. J., Southwick, C., Feng, C., and Correll, J. C. (2011). Characterization of White Rust of Perennial Pepperweed Caused by Albugo candida in California. Plant Dis. 95, 876. doi: 10.1094/PDIS-12-10-0912
Kole, C., Teutonico, R., Mengistu, A., Williams, P. H., and Osborn, T. C. (1996). Molecular mapping of a locus controlling resistance to Albugo candida in Brassica rapa. Phytopathology 86, 367–369. doi: 10.1094/phyto-86-367
Kolte, S. J. (1985). “Diseases of annual edible oilseed crops,” in Rapeseed-Mustard and Sesame Diseases, Vol. 2, (Boca Raton, FL: CRC Press Inc), 135.
Kolte, S. J., Bardoloi, D. K., and Awasthi, R. P. (1991). “Thesearch for resistance to major diseases of rapeseed-mustard in India,” in Proceedings of GCIRC 8th Internationa lRapeseed Congress, Saskatoon.
Kumar, A., Katoch, A., Sharma, P. N., Kumari, V., and Kumar, A. (2014). Pathogenic and genetic variability in Alternaria brassicae infecting rapeseed-mustard and evaluation of resistance sources. Indian Phytopathol. 67, 257–262.
Kumar, D., Shekhar, S., Bisht, S., Kumar, V., and Varma, A. (2015). Ectopic overexpression of lectin in transgenic Brassica juncea plants exhibit resistance to fungal phytopathogen and showed alleviation to salt and drought stress. J. Bioeng. Biomed. Sci. 5:147. doi: 10.4172/2155-9538.1000147
Kumar, S. (2017). Plant secondary metabolites (PSMs) of Brassicaceae and their role in plant defense against insect herbivores -A review. J. Appl. Nat. Sci. 9, 508–519. doi: 10.31018/jans.v9i1.1222
Kumar, S., Sangwan, M. S., Mehta, N., and Kumar, R. (2003). Pathogenic diversity in isolates of Alternaria brassicae infecting rapeseed and mustard. J. Mycol. Pl. Pathol. 33, 59–64.
Kumari, P., and Bhat, S. R. (2019). Allohexaploid H2 (IC0626000; INGR18031), an Indian mustard (Brassica juncea) germplasm with heat tolerance, resistant to Alternaria brassicae. Indian J. Plant Genet. Resourc. 32:439.
Kumari, P., Bisht, D. S., and Bhat, S. R. (2018). Stable, fertile somatic hybrids between Sinapis alba and Brassica juncea show resistance to Alternaria brassicae and heat stress. Plant Cell Tiss. Organ Cult. 133, 77–86. doi: 10.1007/s11240-017-1362-9
Kumari, P., and Singh, K. P. (2019). Characterization of stable somatic hybrids of Sinapis alba and Brassica juncea for Alternaria blight, Sclerotinia sclerotiorum resistance and heat tolerance. Indian Res. J. Ext. Educ. 19, 99–103.
Kumari, P., Singh, K. P., Bisht, D., and Kumar, S. (2020b). Somatic hybrids of Sinapis alba + Brassica juncea: study of backcross progenies for morphological variations, chromosome constitution and reaction to Alternaria brassicae. Euphytica 216:93. doi: 10.1007/s10681-020-02629-3
Kumari, P., Singh, K. P., Kumar, S., and Yadava, D. K. (2020c). Development of a yellow-seeded stable allohexaploid brassica through inter-generic somatic hybridization with a high degree of fertility and resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 11:575591. doi: 10.3389/fpls.2020.575591
Kumari, P., Singh, K. P., and Rai, P. K. (2020a). Draft genome of multiple resistance donor plant Sinapisalba: an insight into SSRs, annotations and phylogenetics. PLoS One 15:e0231002. doi: 10.1371/journal.pone.0231002
Lakra, B. S., and Saharan, G. S. (1989). Correlation of leaf and staghead infection intensities of white rust with yield and yield components of mustard. Indian J. Mycol. Plant Pathol. 19, 279–281.
Larkan, N. J., Lydiate, D. J., Parkin, I. A. P., Nelson, M. N., Epp, D. J., Cowling, W. A., et al. (2013). The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 197, 595–605. doi: 10.1111/nph.12043
Larkan, N. J., Lydiate, D. J., Yu, F., Rimmer, S. R., and Borhan, M. H. (2014). Co-localisation of the blackleg resistance genes Rlm2 and LepR3 on Brassica napus chromosome A10. BMC Plant Biol. 14:387. doi: 10.1186/s12870-014-0387-z
Lee, H., Chawla, H. S., Obermeier, C., Dreyer, F., Abbadi, A., and Snowdon, R. (2020). Chromosome-scale assembly of winter oilseed rape Brassica napus. Front. Plant Sci. 11:496. doi: 10.3389/fpls.2020.00496
Li, C. X., Liu, S. Y., Sivasithamparam, K., and Barbetti, M. J. (2008). New sources of resistance to sclerotinia stem rot caused by Sclerotinia sclerotiorum in Chinese and Australian Brassica napus and Brassica juncea germplasm screened under Western Australian conditions. Austr. Plant Pathol. 38, 149–152.
Li, J., Zhao, Z., Hayward, A., Cheng, H., and Fu, D. (2015). Integration analysis of quantitative trait loci for resistance to Sclerotinia sclerotiorum in Brassica napus. Euphytica 205, 483–489. doi: 10.1007/s10681-015-1417-0
Li, Q., Li, J., Sun, J. L., Ma, X. F., Li, Y., Wang, M. W., et al. (2016). Multiple evolutionary events involved in maintaining homologs of resistance to powdery mildew 8 in Brassica napus. Front. Plant Sci. 7:1065. doi: 10.3389/fpls.2016.01065
Liban, S. H., Cross, D. J., Kutcher, H. R., Peng, G., and Fernando, W. G. D. (2016). Race structure and frequency of avirulence genes in the western Canadian Leptosphaeria maculans pathogen population, the causal agent of blackleg in Brassica species. Plant Pathol. 65, 1161–1169. doi: 10.1111/ppa.12489
Litholdo, J. C. G., Gomes, E. V., Lobo, M., Nasser, L. C. B., and Petrofeza, S. (2011). Genetic diversity and mycelial compatibility groups of the plant-pathogenic fungus Sclerotinia sclerotiorum in Brazil. Genet. Mol. Res. 10, 868–877. doi: 10.4238/vol10-2gmr937
Liu, S. Y., Liu, R. H., Latunde-Dada, A. O., Cools, H. J., Foster, S. J., Huang, Y. J., et al. (2007). Comparison of Leptosphaeria biglobosa-induced and chemically induced systemic resistance to L. maculans in Brassica napus. Chin. Sci. Bull. 52, 1053–1062. doi: 10.1007/s11434-007-0181-5
Lo Presti, L., Lanver, D., Schweizer, G., Tanaka, S., Liang, L., Tollot, M., et al. (2015). Fungal effectors and plant susceptibility. Ann. Rev. Plant Biol. 66, 513–545. doi: 10.1146/annurev-arplant-043014-114623
Loon, L. C., Rep, M., and Pieterse, C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Lucas, J. A., Crute, I. R., Sherriff’t, C., and Gordon, P. L. (1988). The identification of a gene for race-specific resistance to Peronospora parasitica (downy mildew) in Brassica napus var. oleifera (oilseed rape). Plant Pathot. 37, 538–545. doi: 10.1111/j.1365-3059.1988.tb02112.x
Lysak, M. A., Koch, M. A., Pecinka, A., and Schubert, I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15, 516–525. doi: 10.1101/gr.3531105
Ma, L., and Borhan, M. H. (2015). The receptor-like kinase SOBIR1 interacts with Brassica napus LepR3 and is required for Leptosphaeria maculans AvrLm1-triggered immunity. Front. Plant Sci. 6:933. doi: 10.3389/fpls.2015.00933
Mamgain, A., Biswas, M. K., and Dey, N. (2019). Potential role of chemical elicitors in induced systemic resistance for the effective management of Alternaria blight in mustard. J. Pharmacogn. Phytochem. 8, 2246–2250.
