- 1MOE Key Laboratory for Biodiversity Science and Ecological Engineering, College of Life Sciences, Beijing Normal University, Beijing, China
- 2School of Life Sciences, Peking University, Beijing, China
- 3Key Laboratory of Plant Hormones and Development Regulation of Chongqing, School of Life Sciences, Chongqing University, Chongqing, China
- 4Center of Plant Functional Genomics, Institute of Advanced Interdisciplinary Studies, Chongqing University, Chongqing, China
Understanding the roles played by geography and ecology in driving species diversification and in the maintenance of species cohesion is the central objective of evolutionary and ecological studies. The multi-phased orogenesis of Qinghai-Tibetan Plateau (QTP) and global climate changes over late-Miocene has profoundly influenced the environments and evolution of organisms in this region and the vast areas of Asia. In this study, we investigate the lineage diversification of Chrysanthemum-group in subtribe Artemisiinae (tribe Anthemideae, Asteraceae) likely under the effects of climate changes during this period. Using DNA sequences of seven low-copy nuclear loci and nrITS and the coalescent analytical methods, a time-calibrated phylogeny of subtribe Artemisiinae was reconstructed with emphasis on Chrysanthemum-group. The monophyletic Chrysanthemum-group was well resolved into two major clades corresponding to Chrysanthemum and Ajania, two genera which can be well identified by capitulum morphology but have been intermingled in previous plastid and ITS trees. Within Chrysanthemum, a later divergence between Ch. indicum-complex and Ch. zawadskii-complex can be recognized. The time frames of these sequential divergences coincide with the late Cenozoic uplift of the Northern QTP and the concomitant climatic heterogeneity between eastern and inland Asia. Reconstruction of historical biogeography suggested the origin of Chrysanthemum-group in Central Asia, followed by eastward migration of Chrysanthemum and in situ diversification of Ajania. Within Chrysanthemum, Ch. indicum-complex and Ch. zawadskii-complex exhibited contemporary distributional division, the former in more southern and the latter in more northern regions. The geographic structure of the three lineages in Chrysanthemum-group have been associated with the niche differentiation, and environmental heterogenization in Asia interior.
Introduction
Speciation is usually associated with morphological innovation or modification that occurs for internal molecular and/or external environmental reasons. Geographic isolation and ecological segregation are both important external forces for speciation (Coyne and Orr, 2004). Understanding the relative roles of geographic and ecological factors in the increase of species diversity and maintenance of species coherence is one of the central tasks of evolutionary biology (Wiens and Graham, 2005; Rundle and Nosil, 2010, 2012; Anacker and Strauss, 2014; Ebersbach et al., 2018).
In East Asia, the process of species diversification has been greatly influenced by the topographic activities of the Northern Qinghai-Tibet Plateau (QTP) and the aridification in Asian interior (Zheng et al., 2000; An et al., 2001; Guo et al., 2004; Miao et al., 2012; Ge et al., 2013; Li et al., 2015; Shi et al., 2015; Spicer, 2017; Shi et al., 2019; Ding et al., 2020). To what extent have ecogeographical gradients and diverse macrohabitats in this region promoted speciation (He et al., 2010; Qin et al., 2013; Yan et al., 2013)? To gain insight into this issue, comparative phylogeographic analyses of closely related species are required as such studies may help us trace trajectories of lineage splitting and reuniting (if these events happened) and infer evolutionary forces behind rapid speciation (Anacker and Strauss, 2014; Ortiz-Rodriguez et al., 2018; Wang Z. M. et al., 2019; Knope et al., 2020).
Chrysanthemum-group, one of the youngest branches of the largest angiosperm family, Asteraceae, is a proper system for studying the aforementioned problem. This group belongs to subtribe Artemisiinae, tribe Anthemideae of Asteraceae (Bremer and Humphries, 1993; Watson et al., 2002; Kondo et al., 2003; Sanz et al., 2008; Masuda et al., 2009). According to their cladistic analysis of morphological characteristics, Bremer and Humphries (1993) defined 18 genera in subtribe Artemisiinae. This circumscription has been widely accepted with the later incorporation of Hippolytia, Opisthopappus, and Tanacetum tatsienense, which formerly belonged to subtribe Tanacetinae, as well as Leucanthemella and Nipponanthemum, which were formerly in subtribe Leucantheminae. Despite of its uncertain circumscription, the subtribe Artemisiinae is mainly composed of species belonging to two groups, Chrysanthemum-group and Artemisia-group (Bremer and Humphries, 1993; Oberprieler et al., 2007; Sanz et al., 2008; Oberprieler et al., 2009; Sonboli et al., 2012).
Chrysanthemum-group was historically recognized mainly by solitary flower heads or corymbose synflorescences (in contrast to Artemisia-group with paniculate synflorescences), radiate capitula (Chrysanthemum, Arctanthemum, and Brachanthemum) or disciform capitula (Ajania and Phaeostigma), and echinate Anthemis-type pollen grains (except Phaeostigma with microechinate Artemisia-type) (Bremer and Humphries, 1993; Sanz et al., 2008; Pellicer et al., 2010). However, the circumscription and monophyly of the two groups have remained questionable, and the generic relationships within each group have been rather controversial (Bremer and Humphries, 1993; Torrell et al., 1999; Oberprieler et al., 2007; Zhao et al., 2010a, b). These problems are probably due to ongoing speciation, including recent divergence and secondary contacts of lineages (Masuda et al., 2009; Miao et al., 2011; Liu et al., 2012b; Li et al., 2014; Chen et al., 2020).
Within Chrysanthemum-group, there are two major genera, Chrysanthemum and Ajania, both mainly distributed in East Asia and each consisting of 30–35 species (Shih and Fu, 1983; Oberprieler et al., 2006). Poljakov (1955) speculated that Ajania was closely related to Artemisia, but Tzvelev (1961) considered Ajania and Chrysanthemum to be sister lineages derived from a most recent common ancestor that had radiate capitula (Muldashev, 1983; Bremer and Humphries, 1993). Ajania was even once treated as a section under Chrysanthemum (Kitamura, 1978; Kishimoto et al., 2003; Ohashi and Yonekura, 2004). Considering their distinct capitulum morphologies, we have postulated that these two genera must have experienced adaptive divergence associated with differential environmental conditions (Chen et al., 2020). Our recent developmental genetic study revealed that a disciform capitulum may have evolved from a radiated type owing to the dysfunction of a key ray-flower regulator, CYCLOIDEA2g (Chen et al., 2018; Shen et al., 2021). To date, however, no molecular phylogeny has resolved Chrysanthemum and Ajania each as monophyletic (Kondo et al., 2003; Masuda et al., 2009; Zhao et al., 2010a, b; Liu et al., 2012b; Huang et al., 2017). In another aspect, the small genus Phaeostigma (established by Muldashev in 1981) was once described under Ajania (Muldashev, 1981), but was clarified by our previous analysis as a monophyletic group that is probably closer to Artemisia than to Ajania despite its corymbose synflorescence similar to that of the latter (Huang et al., 2017). Brachanthemum, which was supposed to be closely related to Chrysanthemum due to its thin-walled and pappus-lacking achenes (Bremer and Humphries, 1993), was suggested by molecular phylogeny to be closer to Nipponanthemum, Leucanthemella, or Kaschgaria than to Chrysanthemum (Vallès et al., 2003; Sanz et al., 2008; Masuda et al., 2009; Zhao et al., 2010a). As to the monotypic genus Arctanthemum, it has been incorporated into Chrysanthemum in the floristic work of China (Shih and Fu, 1983) thus the name Chrysanthemum arcticum was used instead of Arctanthemum arcticum.
In terms of geography, Chrysanthemum-group is widely distributed in Middle to East Asia, with a few members extending to Central Europe and North America (Shih and Fu, 1983). Two major genera, Ajania and Chrysanthemum, cover most of its full range of distribution, with Ajania in China (from northwestern to northeastern and southwestern parts), Korea, Japan, and the Far East, and Chrysanthemum in the more eastern part of China, Korea, Japan, and Russia (Shih and Fu, 1983; Zhao et al., 2009). Within Chrysanthemum, there are two species complexes corresponding to morphological characteristics and weakly supported by the chloroplast phylogeny—Ch. zawadskii-complex, with relatively larger flower heads and white to purple ray flowers, and Ch. indicum-complex, with smaller flower heads and white or yellow rays (Shimizu, 1961; Lee, 1969; Liu et al., 2012b; Li et al., 2013, Li et al., 2014; Kim et al., 2014; Meng et al., 2020). Considering geographic distribution, Ch. zawadskii-complex is distributed mainly in the northern region of East Asia, while Ch. indicum-complex is relatively in the more southeastern part (Shih and Fu, 1983; Shishkin and Bobrov, 1995; Zhao et al., 2009).
Untangling recent speciation events usually requires a reliable phylogenetic framework. To date, all deep phylogenetic relationships within subtribe Artemisiinae inferred from plastic and nrITS markers have provided limited information (Watson et al., 2000, 2002; Sanz et al., 2008; Pellicer et al., 2010; Zhao et al., 2010a; Liu et al., 2012b). Here, we utilized multilocus nuclear DNA sequences and a coalescent analytical method to update the tree of this subtribe with emphasis on Chrysanthemum-group. Subsequently, we estimated the optimal ancestral distribution and biogeographical history of the major clades in this subtribe. Furthermore, we conducted ecological niche modeling and niche overlap tests to verify the ecological differentiation of lineages within Chrysanthemum-group and to see whether patterns of geographic distribution were linked to environmental conditions. With all these analytical results, we attempted to resolve possible rapid species divergence under macrohabitat differentiation in interior East Asia.
Materials and Methods
Taxon Sampling
Taxon sampling for this study covered all major branches of subtribe Artemisiinae. In total, 101 accessions of 96 species were sampled. Of the 96 species, 53 were of Chrysanthemum-group, including 30 out of approximately 35 Chrysanthemum species, 20 out of approximately 30 Ajania species, one species of the monotypic genus Elachanthemum, and both the two species of Opisthopappus. To clarify generic relationships within subtribe Artemisiinae, 23 species covering all four commonly accepted subgenera of Artemisia and its close allies were included in this study. Also sampled were Phaeostigma, Brachanthemum, and Kaschgaria, which were of uncertain phylogenetic positions in Artemisiinae, as well as Hippolytia, Nipponanthemum, Leucanthemella, and Tanacetum tatsienense, which were later placed in Artemisiinae. Six species of four subtribes that belong to the Eurasian-Mediterranean clade of tribe Anthemideae (Oberprieler et al., 2007) were also sampled and used as outgroups. The detailed sampling information is listed in Supplementary Table 1.
Leaf samples were mostly collected from the wild, while some were obtained from seedlings geminated from seeds from international seed banks1. Voucher specimens were deposited in the herbarium of Peking University, and living seedlings were grown in the greenhouse of Peking University.
DNA Extraction and Gene Isolation
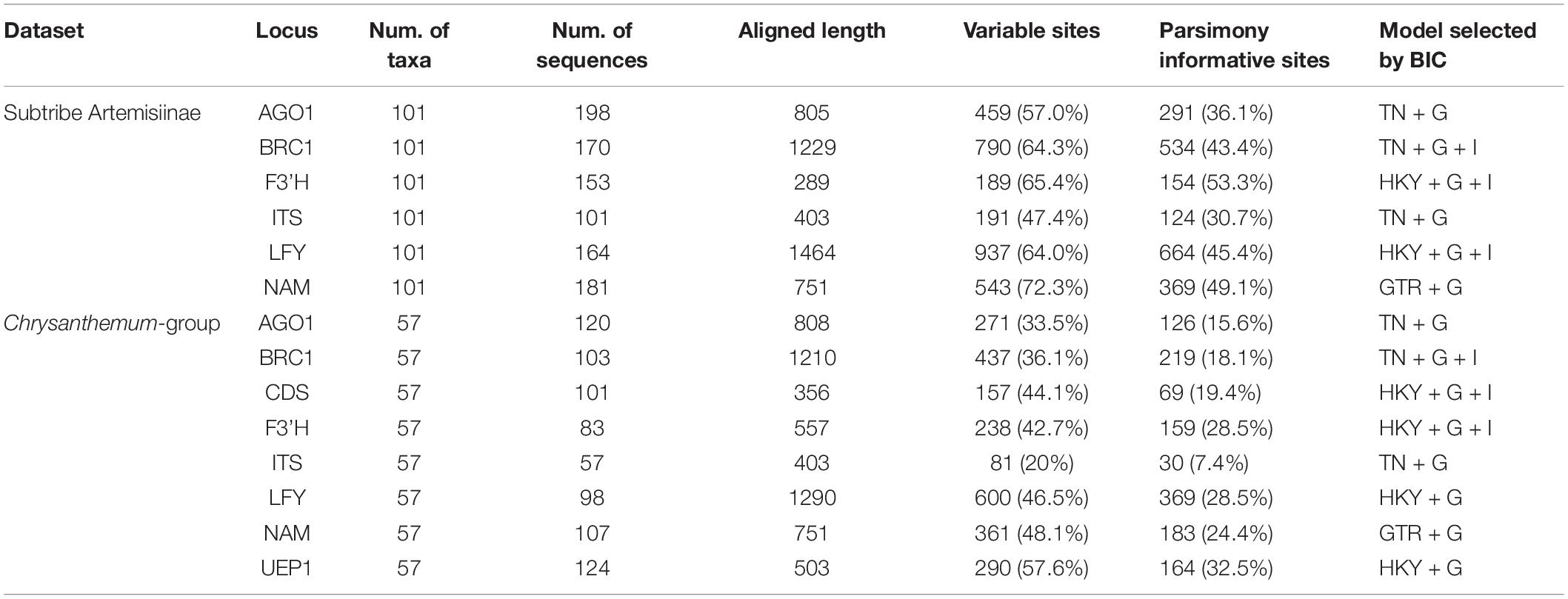
Genomic DNA was extracted from silica gel-dried or fresh leaves with a Plant Genome Extraction Kit (Tiangen Biotech, China) following the manufacturer’s protocol. For better phylogenetic resolution, we utilized nrITS and seven low-copy nuclear genes, AGO1 (ARGONAUTE 1; Zhang et al., 2015), BRC1 (BRANCHED1; Zhou et al., 2012; Wang M. et al., 2019), CDS (chrysanthemyl diphosphate synthase gene; Rivera et al., 2001; Liu et al., 2012a), F3’H (flavonoid3′-hydroxylase gene; Zhao et al., 2013), LFY (LEAFY; Ma et al., 2016), NAM (No Apical Meristem; Sha et al., 2017) and UEP1 (gene of ubiquitin extension protein; Annadana et al., 2002). Polymorphic regions mostly covering introns and 5′UTRs of five of the six genes were amplified using conserved primer pairs that were developed according to sequences acquired from GenBank (Table 1). To isolate AGO1 orthologs from the species of interest, we downloaded the AGO1 sequence of Helianthus tuberosus and then ran a local BLAST in the genome data of Ch. nankingense (Song et al., 2018). The primer pair was designed based on the orthologous sequence of Ch. nankingense (Table 1). All PCR products were ligated into pGEM-T vectors (Promega, United States) and cloned. At least 4–6 positive clones were randomly taken for sequencing. DNA sequences obtained by this study are deposited in GenBank with accession numbers MW344433–MW344631 (AGO1), MW195142–MW195312 (BRC1), MW543604–MW543703 (CDS), MW543450–MW543603 (F3’H), MW011041–MW011206 (LFY), MW195313–MW195497 (NAM), MW344310–MW344432 (UEP1), and MW545598–MW545801 (nrITS).