Mandal, A. K., and Dubey, S. C. (2012). Genetic diversity analysis of Sclerotinia sclerotiorum causing stem rot in chickpea using RAPD, ITS-RFLP, ITS sequencing and mycelial compatibility grouping. World J. Microbiol. Biotechnol. 28, 1849–1855. doi: 10.1007/s11274-011-0981-2
Marcroft, S., Wratten, N., Purwantara, A., Salisbury, P. A., Potter, T. D., Barbetti, M. J., et al. (2002). Reaction of a range of Brassica species under Australian conditions to the fungus, Leptosphaeria maculans, the causal agent of blackleg. Austr. J. Exper. Agric. 42, 587–594. doi: 10.1071/ea01112
Massand, P. P., Yadava, S. K., Sharma, P., Kaur, A., Kumar, A., Arumugam, N., et al. (2010). Molecular mapping reveals two independent loci conferring resistance to Albugo candida in the east European germplasm of oilseed mustard Brassica juncea. Theor. Appl. Genet. 121, 137–145. doi: 10.1007/s00122-010-1297-6
Matheron, M., and Porchas, M. (2005). Influence of soil temperature and moisture on eruptive germination and viability of sclerotia of Sclerotinia minor and S. sclerotiorum. Plant Dis. 89, 50–54. doi: 10.1094/pd-89-0050
Mayerhofer, R., Wilde, K., Mayerhofer, M., Lydiate, D., Bansal, V. K., Good, A. G., et al. (2005). Complexities of chromosome landing in a highly duplicated genome: toward map-based cloning of a gene controlling blackleg resistance in Brassica napus. Genetics 171, 1977–1988. doi: 10.1534/genetics.105.049098
Mazumder, M., Das, S., Saha, U., Chatterjee, M., Bannerjee, K., and Basu, D. (2013). Salicylic acid-mediated establishment of the compatibility between Alternaria brassicicola and Brassica juncea is mitigated by abscisic acid in Sinapis alba. Plant Physiol. Biochem. 70, 43–51. doi: 10.1016/j.plaphy.2013.04.025
McDonald, B. A., and Linde, C. (2002). Pathogen population genetics, evolutionary potential and durable resistance. Annu. Rev. Phytopathol. 40, 349–379. doi: 10.1146/annurev.phyto.40.120501.101443
McDonald, M. R., and Boland, G. J. (2004). Forecasting diseases caused by Sclerotinia spp. in eastern Canada: fact or fiction? Can. J. Plant Pathol. 26, 480–488. doi: 10.1080/07060660409507168
McDowell, J. M., Dhandaydham, M., Long, T. A., Aarts, M. G., Goff, S., Li, S., et al. (1998). Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10, 1861–1874. doi: 10.2307/3870909
McDowell, J. M., Williams, S. G., Funderburg, N. T., Eulgem, T., and Dangl, J. L. (2005). Genetic analysis of developmentally regulated resistance to downy mildew (Hyaloperonospora parasitica) in Arabidopsis thaliana. Mol. Plant Microb. Interact. 18, 1226–1234. doi: 10.1094/MPMI-18-1226
Meena, P. D., Awasthi, R. P., Chattopadhyay, C., Kolte, S. J., and Kumar, A. (2010). Alternaria blight: a chronic disease in rapeseed-mustard. J. Oilseed Brassica 1, 1–11. doi: 10.5455/faa.120733
Meena, P. D., Jambhulkar, S. J., Gupta, R., Meena, H. S., and Singh, D. (2016). Rapid screening technique for alternaria blight resistance in indian mustard (Brassica juncea L.) using cotyledonary leaf method. J. Plant Pathol. 98, 463–469.
Meena, P. D., Thomas, L., and Singh, D. (2014). Assessment of yield losses in Brassica juncea due to downy mildew (Hyaloperonospora brassicae). J. Oilseed Brassica 5, 73–77.
Mehta, N., Sangwan, M. S., Srivastava, M. P., and Kumar, R. (2002). Survival of Alternaria brassicae causing Alternaria blight in rapeseed-mustard. J. Mycol. Plant Pathol. 32, 64–67.
Mei, J., Ding, Y., Lu, K., Wei, D., Liu, Y., Onwusemu, J., et al. (2013). Identification of genomic regions involved in resistance against Sclerotinia sclerotiorum from wild Brassica oleracea. Theor. Appl. Genet. 126, 549–556. doi: 10.1007/s00122-012-2000-x
Mei, J., Shao, C., Yang, R., Feng, Y., Gao, Y., Ding, Y., et al. (2020). Introgression and pyramiding of genetic loci from wild Brassica oleracea into B. napus for improving Sclerotinia resistance of rapeseed. Theor. Appl. Genet. 133, 1313–1319. doi: 10.1007/s00122-020-03552-w
Mei, J., Wei, D., Disi, J. O., Ding, Y., Liu, Y., and Qian, W. (2012). Screening resistance against Sclerotinia sclerotiorum in Brassica crops with use of detached stem assay under controlled environment. Eur. J. Plant Pathol. 134, 599–604. doi: 10.1007/s10658-012-0040-3
Meyers, B. C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R. W. (2003). Genome-Wide Analysis of NBS-LRR-Encoding Genesin Arabidopsis. Plant Cell 15, 809–834. doi: 10.1105/tpc.009308
Mohammed, A. E., You, M. P., Banga, S. S., and Barbetti, M. J. (2018). Resistances to downy mildew (Hyaloperonospora brassicae) in diverse Brassicaceae offer new disease management opportunities for oilseed and vegetable crucifer industries. Eur. J. Plant Pathol. 153, 915–929. doi: 10.1007/s10658-018-01609-7
Mohanty, A., Chrungu, B., Verma, N., and Shivanna, K. R. (2009). Broadening the genetic base of crop Brassicas by production of new intergeneric hybrid. Czech. J. Genet. Plant Breed. 45, 117–122. doi: 10.17221/35/2009-CJGPB
Mondal, K. K., Bhattacharya, R. C., Koundal, K. R., and Chatterjee, S. C. (2007). Transgenic Indian mustard (Brassica juncea) expressing tomato glucanase leads to arrested growth of Alternaria brassicae. Plant Cell Rep. 26, 247–252. doi: 10.1007/s00299-006-0241-3
Mondal, K. K., Chatterjee, S. C., Viswakarma, N., Bhattacharya, R. C., and Grover, A. (2003). Chitinase-mediated inhibitory activity of Brassica transgenic on growth of Alternaria brassicae. Curr. Microbiol. 47, 171–173. doi: 10.1007/s00284-002-3980-6
Monot, C., Pajot, E., Corre, D. L., and Silue, D. (2002). Induction of systemic resistance in broccoli (Brassica oleracea var. botrytis) against downy mildew (Peronospora parasitica) by avirulent isolates. Biol. Control. 24, 75–81. doi: 10.1016/s1049-9644(02)00006-3
Monteiro, A. A., Coelho, P. S., Bahcevandziev, K., and Valerio, L. (2005). Inheritance of downy mildew resistance at cotyledon and adult-plant stages in ‘Couve Algarvia’ (Brassica oleracea var. tronchuda). Euphytica 141, 85–92. doi: 10.1007/s10681-005-5696-8
Morrall, R. A. A., and Dueck, J. (1982). Epidemiology of Sclerotinia stem rot of rapeseed in Saskatchewan. Can. Plant Pathol. 4, 161–168. doi: 10.1080/07060668209501319
Naher, N., Shamsi, S., Ali, M. R., and Bashar, M. A. (2018). Screening of Sclerotinia stem rot resistance in Bangladesh mustard germplasm using cotyledon assay method. Dhaka Univ. J. Biol. Sci. 27, 85–92. doi: 10.3329/dujbs.v27i1.46414
Nanjundan, J., Manjunatha, C., Radhamani, J., and Thakur, A. K. (2020). Identification of new source of resistance to powdery mildew of Indian mustard and studying its inheritance. Plant Pathol. J. 36, 111–120. doi: 10.5423/PPJ.OA.07.2019.0205
Narayani, M., and Srivastava, S. (2018). Elicitation: a stimulation of stress in in vitro plant cell/tissue cultures for enhancement of secondary metabolite production. Phytochem. Rev. 16, 1227–1252. doi: 10.1007/s11101-017-9534-0
Narusaka, M., Hatakeyama, K., Shirasu, K., and Narusaka, Y. (2014). Arabidopsis dual resistance proteins, both RPS4 and RRS1, are required for resistance to bacterial wilt in transgenic Brassica crops. Plant Signal. Behav. 9:e29130. doi: 10.4161/psb.29130
Nashaat, N. I., and Awasthi, R. P. (1995). Evidence for differential resistance to Peronospora parasitica (downy mildew) in accessions of Brassica juncea (mustard) at the cotyledon stage. J. Phytopathol. 143, 157–159. doi: 10.1111/j.1439-0434.1995.tb00250.x
Nashaat, N. I., Heran, A., Awasthi, R. P., and Kolte, S. J. (2004). Differential response and genes for resistance to Peronospora parasitica (downy mildew) in Brassica juncea (mustard). Plant Breed. 123, 512–515. doi: 10.1111/j.1439-0523.2004.01037.x
Nashaat, N. I., Heran, A., Mitchell, S. E., and Awasthi, R. P. (1997). New genes for resistnace to downy mildew (Peronospora parasitica) in oilseed rape (Brassica napus ssp. oleifrea). Plant Pathol. 46, 964–968. doi: 10.1046/j.1365-3059.1997.d01-76.x
Nayanakantha, N. M. C., Rawat, S., Ali, S., and Grover, A. (2016). Differential expression of defense-related genes in Sinapis alba and Brassica juncea upon the infection of Alternaria brassicae. Trop. Agric. Res. 27, 123–136. doi: 10.4038/tar.v27i2.8161
Neerja, S., and Sohal, B. S. (2012). Induced resistance in Brassica juncea L. against Alternaria blight by foliar application of arachidonic acid. Plant Dis. Res. 27, 153–157.
Neik, T. X., Barbetti, M. J., and Batley, J. (2017). Current status and challenges in identifying disease resistance genes in Brassica napus. Front. Plant Sci. 8:1788. doi: 10.3389/fpls.2017.01788
Neukermans, J., Inzé, A., Mathys, J., Coninck, B. D., Cotte, B. V., Bruno, P. A., et al. (2015). ARACINs, Brassicaceae-specific peptides exhibiting antifungal activities against necrotrophic pathogens in Arabidopsis. Plant Physiol. 167, 1017–1029. doi: 10.1104/pp.114.255505
Nishimura, S., and Kohmoto, K. (1983). Host-specific toxins and chemical struc-tures from Alternaria species. Annu. Rev. Phytopathol. 21, 87–116. doi: 10.1146/annurev.py.21.090183.000511
Nourani, S. L., Minassian, V., and Safaie, N. (2008). Identification, pathogenicity and distribution of Alternaria spp. of canola in Iran. Iran. J. Plant Path. 44, 33–36.
Nowicki, M., Nowakowska, M., Niezgoda, A., and Kozik, E. U. (2012). Alternaria black spot of crucifers: symptoms, importance of disease, and perspectives of resistance breeding. Veget. Crops Res. Bull. 76, 5–19. doi: 10.2478/v10032-012-0001-6
Osbourn, A. E. (1996). Preformed antimicrobial compounds and plant defense against fungal attack. Plant Cell 8, 1821–1831. doi: 10.1105/tpc.8.10.1821
Pan, Y., Wei, J., Yao, C., Reng, H., and Gao, Z. (2018). SsSm1, a Cerato-platanin family protein, is involved in the hyphal development and pathogenic process of Sclerotinia sclerotiorum. Plant Sci. 270, 37–46. doi: 10.1016/j.plantsci.2018.02.001
Parada, R. Y., Sakuna, E., Mori, N., Oka, K., Egusa, M., Kodoma, M., et al. (2008). Alternaria brassicae produces a host-specific protein toxin from germinating spores on host leaves. Phytopathology 98, 458–463. doi: 10.1094/PHYTO-98-4-0458
Paritosh, K., Yadava, S. K., Singh, P., Bhayana, L., Mukhopadhyay, A., Gupta, V., et al. (2020). A chromosome-scale assembly of allotetraploid Brassica juncea (AABB) elucidates comparative architecture of the A and B genomes. Plant Biotechnol. J. doi: 10.1111/pbi.13492
Park, Y. S., Jeon, M. H., Lee, S. H., Moon, J. S., Cha, J. S., Kim, H. Y., et al. (2005). Activation of defense responses in chinese cabbage by a nonhost pathogen, Pseudomonas syringae pv. tomato. J. Biochem. Mol. Biol. 38, 748–754. doi: 10.5483/bmbrep.2005.38.6.748
Parker, J. E., Coleman, M. J., Szabò, V., Frost, L. N., Schmidt, R., Wang, L., et al. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell 9, 879–894. doi: 10.1105/tpc.9.6.879
Parker, J. E., Holub, E. B., Frost, L. N., Falk, A., Gunn, N. D., and Danielsa, M. J. (1996). Characterization of edsl, a mutation in Arabidopsis suppressing resistance to Peronospora parasíiíca specified by several different RPP genes. Plant Cell 8, 2033–2046.
Pedras, M. C., and Zaharia, L. I. (2000). Sinalbins A and B, phytoalexins from Sinapis alba:elicitation, isolation, and synthesis. Phytochemistry 55, 213–216. doi: 10.1016/s0031-9422(00)00277-6
Pedras, M. S., and Smith, K. C. (1997). Sinalexin, a phytoalexin from white mustard elicited by destruxin B and Alternaria brassicae. Phytochemistry 46, 833–837. doi: 10.1016/s0031-9422(97)00362-2
Peltier, A. J., Bradley, C. A., Chilvers, M. I., Malvick, D. K., Mueller, D. S., Wise, K. A., et al. (2012). Biology, yield loss and control of Sclerotinia stem rot of soybean. J. Integ. Pest Mngmt. 3, 1–7. doi: 10.1603/IPM11033
Perumal, S., Koh, C. S., Jin, L., Sankoff, D., Robinson, S. J., Ian, L., et al. (2020). A high-contiguity Brassica nigra genome localizes active centromeres and defines the ancestral Brassica genome. Nat. Plants 6, 929–941. doi: 10.1038/s41477-020-0735-y
Perwaiz, M. S., Moghal, S. M., and Kamal, M. (1969). Studies on the chemical control of white rust and downy mildew of rape (Sarsoon). West Pak. J. Agric. Res. 7, 71–75.
Petrie, G. A. (1973). Diseases of Brassica species in Saskatchewan, 1970-72. I. Staghead and aster yellows. Can. Plant Dis. Surv. 53, 19–25.
Petrie, G. A. (1978). Occurrence of a highly virulent strain of blackleg (Leptosphaeria maculans) on rape in Saskatchewan (1975-77). Can. Plant Dis. Surv. 58, 21–25.
Petrie, G. A. (1986). Consequences of survival of Leptosphaeria maculans (blackleg) in canola stubble residue through an entire crop rotation sequence. Can. J. Plant Pathol. 8:353.
Petrie, G. A. (1988). Races of Albugo candida (white rust and staghead) on cultivated Cruciferae in Saskatchewan. Can. J. Plant Pathol. 10, 142–150. doi: 10.1080/07060668809501746
Petrie, G. A. (1995). Long-term survival and sporulation of Leptosphaeria maculans (blackleg) on naturally-infected rapeseed/canola stubble in Saskatchewan. Can. Plant Dis. Surv. 75, 23–34. doi: 10.1071/app9790023
Petrie, G. A., and Vanterpool, T. C. (1974). Fungi associated with hypertrophies caused by infection of Cruciferae by Albugo cruciferatum. Can. Plant Dis. Surv. 54, 37–42.