Phylogenetic Analysis
To guarantee that the fragments we sequenced for each gene were orthologous, reciprocal BLAST of the obtained sequences was conducted (Wall and Deluca, 2007). Then, alignment was performed using the online toolkit MAFFT v.72 with default parameters followed by manual checking in MEGA 7 (Kumar et al., 2016). Highly polymorphic positions including multiple gaps that made alignment ambiguous were removed from the alignment. For each nrITS sequence, ITS1 and ITS2 were tandemly linked with 5.8S-rRNA excluded. Subsequently, we used DAMBE v. 6.4.101 (Xia, 2017) to merge identical sequences within each species. The aligned data matrix of each of six low-copy genes was subjected to a prior Bayesian tree with which we found no evidence showing paralogous copies (Supplementary Figure 1) [As CDS gene was previously proved as single-copy (Rivera et al., 2001; Liu et al., 2012a), it was not shown in Supplementary Figure 1]. To construct the Bayesian tree, the best-fit evolutionary model of each dataset was selected using jModeltest 2.1.7 (integrated in online bioinformatic platform CIPRES3) according to the Bayesian informatic criterion (BIC) values (Posada, 2008; Table 2).
The tree of the whole subtribe Artemisiinae was constructed based on six of the eight loci, as the targeted fragments of CDS and UEP1 could not be amplified from some Artemisia species and the outgroups. The phylogenetic reconstruction of subtribe Artemisiinae followed a coalescent strategy using ∗BEAST embedded in BEAST v. 1.8.4 by selecting the Yule tree prior and the Piecewise linear and constant root of population size model (Drummond et al., 2012a, b; Liu et al., 2015; Edwards et al., 2016). The divergence times of major nodes were estimated under uncorrelated relaxed clock by setting two secondary calibration points with normal distributions: The first was the divergence time of the Eurasian-Mediterranean clade and the Asian-southern African clade (17.3 Ma ± 1 SD), and the second was the age of the crown clade of Artemisiinae (9.7 Ma ± 1 SD) according to Oberprieler (2005). To test the influence of different time-point settings on molecular dating, the secondary calibration point was set according to Tomasello et al. (2015) (the divergence time of Artemisiinae and Santolininae as 18 Ma ± 1 SD; or the divergence time of Artemisiinae-Santolininae-Glebionidinae and other Eurasian lineages as 22.5 Ma ± 1 SD). Moreover, the fossil records of Artemisia-like pollens were also considered for calibration. We set tmrca prior at 13 Ma ± 1 SD for the crown lineage of Artemisiinae (the stem node of lineages sharing Artemisia-type pollens, e.g., Artemisia, Elachanthemum, and Phaeostigma) according to the records of the commonly occurred Artemisia-like pollen fossils (Wang and Zhang, 1990; Ma et al., 2005). Trees were sampled every 10000 generations for a total of 400 million generations. We checked for topological convergence and adequate ESS (>200) using Tracer v.1.7.1 (Rambaut et al., 2018). The consensus tree was exported using TreeAnnotator v. 1.8.4, discarding the first 30% of trees as burn-in.
Then, all eight loci were applied to construct a tree of Chrysanthemum-group to better resolve relationships within this particular group following the same analytical method specified above.
Estimation of Diversification Dynamics
Subsequently, 5000 trees from the ∗BEAST analysis were resampled to construct a lineage-through-time (LTT) plot using the R package ape v. 5.0 (Paradis and Schliep, 2019). Then, 1000 random trees were simulated under birth-death (BD) and pure-birth (PB) models using the R package geiger to avoid errors due to incomplete sampling (Pennell et al., 2014). These simulations were used to establish 95% confidence intervals on the LTTs for each model for comparison with the empirical dataset. The γ statistic for the optimal ∗BEAST phylogeny was calculated using ape v. 5.0. Speciation rates were compared with the null hypothesis that a clade diversified at a constant rate using 2∗[1-pnorm(abs(gammaStat(tree)))] provided by the R package ape v. 5.0 for a two-tailed test (Paradis and Schliep, 2019). A significantly positive or negative γ value meant an accelerated/decelerated rate of speciation toward the present.