Pickar-Oliver, A., and Gersbach, C. A. (2019). The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell. Biol. 20, 490–507. doi: 10.1038/s41580-019-0131-5
Plieske, J., and Struss, D. (2001). STS markers linked to Phoma resistance genes of the Brassica B-genome revealed sequence homology between Brassica nigra and Brassica napus. Theor. Appl. Genet. 102, 483–488. doi: 10.1007/s001220051671
Pokotylo, I., Janda, M., Kalachova, T., Zachowski, A., and Ruelland, E. (2017). “Phosphoglycerolipid signaling in response to hormones under stress,” in Mechanism of Plant Hormone Signaling Under Stress-II, Part II: Interaction of Other Components with Phytohormones, ed. G. K. Pandey (Cham: Springer), 91–126. doi: 10.1002/9781118889022.ch22
Poveda, J., Hermosa, R., Monte, E., and Nicolás, C. (2019). The Trichoderma harzianum kelch protein ThKEL1 plays a key role in root colonization and the induction of systemic defense in Brassicaceae plants. Front. Plant Sci. 10:1478. doi: 10.3389/fpls.2019.01478
Prabhu, K. V., Somers, D. J., Rakow, G., and Gugel, R. K. (1998). Molecular markers linked to white rust resistance in mustard Brassica juncea. Theor. Appl. Genet. 97, 865–870. doi: 10.1007/s001220050966
Pradhan, A. K., Prakash, S., Mukhopadhyay, A., and Pental, D. (1992). Phylogeny of Brassica and allied genera based on the variation in chloroplast and mitochondrial DNA patterns. Molecular and taxo-nomic classifi cations are incongruous. Theor. Appl. Genet. 85, 331–340. doi: 10.1007/bf00222878
Prakash, S., Bhat, S. R., Quiros, C. F., Kirti, P. B., and Chopra, V. L. (2009). Brassica and its close allies: cytogenetics and evolution. Plant Breed. Rev. 31, 21–187. doi: 10.1002/9780470593783.ch2
Prakash, S., and Hinata, K. (1980). Taxonomy, cytogenetics and origin of crop Brassicas, a review. Operar. Bot. 55, 1–57. doi: 10.1016/b978-1-891127-79-3.50001-9
Primard, C., Vedel, F., Mathieu, C., Pelletier, G., and Chevre, A. M. (1988). Interspecific somatic hybridization between Brassica napus and Brassica hirta (Sinapis alba L.). Theory Appl. Genet. 75, 546–552. doi: 10.1007/BF00289119
Purdy, L. H. (1979). Sclerotinia sclerotiorum: history, diseases and symptomatology, host range, geographic distribution, and impact. Phytopathology 69, 875–880. doi: 10.1094/phyto-69-875
Purnamasari, M. I., Erskine, W., Croser, J. S., You, M. P., and Barbetti, M. J. (2019). Comparative reaction of Camelina sativa to Sclerotinia sclerotiorum and Laptosphaeria maculans. Plant Dis. 103, 2884–2892. doi: 10.1094/PDIS-03-19-0664-RE
Qasim, M. U., Zhao, Q., Shahid, M., Samad, R. A., Ahmar, S., Wu, J., et al. (2020). Identification of QTLs containing resistance genes for Sclerotinia stem rot in Brassica napus using comparative transcriptomic studies. Front. Plant Sci. 11:776. doi: 10.3389/fpls.2020.00776
Rajarammohan, S., Kumar, A., Gupta, V., Pental, D., Pradhan, A. K., and Kaur, J. (2017). Genetic architecture of resistance to Alternaria brassicae in Arabidopsis thaliana: QTL mapping reveals two major resistance-conferring loci. Front. Plant Sci. 8:260. doi: 10.3389/fpls.2017.00260
Rajarammohan, S., Paritosh, K., Pental, D., Barbulescu, D. M., Qiu, Y., Liu, S., et al. (2019a). Comparative genomics of Alternaria species provides insights into the pathogenic lifestyle of Alternaria brassicae - a pathogen of the Brassicaceae family. BMC Genom. 20:1036. doi: 10.1186/s12864-019-6414-6
Rajarammohan, S., Pental, D., and Kaur, J. (2019b). Near-complete genome assembly of Alternaria brassicae-A necrotrophic pathogen of Brassica crops. MPMI 32, 928–930. doi: 10.1094/MPMI-03-19-0084-A
Raman, H., McVittie, B., Pirathiban, R., Raman, R., Zhang, Y., Barbulescu, D. M., et al. (2020). Genome-wide association mapping identifies novel loci for quantitative resistance to blackleg disease in canola. Front. Plant Sci. 11:1184. doi: 10.3389/fpls.2020.01184
Raman, H., Raman, R., Diffey, S., Qiu, Y., McVittie, B., Barbulescu, D. M., et al. (2018). Stable quantitative resistance loci to blackleg disease in canola (Brassica napus L.) over continents. Front. Plant Sci. 9:1622. doi: 10.3389/fpls.2018.01622
Raman, R., Taylor, B., Lindbeck, K., Coombes, N., Barbulescu, D., Salisbury, P., et al. (2012a). Molecular mapping and validation of Rlm1 gene for resistance to Leptosphaeria maculans in canola (Brassica napus L.). Crop Past. Sci. 63, 1007–1017. doi: 10.1071/CP12255
Raman, R., Taylor, B., Marcroft, S., Stiller, J., Eckermann, P., Coombes, N., et al. (2012b). Molecular mapping of qualitative and quantitative loci for resistance to Leptosphaeria maculans causing blackleg disease in canola (Brassica napus L.). Theor. Appl. Genet. 125, 405–418. doi: 10.1007/s00122-012-1842-6
Rana, K., Atri, C., Akhatar, J., Kaur, R., Goyal, A., Singh, M. P., et al. (2019). Detection of first marker trait associations for resistance against Sclerotinia sclerotiorum in Brassica juncea-Erucastrum cardaminoides introgression lines. Front. Plant Sci. 10:1015. doi: 10.3389/fpls.2019.01015
Rana, K., Atri, C., Gupta, M., Akhatar, J., Sandhu, P., Kumar, N., et al. (2017). Mapping resistance to sclerotina infestation in introgression lines of Brassica juncea carring genomic segments from wild Brassicaceae B. fruticulosa. Sci. Rep. 7:5904. doi: 10.1038/s41598-017-05992-9
Rayss, T. (1938). Nouvelle contribution altetude de la mycofbre de Palestine. Palestian J. Bot. 1, 143–160.
Rejeb, I. B., Pastor, V., and Mauch-Mani, B. (2014). Plant responses to simultaneous biotic and abiotic stress: molecular mechanisms. Plants 3, 458–475. doi: 10.3390/plants3040458
Ren, L., Xu, L., Liu, F., Chen, K., Sun, C., Li, J., et al. (2016). Host range of Plasmodiophora brassicae on cruciferous crops and weeds in China. Plant Dis. 100, 933–939. doi: 10.1094/PDIS-09-15-1082-RE
Rimmer, S. R., and Van-den Berg, C. G. J. (1992). Resistance of oilseed Brassica spp. to blackleg caused by Leptosphaeria maculans. Can. J. Plant Pathol. 14, 56–66.
Ripley, V., Thorpe, M., Iler, S., Mizier, K., and Beversdorf, W. D. (1992). Isozyme analysis as a tool for introgression of Sinapis albagerm plasm into Brassica napus. Theor. Appl. Genet. 84, 403–410. doi: 10.1007/BF00229500
Rouxel, T., and Balesdent, M. H. (2005). The stem canker (blackleg) fungus, Leptosphaeria maculans, enters the genomic era. Mol. Plant Pathol. 6, 225–241. doi: 10.1111/j.1364-3703.2005.00282.x
Rouxel, T., Penaud, A., Pinochet, X., Brun, H., Gout, L., Delourme, R., et al. (2003). A 10-year survey of populations of Leptosphaeria maculans in France indicates a rapid adaptation towards the rlm1 resistance gene of oilseed rape. Eur. J. Plant Pathol. 109, 871–881.
Rustagi, A., Kumar, D., Shekhar, S., Yusuf, M. A., Misra, S., and Sarin, N. B. (2014). Transgenic Brassica juncea plants expressing MsrA1, a synthetic cationic antimicrobial peptide, exhibit resistance to fungal phytopathogens. Mol. Biotechnol. 56, 535–545. doi: 10.1007/s12033-013-9727-8
Saal, B., Brun, H., Glais, I., and Struss, D. (2004). Identification of a Brassica juncea-derived recessive gene conferring resistance to Leptosphaeria maculans in oilseed rape. Plant Breed. 123, 505–511. doi: 10.1111/j.1439-0523.2004.01052.x
Saal, B., and Struss, D. (2005). RGA- and RAPD-derived SCAR markers for a Brassica B-genome introgression conferring resistance to blackleg in oilseed rape. Theoret. Appl. Genet. 111, 281–290. doi: 10.1007/s00122-005-2022-8
Saccardo, P. A. (1886). “Hyphomyceteae,” in Sylloge Fungorum Omninum Hucusque Cognitorum, Vol. 4, (Pavia: BHL), 807.
Saharan, G. S., and Kadian, A. K. (1983). Physiologic specialization in Alternaria brassicae. Crucif. News Lett. 8, 32–33.
Saharan, G. S., and Mehta, N. (2008). Sclerotinia Diseases of Crop Plants: Biology, Ecology and Disease Management. Netherlands: Springer, 485.
Saharan, G. S., and Verma, P. R. (1992). White Rust. A Review of Economically Important Species. Ottawa, ON: International Development Research Centre, 65. IDRC-MR315e, IV.
Sangwan, M. S., and Mehta, N. (2007). Pathogenic variability in isolates of Alternaria brassicae (Berk.) Sacc. from different agro-climatic zones of India. Plant Dis. Res. 22, 101–107.
Savulescu, O. (1946). Study of the species of Cystopus (Pers.) Lev. Bucharest, 1946. Anal. Acad. Rous. Mem. Sect. Stimtiface. Soc. 21:13.
Schardl, C. L., and Craven, K. D. (2003). Interspecific hybridization in plant associated fungi and oomycetes: a review. Mol. Ecol. 12, 2861–2873. doi: 10.1046/j.1365-294x.2003.01965.x
Sexton, A. C., Whitten, A. R., and Howlett, B. J. (2006). Population structure of Sclerotinia sclerotiorum in an Australian canola field at flowering and stem-infection stages of the disease cycle. Genome 49, 1408–1415. doi: 10.1139/g06-101
Sharma, G., Kumar, V. D., Haque, A., Bhat, S. R., Prakash, S., and Chopra, V. L. (2002). Brassica coenospecies: a rich reser-voir for genetic resistance to leaf spot caused by Alternaria brassicae. Euphytica 125, 411–417. doi: 10.1023/A:1016050631673
Sharma, P., Deep, S., Sharma, M., and Bhati, D. S. (2013). Genetic variation of Alternaria brassicae (Berk.) Sacc., causal agent of dark leaf spot of cauliflower and mustard in India. J. Gen. Plant Pathol. 79, 41–45. doi: 10.1007/s10327-012-0417-3
Sharma, T. R., and Singh, B. M. (1992). Transfer of resistance to Alternaria brassicae in Brassica juncea through interspecific hybridization among Brassicas. J. Genet. Breed. 46, 373–378.
Sharma, T. R., and Tewari, J. P. (1998). RAPD analysis of three Alternaria species pathogenic to crucifers. Mycol. Res. 102, 807–814. doi: 10.1017/S0953756297005479
Shoemaker, R. A., and Brun, H. (2001). The teleomorph of the weakly aggressive segregate of Leptosphaeria maculans. Can. J. Bot. 79, 412–419. doi: 10.1139/cjb-79-4-412
Shrestha, S. K., Munk, L., and Mathur, S. B. (2005). Role of weather on Alternaria leaf blight disease and its effect on yield and yield component of mustard. Nepal Agric. Res. J. 6, 62–72. doi: 10.3126/narj.v6i0.3366
Shukla, A. K. (2005). Estimation of yield losses to Indian mustard (Brassica juncea) due to Sclerotinia stem rot. J. Phytol. Res. 18, 267–268.
Sinapidou, E., Williams, K., Nott, L., Bahkt, S., and Tör, M. (2004). Two TIR: NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 38, 898–909.
Singh, B. K., Nandan, D., Supriya, A., Ram, B., Kumar, A., Singh, T., et al. (2015). Validation of molecular markers for marker-assisted pyramiding of white rust resistance loci in Indian mustard (Brassica juncea L.). Can. J. Plant Sci. 95, 939–945. doi: 10.4141/cjps-2014-215
Singh, H. V. (2000). Biochemical basis of resistance in Brassica species against downy mildew and white rust of mustard. Plant Dis. Res. 15, 75–77.
Singh, R., Singh, D., Barbetti, M. J., Singh, H., Caixia, L., Sivasithamparam, K., et al. (2008). Sclerotinia rot tolerance in oilseed Brassica. J. Oilseeds Res. 25, 223–225.
Singh, S., Sharma, S. R., Kalia, P., Deshmukh, R., Kumar, V., Sharma, P., et al. (2012). Molecular mapping of the downy mildew resistance gene Ppa3 in cauliflower (Brassica oleracea var. botrytis L.). J. Hortic. Sci. Biotechnol. 87, 137–143. doi: 10.1080/14620316.2012.11512844
Singh, U. S., Nashaat, N. I., Doughty, K. J., and Awasthi, R. P. (2002). Altered phenotypic response to Peronospora parasitica in Brassica juncea seedlings following prior inoculation with an avirulent or virulent isolate of Albugo candida. Eur. J. Plant Pathol. 108, 555–564. doi: 10.1023/A:1019937115378
Sirhindi, G., Mushtaq, R., Sharma, P., Kaur, H., and Ahmad, M. M. (2017). “Jasmonate signaling and stress management in plants,” in Mechanism of Plant Hormone Signaling under Stress-II, Part I: Action of Phytohormones in Stress, ed. G. K. Pandey (Hoboken, NY: Wiley), 43–171. doi: 10.1002/9781118889022.ch7
Sjodin, C., and Glimelius, K. (1988). Screening for resistance to blackleg Phoma lingam (Tode ex Fr.) Desm. within Brassicaceae. J. Phytopathol. 123, 322–332. doi: 10.1111/j.1439-0434.1988.tb04484.x
Sodelade, M., Pedras, C., and Khallaf, I. (2012). Molecular interactions of the phytotoxins destruxin B and sirodesmin PL with crucifers and cereals. Metabolism and elicitation of plant defenses. Phytochemistry 77, 129–139. doi: 10.1016/j.phytochem.2012.02.010
Somers, D. J., Rakow, G., and Rimmer, S. R. (2002). Brassica napus DNA markers linked to white rust resistance in Brassica juncea. Theor. Appl. Genet. 104, 1121–1124. doi: 10.1007/s00122-001-0812-1
Song, J., Guan, Z., Hu, J., et al. (2020). Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 6, 34–45. doi: 10.1038/s41477-019-0577-7
Sperschneider, J., Dodds, P. N., Gardiner, D. M., Agarwal, P., Howlett, B. J., Li, Y., et al. (2015). Advances and challenges in computational prediction of effectors from plant pathogenic fungi. PLoS Pathg. 11:e1004806. doi: 10.1371/journal.ppat.1004806
Sprague, S. J., Balesdent, M. H., Brun, H., Hayden, H. L., Marcroft, S. J., Pinochet, X., et al. (2006a). Major gene resistance in Brassica napus (oilseed rape) is overcome by changes in virulence of populations of Leptosphaeria maculans in France and Australia. Eur. J. Plant Pathol. 114, 33–40. doi: 10.1007/1-4020-4525-5_3
Sprague, S. J., Marcroft, S. J., Hayden, H. L., and Howlett, B. J. (2006b). Major gene resistance to blackleg in Brassica napus overcome within three years of commercial production in southeastern Australia. Plant Dis. 90, 190–198. doi: 10.1094/pd-90-0190
Stergiopoulos, I., and de Wit, P. J. G. M. (2009). Fungal effector proteins. Ann. Rev. Phytopathol. 47, 233–263. doi: 10.1146/annurev.phyto.112408.132637
Stotz, H. U., Mitrousia, G. K., de Wit, P. J. G. M., and Fitt, B. D. L. (2014). Effector-triggered defence against apoplastic fungal pathogens. Trends Plant Sci. 19, 491–500. doi: 10.1016/j.tplants.2014.04.009
Taj, G., Agarwal, P., Grant, M., and Kumar, A. (2011). Co-expression and in-silico interaction studies for inter-linking the activation of MAPK3 and LOX genes during pathogenesis of Alternaria brassicae in Brassica juncea. J. Oilseed Brassica 2, 13–20.