Reconstruction of Ancestral Distributions and Character States
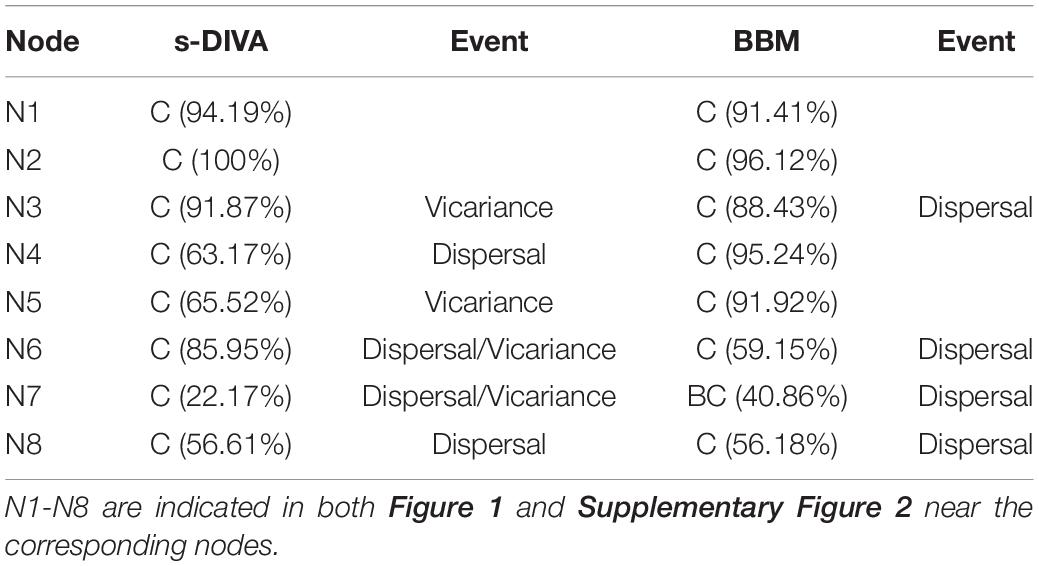
To infer the biogeographic history of lineages in subtribe Artemisiinae, dispersal and vicariance analysis (s-DIVA), Bayesian binary MCMC analysis (BBM) and dispersal-extinction-cladogenesis (DEC, s-DEC) were conducted to estimate the optimized geographical distributions of internal nodes using RASP 4.0 (Yu et al., 2015). The distribution information of all samples was obtained from floristic works (Shih and Fu, 1983; Shishkin and Bobrov, 1995; Zhao et al., 2009) and herbarium specimens. Based on this information and major biogeographic boundaries, eight geographic units were defined: (A) Europe and the Mediterranean coast; (B) Middle Asia; (C) Central Asia; (D) the QTP region; (E) the southern part of East Asia; (F) the northern part of East Asia; (G) Korea-Japan; and (H) the pan-Arctic region (Spencer and Robinson, 1968; Xie et al., 2002; Cowan, 2007; Lu et al., 2018). The ranges of geographic units are as shown in Supplementary Figure 2. The current distribution of each species is marked by a particular color before the species name. For s-DIVA, s-DEC and DEC, 10000 trees that were resampled from the ∗BEAST phylogenies after burn-in and the consensus tree were loaded into RASP to estimate the likelihoods of ancestral states at each internal node of the consensus tree. BBM was run with F81 state frequencies using gamma variation for 1,000,000 iterations. The reconstructed state was sampled every 1000 generations, and the first 10% was discarded as burn-in.
To estimate the ancestral states of flowerhead architectures, character states were mapped onto terminal branches of the consensus tree resulting from the ∗BEAST analysis, and then searches for optimized states at the internal nodes were run with the likelihood model in Mesquite v. 3.5.2 and with the continuous-time Markov model using the phytools v. 0.7-70 package in R (Maddison and Maddison, 2009; Revell, 2012).
Ecological Niche Modeling, Lineage Distribution Models and Niche Overlap Tests
To analyze niche differentiation and to understand how the distributions of three major clades of Chrysanthemum-group changed during the last glacial maximum (LGM), niche modeling was conducted under recent climatic scenarios and LGM scenarios from the CCSM4 model (current climate at a 30 arcsec resolution and LGM at a 2.5 arcminute resolution). Ecological niche data including temperature and precipitation (19 bioclimatic variables) data that were drawn as climate layers were downloaded from WorldClim 1.44. Coordinate information was collected from occurrence records in the Global Biodiversity Information Facility (GBIF5) and from herbarium specimen information acquired from NSII6. For every two occurrences, only one was kept if less than 1 km in distance. All of the coordinates used for inferring lineage distribution modes are listed in Supplementary Table 2. In total, the current environmental data of 1249 occurrence records, including 529 of Ajania, 529 of Ch. indicum-complex and 191 of Ch. zawadskii-complex were sampled. To avoid collinearity, Pearson pairwise correlation analysis of environmental factors was conducted. For each pair, one factor with a correlation value (| R|) higher than 0.75 was eliminated, and the result was visualized in SPSS 22 and Heml (Deng et al., 2014). We used lineage geographic distribution spots to estimate the environmental space of each of the three main subclades of Chrysanthemum-group with the PCA-env approach in SPSS 22 and R (R Core Team, 2013). Species distribution models (SDMs) were built under the maximum-entropy method implemented in MAXENT 3.3 following Papeş and Gaubert (2007) for parameter settings (25% of occurrence records were used as testing data) (Papeş and Gaubert, 2007; Phillips and Dudík, 2008). The area under the receiver operating characteristic curve (AUC) was used to evaluate the prediction performance of the models (Phillips et al., 2006). AUC values > 0.75 indicate good predictions, whereas values < 0.5 suggest poor predictions (not better than random). We also used the true skill statistics (TSS) to evaluate the accuracy of the resulting distribution models (Liu et al., 2005; Allouche et al., 2006), where TSS values ranging from 0.4 to 0.8 are indicative of good model performance (Landis and Koch, 1977; Fielding and Bell, 1997). The 10-percentile training presence threshold was used to generated binary map. Subsequently 10000 random geographic points were extracted to calculated TSS values for each niche model in R. Moreover, DIVA-GIS v. 7.5 was applied to model potential distributions under the current scenario to ensure the reliability of SDMs (Hijmans et al., 2001). To quantify climatic niche overlap, Schoener’s D-index (Schoener, 1970) and the modified Hellinger distance I-index (Van der Vaart, 2000) were calculated in R. Subsequently, niche equivalency and similarity tests (Evans et al., 2009; Warren et al., 2008) were performed between subclades using 50 permutations in the R packages phyloclim v. 0.9.5 and ENMtools v. 1.0.2 (Warren et al., 2010; Heibl and Calenge, 2013). To analyze pairwise differentiation in detail on every explanatory bioclimatic variable resulting from analysis by PCA-env, the profiling niche occupancy (PNO) of each climatic factor was computed, and then the D-index and the modified I-index were calculated using the phyloclim v. 0.9.5 package in R (Heibl and Calenge, 2013).