Taylor, I. D., Conway, S., Roberts, S. J., Astley, D., and Vicente, I. G. (2002). Sources and origin of resistance to Xanthomonas campestris pv. campestris in Brassica genomes. Phytopathology 92, 105–111. doi: 10.1094/PHYTO.2002.92.1.105
Terras, F. R. G., Eggermont, K., Kovaleva, V., Raikhel, N. V., Osborn, R. W., Kester, A., et al. (1995). Small cysteine-rich antifungal proteins from radish: their role in host defense. Plant Cell 7, 573–588. doi: 10.1105/tpc.7.5.573
Terras, F. R. G., Torrekens, S., Leuven, F. V., Osborn, R. W., Vanderleyden, J., Cammuea, B. P. A., et al. (1993). A new family of basic cysteine-rich plant antifungal proteins from Brassicaceae species. FEBS Lett. 316, 233–240. doi: 10.1016/0014-5793(93)81299-f
Tewari, J. P. (1983). Cellular alterations in the blackspot of rapeseed caused by Alternaria brassicae. Phytopathology 73:831.
Tewari, J. P. (1991b). Structural and biochemical bases of the black spot diseases of crucifers. Adv. Struc. Biol. 1, 25–34.
Tewari, J. P., and Conn, K. L. (1993). Reactions of some wild crucifers to Alternaria brassicae. Bull. OILS SROP 16, 53–58.
Thakur, M., Sohal, B. S., and Sandhu, P. S. (2014). Impact of elicitor spray on Alternaria blight severity and yield of Brassica juncea and Brassica napus species. J. Oilseed Brassica 5, 78–82.
Thomma, B. P. H. J., Eggermont, K., Broekaert, W. F., and Cammue, B. P. A. (2000). Disease development of several fungi on Arabidopsis can bereduced by treatment with methyl jasmonate. Plant Physiol. Biochem. 38, 421–427. doi: 10.1016/s0981-9428(00)00756-7
Thomma, B. P. H. J., Eggermont, K., Penninckx, I. A. M. A., Mauch-Mani, B., Vogelsang, R., Cammue, B. P. A., et al. (1998). Separate jasmonate-dependent and salicylate-dependent defense response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. U.S.A. 95, 15107–15111. doi: 10.1073/pnas.95.25.15107
Thomma, B. P. H. J., Eggermont, K., Tierens, F. M. J., and Broekaert, W. F. (1999a). Requirement of functional EIN2 (ethylene insensitive 2) gene for efficient resistance of Arabidopsis thaliana to infection by Botrytis cinerea. Plant Physiol. 121, 1093–1101. doi: 10.1104/pp.121.4.1093
Thomma, B. P. H. J., Nelissen, I., Eggermont, K., and Broekaert, W. F. (1999b). Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 19, 163–171. doi: 10.1046/j.1365-313x.1999.00513.x
Thomma, B. P. H. J., Penninckx, I. A. M. A., Broekaert, W. F., and Cammue, B. P. A. (2001). The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13, 63–68. doi: 10.1016/s0952-7915(00)00183-7
Thukral, S. K., and Singh, H. (1986). Inheritance of white rust resistance in Brassica juncea. Plant Breed. 97, 75–77. doi: 10.1111/j.1439-0523.1986.tb01304.x
Tiwari, A. S., Petrie, G. A., and Downey, R. K. (1988). Inheritance of resistance to Albugo candida race 2 in mustard [Brassica juncea (L.) Czem.]. Can. J. Plant Sci. 68, 297–300. doi: 10.4141/cjps88-039
Tollenaere, R., Hayward, A., Dalton-Morgan, J., Campbell, E., Lee, J. R. M., Lorenc, M. T., et al. (2012). Identification and characterization of candidate Rlm4 blackleg resistance genes in Brassica napus using next-generation sequencing. Plant Biotechnol. J. 10, 709–715. doi: 10.1111/j.1467-7652.2012.00716.x
Tonguc, M., and Griffiths, P. D. (2004). “Transfer of powdery mildew resistance from Brassica carinata to B. oleracea,” in Proceeings of the 101st International conference of American Society of Horticulture Science, Austin, TX.
Tosi, L., and Zezzerini, A. (1985). Septoria helianthi Ell. & Kell. nuovo parassita del girasole in Italia. Inform. Fitopatol. 39, 43–44.
Tripathi, N. N., and Kaushik, C. D. (1984). Studies on the survival of Alternaria brassicae the causal organism of leaf spot of rapeseed and mustard. Madras Agric. J. 71, 237–241.
Tylkowska, K., Korbas, M., Kurzawińska, H., and Rataj-Guranowska, M. (2004). “Objawy chorób powodowanych przez grzyby rodzaju Alternaria,” in Kompendium Symptomów Cho-rób Roślin Oraz Morfologii Ich Sprawców, ed. M. Rataj-Guranowska (Poznań: Bogucki Wydawnictwo Nau-kowe), 3–11.
Uloth, M., You, M. P., Finnegan, P. M., Banga, S. S., Yi, H., and Barbetti, M. J. (2014). Seedling resistance to Sclerotinia sclerotiorum as expressed across diverse cruciferous species. Plant Dis. 98, 184–190. doi: 10.1094/pdis-06-13-0612-re
Uloth, M. B., You, M. P., Finnegan, P. M., Banga, S. S., Banga, S. K., Sandhu, P. S., et al. (2013). New sources of resistance to Sclerotinia sclerotiorum for crucifer crops. Field Crops Res. 154, 40–52. doi: 10.1016/j.fcr.2013.07.013
USDA (2020). Foreign Agriculture Service. United States Department of Agriculture. Available online at: https://www.fas.usda.gov/regions/india.
Van Der Biezen, E. A., Freddie, C. T., Kahn, K., Parker, J. E., and Jones, J. D. (2002). Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29, 439–451. doi: 10.1046/j.0960-7412.2001.01229.x
Van-de Wouw, A., Lowe, R. G. T., Elliott, C. E., Dubois, D. J., and Howlett, B. J. (2014). An avirulence gene, AvrLmJ1, from the blackleg fungus, Leptosphaeria maculans, confers avirulence to Brassica juncea cultivars. Mol. Plant Pathol. 15, 523–530. doi: 10.1111/mpp.12105
Van-de Wouw, A. P., Howlett, B. J., and Idnurm, A. (2017). Changes in allele frequencies of avirulence genes in the blackleg fungus, Leptosphaeria maculans, over two decades in Australia. Crop Pasture Sci. 69, 20–29. doi: 10.1071/cp16411
Varshney, A., Mohapatra, T., and Sharma, R. P. (2004). Development and validation of CAPS and AFLP markers for white rust resistance gene in Brassica juncea. Theor. Appl. Genet. 109, 153–159. doi: 10.1007/s00122-004-1607-y
Vellios, E., Karkanis, A., and Bilalis, D. (2017). Powdery mildew (Erysiphe cruciferarum) infection on camelina (Camelina sativa) under Mediterranean conditions and the role of wild mustard (Sinapis arvensis) as alternative host of this pathogen. Emirat. J. Food Agric. 29, 639–642. doi: 10.9755/ejfa.2017-02-493
Verma, A., and Singh, Y. (2018). Inheritance of downy mildew resistance and its relationship with biochemical traits in cauliflower (Brassica oleracea L. var. botrytis). Crop Protect. 106, 132–138. doi: 10.1016/j.cropro.2017.12.024
Verma, P. R., and Saharan, G. S. (1994). Monograph on Alternaria diseases of crucifers. Agric. Can. Res. Stat. 6:162.
Verma, S. S., Yajima, W. R., Rahman, M. H., Shah, S., Liu, J., Ekramoddoullah, A. K. M., et al. (2012). A cysteine-rich antimicrobial peptide from Pinus monticola (PmAMP1) confers resistance to multiple fungal pathogens in canola (Brassica napus). Plant Mol. Biol. 79, 61–74. doi: 10.1007/s11103-012-9895-0
Verma, V. D., and Rai, B. (1980). Note on induced mutagenesis for spotting out the sources of resistance to Alternaria leaf spot in Indian mustard. Indian J. Agric. Sci. 50, 278–280.
Vicente, J. G., Gunn, N. D., Bailey, L., Pink, D. A. C., and Holub, E. B. (2012). Genetics of resistance to downy mildew in Brassica oleracea and breeding towards durable disease control for UK vegetable production. Plant Pathol. 61, 600–609. doi: 10.1111/j.1365-3059.2011.02539.x
Viegas, A. P., and Teixeira, A. R. (1943). Alguns fungos do Brasil. Bragantia Sao Paulo 3, 223–269. doi: 10.1590/s0006-87051943000800003
Vincenot, L., Balesdent, M., Li, H., Barbetti, M. J., Sivasithamparam, K., Gout, L., et al. (2008). Occurrence of a new subclade of Leptosphaeria biglobosa in Western Australia. Phytopathology 98, 321–329. doi: 10.1094/phyto-98-3-0321
Vishwanath, and Kolte, S. J. (1997). Variability in Alternaria brassicae: Response to host genotypes, toxin production and fungicides. Indian Phytopathol. 50, 373–381.
Vishwanath, Kolte, S. J., Singh, M. P., and Awasthi, R. P. (1999). Induction of resistance in mustard (Brassica juncea) against Alternaria black spot with an avirulent Alternaria brassicae isolate-D. Eur. J. Plant Pathol. 105, 217–220.
Vleeshouwers, V. G. A. A., Rietman, H., Krenek, P., Champouret, N., Young, C., Oh, S. K., et al. (2008). Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora Infestans avirulence genes. PLoS One 3:e2875. doi: 10.1371/journal.pone.0002875
Vogel, J., and Somerville, S. (2000). Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. U.S.A. 97, 1897–1902. doi: 10.1073/pnas.030531997
Volkan, C., Freddy, B., Wiebke, A., Alexandre, R.-S., Oliver, J. F., Amey, R., et al. (2019). Transgressive segregation reveals mechanisms of Arabidopsis immunity to Brassica-infecting races of white rust (Albugo candida). Proc. Natl. Acad. Sci. 116, 2767–2773. doi: 10.1073/pnas.1812911116
Waalen, W. M., Tanino, K. K., Olsen, J. E., Eltun, R., Rognli, O. A., and Gusta, L. V. (2011). Freezing tolerance of winter canola cultivars is best revealed by a prolonged freeze test. Crop Sci. 51, 1988–1996. doi: 10.2135/cropsci2011.02.0098
Wang, M., Farnham, M. W., and Thomas, C. E. (2001). Inheritance of true leaf stage downy mildew resistance in broccoli. J. Amer. Soc. Hort. Sci. 126, 727–729. doi: 10.21273/jashs.126.6.727
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). Brassica rapa genome sequencing project consortium. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Wang, Y. P., Sonntag, K., Rudloff, E., and Chen, J. (2005). Intergeneric somatic hybridization between Brassica napus L. and Sinapis alba L. J. Integr. Plant Biol. 47, 84–91. doi: 10.1111/j.1744-7909.2005.00009.x
Wang, Z., Ma, L. Y., Cao, J., Li, Y. L., Ding, L. N., Zhu, K. M., et al. (2019). Recent Advances in Mechanisms of Plant Defense to Sclerotinia sclerotiorum. Front. Plant Sci. 10:1314. doi: 10.3389/fpls.2019.01314
Warwick, S. I. (2011). “Brassicaceae in agriculture,” in Genetics and Genomics of the Brassicaceae. Plant Genetics and Genomics: Crop Models 9, eds R. Schmidt and I. Bancroft (Berlin: Springer Science Business Media), 33–65. doi: 10.1007/978-1-4419-7118-0_2
Warwick, S. I., and Black, L. D. (1991). Molecular systematics of Brassica and allied genera (Subtribe Brassicinae Brassicae) chloroplast genome and cytodeme congruence. Theor. Appl. Genet. 82, 81–92. doi: 10.1007/bf00231281
Warwick, S. I., Francis, A., and Al-Shehbaz, I. A. (2006). Brassicaceae: species checklist and database on CD-Rom. Plant Syst. Evol. 259, 249–258. doi: 10.1007/s00606-006-0422-0
Wei, D., Mei, J., Fu, Y., Disi, J. O., Li, J., and Qian, W. (2014). Quantitative trait loci analyses for resistance to Sclerotinia sclerotiorum and flowering time in Brassica napus. Mol. Breed. 34, 1797–1804. doi: 10.1007/s11032-014-0139-7
Wei, Z., Wang, M., Chang, S., Wu, C., Liu, P., Meng, J., et al. (2016). Introgressing subgenome components from Brassica rapa and B. carinata to B. juncea for broadening its genetic base and exploring intersubgenomic heterosis. Front. Plant Sci. 7:1677. doi: 10.3389/fpls.2016.01677
Werner, S., Diederichsen, E., Frauen, M., Schondelmaier, J., and Jung, C. (2007). Genetic mapping of clubroot resistance genes in oilseed rape. Theor. Appl. Genet. 116, 363–372. doi: 10.1007/s00122-007-0674-2
West, J. S., Kharbanda, P., Barbetti, M. J., and Fitt, B. D. L. (2001). Epidemiology and management of Leptosphaeria maculans (phoma stem canker) in Australia, Canada and Europe. Plant Pathol. 50, 10–27. doi: 10.1046/j.1365-3059.2001.00546.x
Westman, A. L., Kresovich, S., and Dickson, M. H. (1999). Regional variation in Brassica nigra and other weedy crucifers for disease reaction to Alternaria brassicicola and Xanthomonas campestris pv. campestris. Euphytica 106, 253–259. doi: 10.1023/A:1003544025146
Wight, W. D., Kim, K. H., Lawrence, C. B., and Walton, J. D. (2009). Biosynthesis and role in virulence of the histone deacetylase inhibitor depudecin from Alternaria brassicicola. MPMI 22, 1258–1267. doi: 10.1094/MPMI-22-10-1258
Williams, P. H., and Saha, L. R. (1993). Diseases of Mustard (Brassica juncea). Available online at: https://www.Apsnet.org/edcenter/resources/commonnames/Pages/Mustard.Aspx (accessed June 28, 2020).
Winter, H., Diestel, A., Gärtig, S., Wu, J., Chen, P., Zhao, Q., et al. (2003). “Transfer of new blackleg resistances into oilseed rape,”in Proceedings of GCIRC 11th International Rapeseed Congress, Saskatoon, Vol. 1, 19–21.