Results
Sequence Characteristics
Despite repeated amplifications, we failed to isolate the target CDS fragment from some species, e.g., Artemisia frigida and Seriphidium finitum, and we failed to isolate its complete open reading frame in Nipponanthemum nipponicum, Hippolytia spp. We were not able to obtain UEP1 from Stilpnolepis centiflora and the outgroup taxa. Therefore, the analyses were conducted separately with two datasets, with one excluding CDS and UEP1 for all the sampled taxa of subtribe Artemisiinae and the other including all eight loci but limited to Chrysanthemum-group. For each of the datasets, the number of species/taxa included, the number of haplotypes obtained at each locus and other information related to phylogenetic analyses are given in Table 2.
Phylogeny of Subtribe Artemisiinae and the Estimation of Diversification Dynamics
The coalescent species tree of Artemisiinae overall was inferred from six of the eight markers (Figure 1). Rooted by outgroup species from the Eurasian-Mediterranean clade of tribe Anthemideae (Oberprieler et al., 2007), the tree showed subtribe Artemisiinae as monophyletic. Except for the basal monotypic genus Stilpnolepis, two main clades, Clades I and II, were found (Figure 1: posterior probabilities PP = 0.78 and 1, respectively). These two clades roughly corresponded to but differed slightly in circumscriptions from the Artemisia-group and Chrysanthemum-group traditionally defined by morphological characteristics. The species of Artemisia and its allies, e.g., Filifolium and Crossostephium, were found in Clade I. However, the broadly defined Artemisia was not supported as monophyletic. Several genera with radiate capitula but uncertain phylogenetic positions, such as Leucanthemella, Nipponanthemum, Hippolytia, Brachanthemum, and Tanacetum tatsienense, fell into this clade. Two subclades, Ia and Ib, were further recognized with high posterior probability (PP = 1). Notably, the present data showed that species of Phaeostigma were clustered in subclade Ia, supporting the proposal that they were independent of Ajania but were a lineage within Artemisia (Figure 1). Subclade Ib contained species formerly described under Artemisia subgenus Dracunculus, as well as Kaschgaria and Brachanthemum (with radiate capitula) (Figure 1).
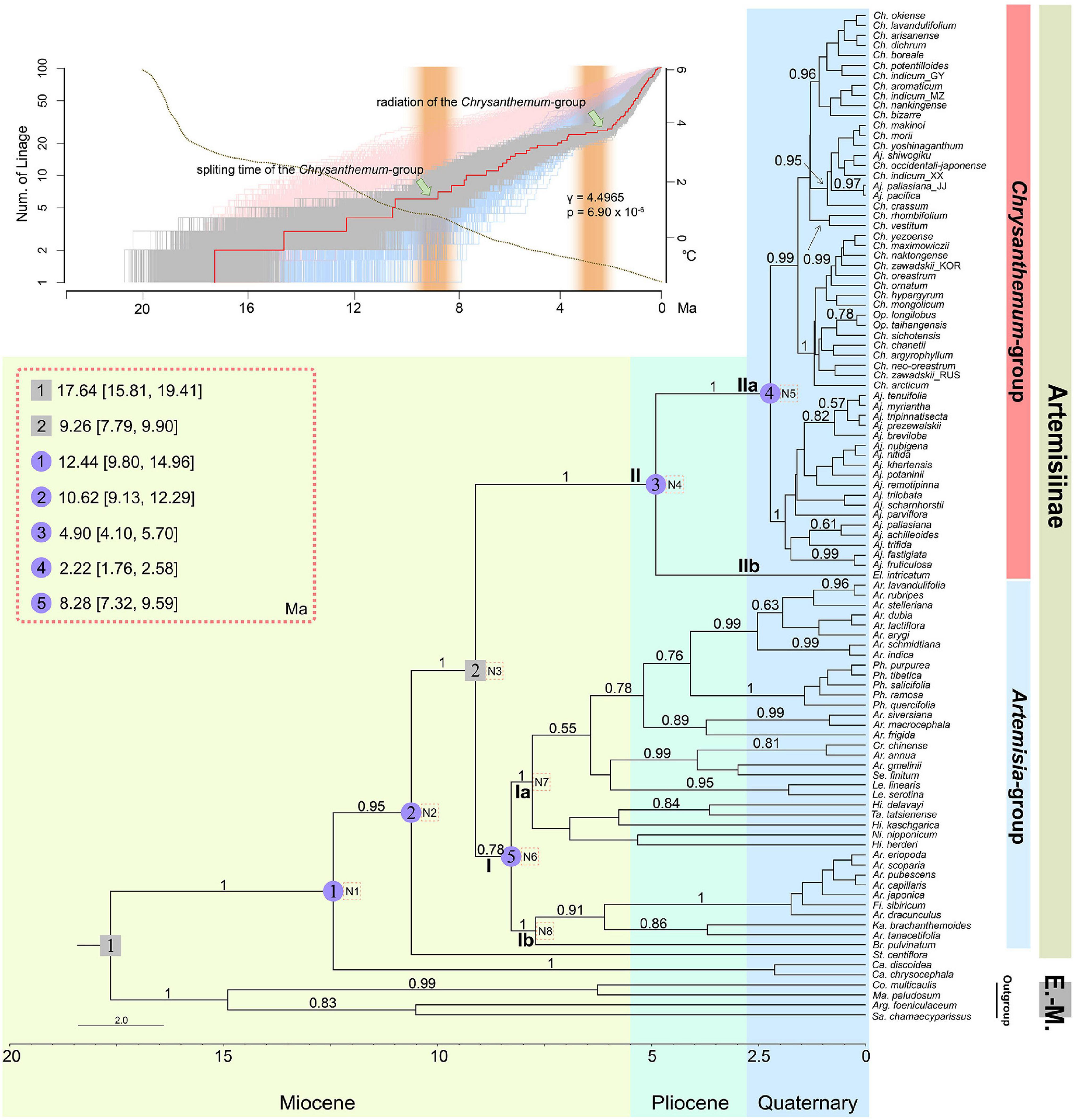
Figure 1. Coalescent species tree inferred from six nuclear gene sequences and the lineage through time (LTT) plot of subtribe Artemisiinae. The tree was constructed in *BEAST and rooted by four Eurasian-Mediterranean (E.-M.) species of tribe Anthemideae. Bayesian posterior probabilities (>0.5) are indicated above branches. Two secondary calibration time points used for estimating divergence times are marked with gray squares. Five major clades are highlighted by blue circled numbers, and their estimated divergence times are noted in the upper-left box. Blue, green, and yellow blocks indicate different geologic ages. The dotted square with N1–N8 inside indicates the nodes analyzed for biogeographic history, as shown in Table 3. Abbreviations of genus names: Ch., Chrysanthemum; Op., Opisthopappus; Aj., Ajania; El., Elachanthemum; Ar., Artemisia; Ph., Phaeostigma; Cr., Crossostephium; Se., Seriphidium; Le., Leucanthemella; Ni., Nipponanthemum; Hi., Hippolytia; Ta., Tanacetum; Fi., Filifolium; Ka., Kaschgaria; Br., Brachanthemum; St., Stilpnolepis; Ca., Cancrinia; Co., Coleostephus; Ma., Mauranthemum; Sa., Santolina; Arg., Argyranthemum. The LTT plot using the uncorrected relaxed clock model in *BEAST is shown in the upper-left corner in gray, where the red line indicates the LTT plot of the maximum credibility tree for the *BEAST analysis. Lineages accumulated over time under the pure-birth model are marked in blue, and those under birth-death are marked in pink. The thin dotted line represents the decrease in the environmental temperature during the time frame of diversification of subtribe Artemisiinae, referring to Hansen et al. (2013). Statistic γ takes on positive values when there are accelerated speciation rates toward the present; p indicates the significance value (< 0.001). Two brown columns mark the periods of uplift of the Northern QTP during the late Miocene.
Clade II included Chrysanthemum and Ajania and two small genera, Elachanthemum and Opisthopappus. Two sister branches, IIa and IIb were also found in Clade II: Elachanthemum, the only member of IIb, is different from Chrysanthemum and Ajania by having a discoid capitulum and an annual life form. Interestingly, Opisthopappus, which was formerly treated in subtribe Tanacetinae and then were incorporated into Artemisiinae (Bremer and Humphries, 1993; Oberprieler et al., 2009), was clustered in IIa; morphologically, this small genus is indeed similar to Chrysanthemum by having radiate capitulum and myxogenic achenes (Figures 1, 2).
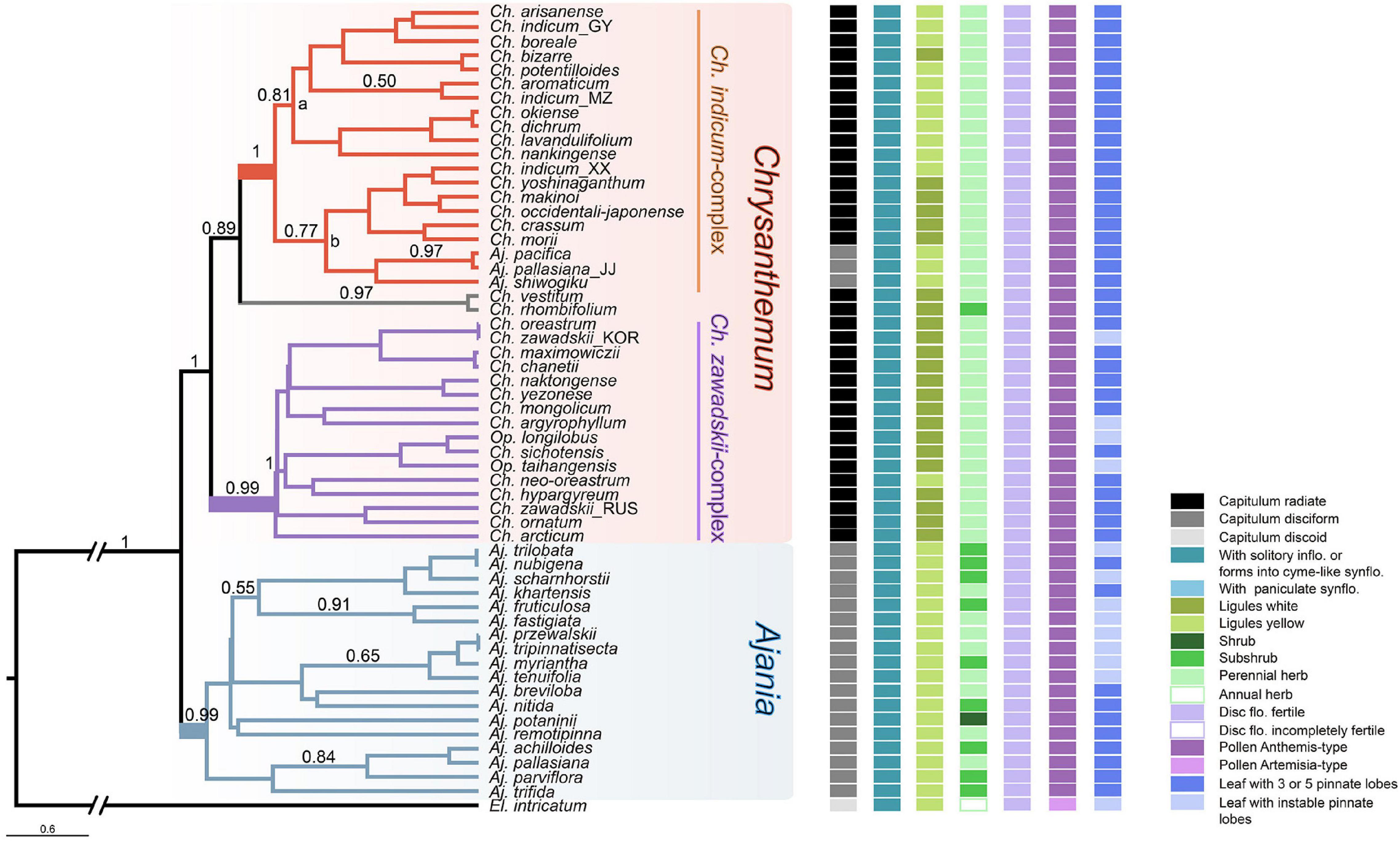
Figure 2. Coalescent tree of the Chrysanthemum-group inferred from eight nuclear gene sequences. Bayesian posterior probabilities higher than 0.5 are indicated on branches. States of important morphological characters (represented by small squares in different colors) are mapped at the right side of terminal nodes. In this tree, two subclades, Ch. indicum-complex and Ch. zawadskii-complex, can be recognized within Chrysanthemum. Naming of the two Chrysanthemum subclades follows Liu et al. (2012b).
The chronograms achieved using relaxed molecular clock analyses showed the time frames were largely similar among four different calibration settings (Supplementary Figure 3). According the chronogram inferred from setting two secondary calibration time points suggested that subtribe Artemisiinae began to diverge at approximately 10 Ma ago, and most of its major lineages diversified during the late Miocene, particularly densely at approximately 7-9 Ma ago (Figure 1). Artemisia-group split from Chrysanthemum-group at approximately 9 Ma ago and then diversified immediately, while the latter group diversified much later. Within Chrysanthemum-group, the monotypic genus Elachanthemum diverged much earlier during the mid-Pliocene (Figure 1, median age 4.90 Ma, 95% HPD 4.10-5.70 Ma), while other species seemed to have undergone evolutionary radiation since the boundary between the Tertiary and Quaternary (median age 2.22 Ma, 95% HPD 1.76-2.58 Ma). The LTT plots highlighted a significant speedup of diversification of subtribe Artemisiinae (γ = 4.50, p < 0.001), which was largely correlated with the cooling climate estimated by the δ18 O content (Figure 1; Hansen et al., 2013). The time frame of evolution of Chrysanthemum-group was in concordance with the intensive aridification of Middle to Central Asia that was probably due to late-Miocene to Pliocene geographic activity of the northern region of QTP and the global climatic changes (Figure 1).