Winter, M., and Koopmann, B. (2016). Race spectra of Leptosphaeria maculans, the causal agent of blackleg disease of oilseed rape, in different geographic regions in northern Germany. Eur. J. Plant Pathol. 145, 629–641. doi: 10.1007/s10658-016-0932-8
Wu, J., Cai, G. Q., Tu, J. Y., Li, L. X., Liu, S., Luo, X. P., et al. (2013). Identification of QTLs for resistance to Sclerotinia stem rot and BnaC.IGMT5. A as a candidate gene of the major resistant QTL SRC6 in Brassica napus. PLoS One 8:e67740. doi: 10.1371/journal.pone.0067740
Wu, J., Chen, P., Zhao, Q., Cai, G., Hu, Y., Xiang, Y., et al. (2019). Co-location of QTL for Sclerotinia stem rot resistance and flowering time in Brassica napus. Crop J. 7, 227–237. doi: 10.1016/j.cj.2018.12.007
Wu, J., Zhao, Q., Yang, Q., Liu, H., Li, Q., Yi, X., et al. (2016). Comparative transcriptomic analysis uncovers the complex genetic network for resistance to Sclerotinia sclerotiorum in Brassica napus. Sci. Rep. 6:19007. doi: 10.1038/srep19007
Xiao, S., Calis, O., Patrick, E., Zhang, G., Charoenwattana, P., Muskett, P., et al. (2005). The atypical resistance gene, RPW8, recruits components of basal defense for powdery mildew resistance in Arabidopsis. Plant J. 42, 95–110. doi: 10.1111/j.1365-313X.2005.02356.x
Xiao, S., Charoenwattana, P., Holcombe, L., and Turner, J. G. (2003). The Arabidopsis genes RPW8.1 and RPW8.2 confer induced resistance to powdery mildew diseases in tobacco. MPMI 16, 289–294. doi: 10.1094/mpmi.2003.16.4.289
Xiao, S., EIIwood, S., Findlay, K., Oliver, R. P., and Turner, J. G. (1997). Characterization of three loci controlling resistance of Arabidopsis thaliana accession Ms-0 to two powdery mildew diseases. Plant J. 12, 757–768. doi: 10.1046/j.1365-313x.1997.12040757.x
Yang, J., Liu, D., Wang, X., Ji, C., Cheng, F., Liu, B., et al. (2016). The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 48, 1225–1232. doi: 10.1038/ng.3657
Yang, J., Liu, D., Wang, X., Ji, C., Cheng, F., Liu, B., et al. (2018). The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 50:1616. doi: 10.1038/s41588-018-0227-4
Yin, X., Yi, B., Chen, W., Liang, X., Yu, O., and Zhang, Y. et al. (2010). Mapping of QTLs detected in a Brassica napus DH population for resistance to Sclerotinia sclerotiorum in multiple environments. Euphytica 173, 25–35. doi: 10.1007/s10681-009-0095-1
You, M. P., Uloth, M. B., Li, X. X., Banga, S. S., Banga, S. K., and Barbetti, M. J. (2016). Valuable new resistances ensure improved management of Sclerotinia Stem Rot (Sclerotinia sclerotiorum) in horticultural and oilseed Brassica species. J. Phytopathol. 164, 291–299. doi: 10.1111/jph.12456
Yu, F., Gugel, R. K., Kutcher, H. R., Peng, G., and Rimmer, S. R. (2013). Identification and mapping of a novel blackleg resistance locus LepR4 in the progenies from Brassica napus × B. rapa subsp. sylvestris. Theor. Appl. Genet. 126, 307–315. doi: 10.1007/s00122-012-1919-2
Yu, F., Lydiate, D., and Rimmer, S. R. (2005). Identification of two novel genes for blackleg resistance in Brassica napus. Theo. Appl. Genet. 110, 969–979. doi: 10.1007/s00122-004-1919-y
Yu, F., Lydiate, D., and Rimmer, S. R. (2008). Identification and mapping of a third blackleg resistance locus in Brassica napus derived from B. rapa subsp. sylvestris. Genome 51, 64–72. doi: 10.1139/g07-103
Yu, J., Tehrim, S., Zhang, F., Tong, C., Huang, J., Cheng, X., et al. (2014). Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genom. 15:3. doi: 10.1186/1471-2164-15-3
Yu, S., Su, T., Zhi, S., Zhang, F., Wang, W., Zhang, D., et al. (2016). Construction of a sequence-based bin map and mapping of QTLs for downy mildew resistance at four developmental stages in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Mol. Breed. 36:44. doi: 10.1007/s11032-016-0467-x
Yu, S., Zhang, F., Zhao, X., Yu, Y., and Zhang, D. (2011). Sequence-characterized amplified region and simple sequence repeat markers for identifying the major quantitative trait locus responsible for seedling resistance to downy mildew in Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Breed. 130, 580–583.
Zeilmaker, T., Ludwig, N. R., Elberse, J., Seidl, M. F., Berke, L., Doorn, A. V., et al. (2015). DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 81, 210–222. doi: 10.1111/tpj.12719
Zhang, H., Wu, Q., Cao, S., Zhao, T., Chen, L., Zhuang, P., et al. (2014). A novel protein elicitor (SsCut) from Sclerotinia sclerotiorum induces multiple defense responses in plants. Plant Mol. Biol. 86, 495–511.
Zhang, L., Cai, X., Wu, J., Berke, L., Doorn, A. V., Cao, S., et al. (2018). Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 5:50. doi: 10.1038/s41438-018-0071-9
Zhang, W., Fraiture, M., Kolb, D., Loffelhardt, B., Desaki, Y., Boutrot, F. F., et al. (2013). Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25, 4227–4241. doi: 10.1105/tpc.113.117010
Zhang, X., and Fernando, W. G. D. (2018). Insights into fighting against blackleg disease of Brassica napus in Canada. Crop Pasture Sci. 69, 40–47. doi: 10.1071/cp16401
Zhang, X., Peng, G., Kutcher, H. R., Balesdent, M. H., Delourme, R., and Fernando, W. G. D. (2016). Breakdown of Rlm3 resistance in the Brassica napus-Leptosphaeria maculans pathosystem in western Canada. Eur. J. Plant Pathol. 145, 659–674. doi: 10.1007/s10658-015-0819-0
Zhang, Z. Y., Wang, Y. X., and Liu, Y. L. (1984). Taxonomic studies of the family Albuginaceae of China. II. A new species of Albugo on Acanthaceae and known species of Albugo on cruciferae. Acta Mycol. Sin. 3, 65–71.
Zhao, J., and Meng, J. (2003). Genetic analysis of loci associated with partial resistance to Sclerotinia sclerotiorum in rapeseed (Brassica napus L.). Theor. Appl. Genet. 106, 759–764. doi: 10.1007/s00122-002-1171-2
Zhao, J., Udall, J. A., Quijada, P. A., Grau, C. R., Meng, J., and Osborn, T. C. (2006). Quantitative trait loci for resistance to Sclerotinia sclerotiorum and its association with a homeologous non-reciprocal transposition in Brassica napus L. Theor. Appl. Genet. 112, 509–516. doi: 10.1007/s00122-005-0154-5
Zheng, L., Zhao, J., Liang, X., Zhan, G., Jiang, S., and Kang, Z. (2017). Identification of a novel Alternaria alternata strain able to hyperparasitize Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust. Front. Microbiol. 8:71. doi: 10.3389/fmicb.2017.00071
Keywords: B. juncea, diseases, resistant sources, quantitative trait loci, improvement strategies
Citation: Singh KP, Kumari P and Rai PK (2021) Current Status of the Disease-Resistant Gene(s)/QTLs, and Strategies for Improvement in Brassica juncea. Front. Plant Sci. 12:617405. doi: 10.3389/fpls.2021.617405
Received: 14 October 2020; Accepted: 08 February 2021;
Published: 03 March 2021.
Edited by:
Harsh Raman, New South Wales Department of Primary Industries, AustraliaReviewed by:
Angela Mehta, Embrapa Genetic Resources and Biotechnology, BrazilJose Donoso, Institute of Agricultural Research, Chile
Copyright © 2021 Singh, Kumari and Rai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaushal Pratap Singh, kaushalmpi1978@gmail.com
†These authors share first authorship
 Kaushal Pratap Singh
Kaushal Pratap Singh Preetesh Kumari
Preetesh Kumari Pramod Kumar Rai1
Pramod Kumar Rai1