Internal Relationships Within the Chrysanthemum-Group
To better resolve relationships within Chrysanthemum-group, all eight nuclear gene sequences were used. As a result, two major genera, Chrysanthemum and Ajania were well separated into two clades (PP > 0.99) with a few exceptions (Figure 2), which provided important support for traditional taxonomy that treats them as independent genera mainly according to capitulum morphology. In Chrysanthemum, two Chinese endemic species with white ray flowers, Ch. rhombifolium and Ch. vestitum, formed a small clade sister to Ch. indicum-complex with yellow or white ray flowers (Figure 2). In the clade of Ch. indicum-complex, two branches, a and b, were recognized—species in ‘b’ basically all had island distributions, while species in ‘a’ were mostly distributed in mainland China (Figure 2). Another relatively large group is the so-called Ch. zawadskii-complex (purple-colored in the tree, PP = 0.99, Figure 2). This group is characterized by white to purple ray flowers with continuous color variation. Species of Opisthopappus, which were shown at the basal position of Chrysanthemum-group in the ITS and plastid DNA tree, fell into this clade. In contrast to Chrysanthemum, relationships within Ajania were still ambiguous, with most branches being poorly supported.
Reconstructions of Ancestral Distributions of Chrysanthemum-Group
To explore the historical process of diversification of Chrysanthemum-group, the ancestral distribution state of each of the major nodes was estimated based on their recent distributions acquired from floristic works and specimen information. The Bayesian BBM analysis, s-DIVA analysis and DECs produced similar results regarding the ancestral area of Artemisiinae (Table 3 and Supplementary Table 3). Eight geographic units and optimal area reconstruction at each major node are summarized in Supplementary Figure 2. In RASP, both s-DIVA and BBM favored ancestral distributions of most of the major clades of Artemisiinae (N1-N8) in Central Asia (region C; >50%) (Table 3 and Supplementary Figure 2). Subsequent vicariance between Central Asia and other regions of Asia was reconstructed by s-DIVA for node N3 and N5, and dispersal from Central Asia to the QTP and eastern parts of Asia was reconstructed for nodes N4, N6-N8 and Chrysanthemum-group (Table 3). The time-effect curve reconstructed with BBM showed that the dispersal curve had extremely high peaks over the last 3 Ma (Supplementary Figure 2).
SDMs and Niche Differentiation Among Lineages Within Chrysanthemum-Group
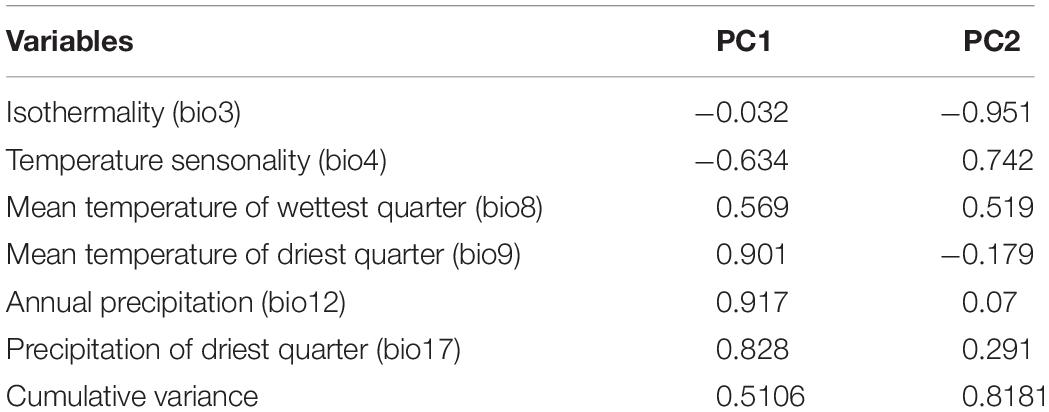
To reduce multicollinearity, one factor of each pair of environmental factors was eliminated when the correlation coefficient of that pair was larger than 0.75. Thus, six factors were finally selected: isothermality (bio3), temperature seasonality (bio4), mean temperature of the wettest quarter (bio8), mean temperature of the driest quarter (bio9), annual precipitation (bio12) and the precipitation of the driest quarter (bio17) (Figure 3 and Supplementary Figure 4A). The variable loadings for PCA-env are shown in Table 4. The first two PCs explained 81.8% of the niche variation among subclades (51.1 and 30.7%, respectively). PC1 is dominated by the absolute temperature and precipitation variables (bio9, bio12, bio17) while PC2 by the two variables describing oscillations of temperature (bio3, bio4) (Table 4). The PCA-env assay suggested great variability in the environmental space inhabited by the different subclades within Chrysanthemum-group, especially between Ajania and Ch. indicum-complex (Figure 3 and Supplementary Figure 4).
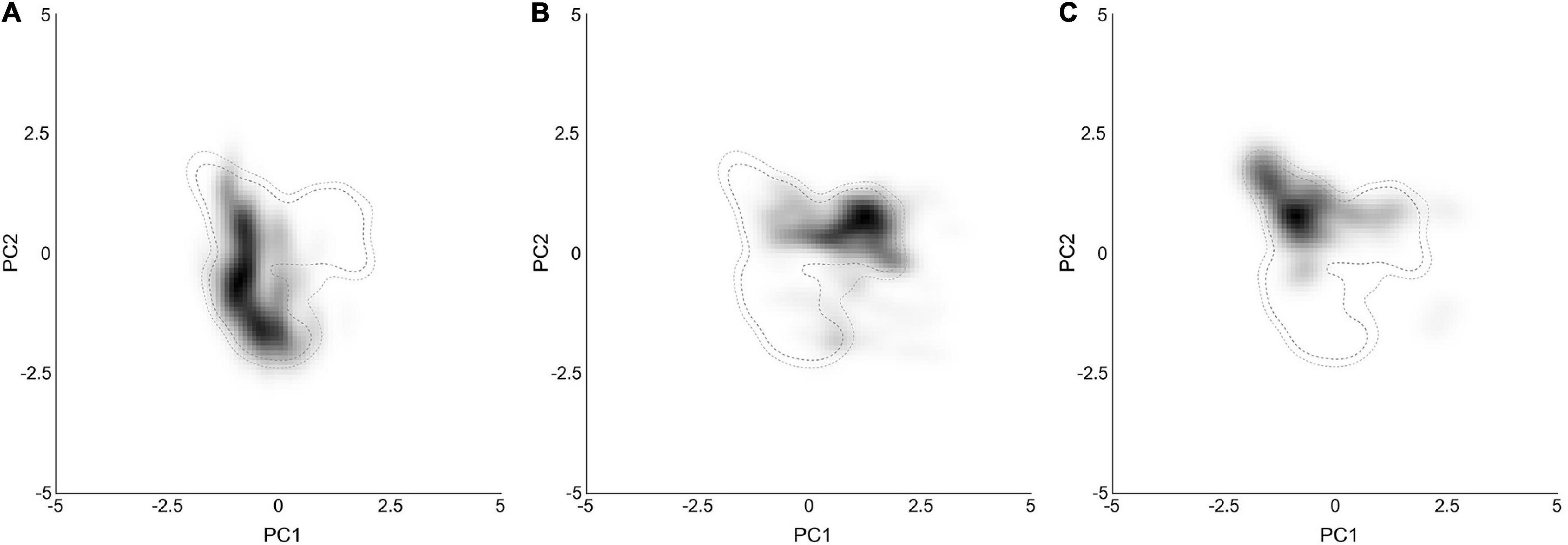
Figure 3. Ecological niches of the three analyzed subclades within the Chrysanthemum-group in environmental space produced by the principal component analysis method. The PCA results of (A–C) represent the niches of the lineages in the two main axes with the environmental conditions of sampled occurrences for Ajania, Ch. indicum-complex and Ch. zawadskii-complex, respectively. The gray-to-black shading shows the density of the occurrences by cell. The outer and inner dashed contour lines illustrate 100 and 95% of the available (background) environment, respectively.
Furthermore, the potential distribution of subclades within Chrysanthemum-group was predicted through MAXENT and DIVA-GIS. The results from both approaches were, in principle, similar, with the area predicted by DIVA-GIS being slightly restricted (Figure 4 and Supplementary Figures 5A-C). The AUC values for the replicate runs in MAXENT were 0.905 ± 0.003, 0.930 ± 0.003, and 0.907 ± 0.009 for Ajania, Ch. indicum-complex and Ch. zawadskii-complex, respectively. Establishing the threshold probability for niche models using TSS of each of three subclades resulted in 0.72 ± 0.055, 0.77 ± 0.047, and 0.70 ± 0.092. Lineage distribution models indicated that each lineage occurred over different geographical areas with more or less contact areas at margins (Figure 4). The suitability map reconstructed based only on the occurrences of three aberrant Ajania species of Korea-Japan distribution was congruent with those of other Ch. indicum-complex species (Figures 2, 4 and Supplementary Figure 5D).
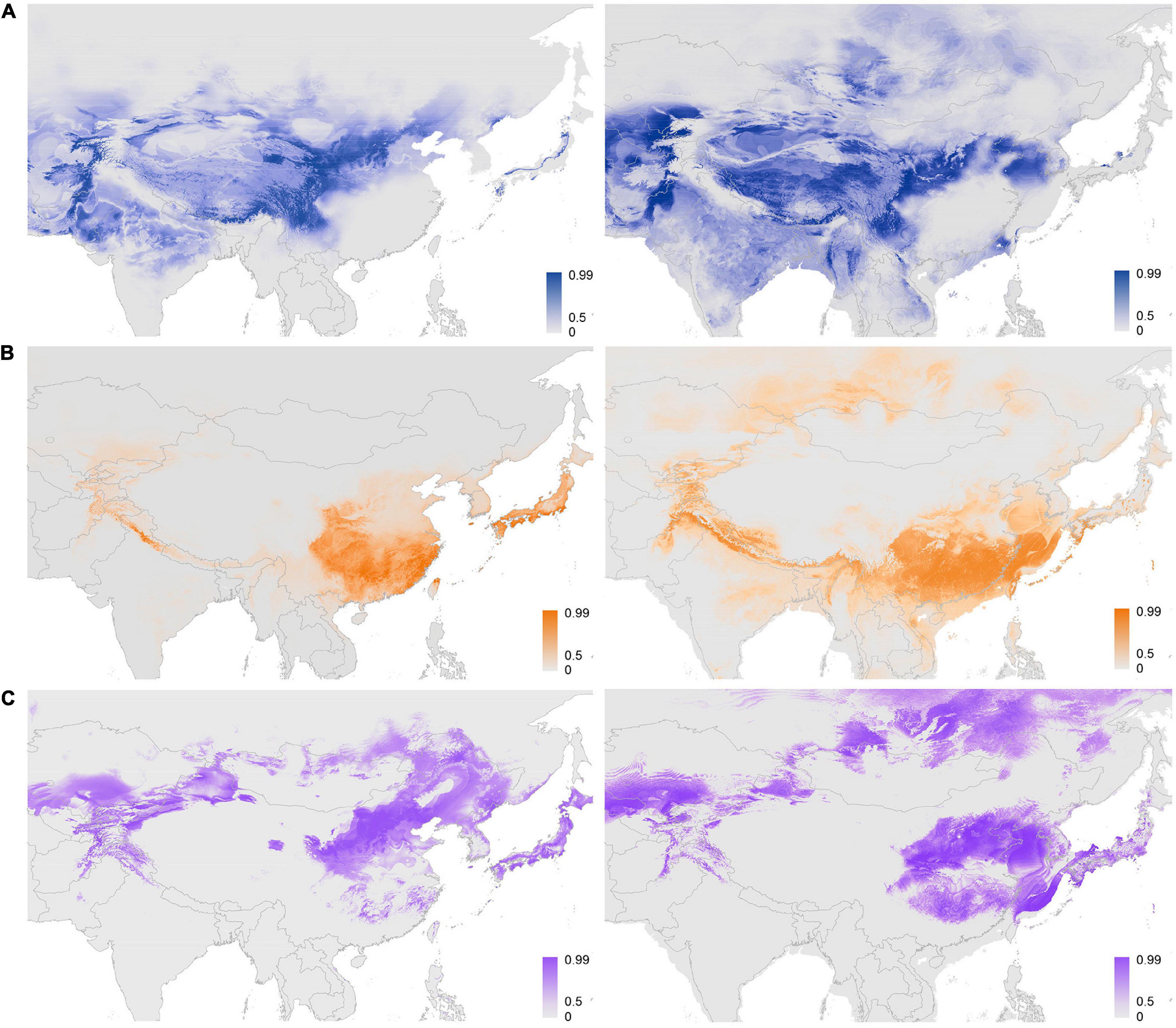
Figure 4. Potential distribution area of Ajania and two subclades of Chrysanthemum predicted by the maximum-entropy model. (A–C) Represent the predicted distributions of Ajania, Ch. indicum-complex and Ch. zawadskii-complex, respectively, during the current period (left) and during the Last Glacial Maximum (LGM) (right). Shadings in color show the probability of geographic occurrences. The prediction for the Korea–Japan distributed Ajania species is shown in Supplementary Figure 5.
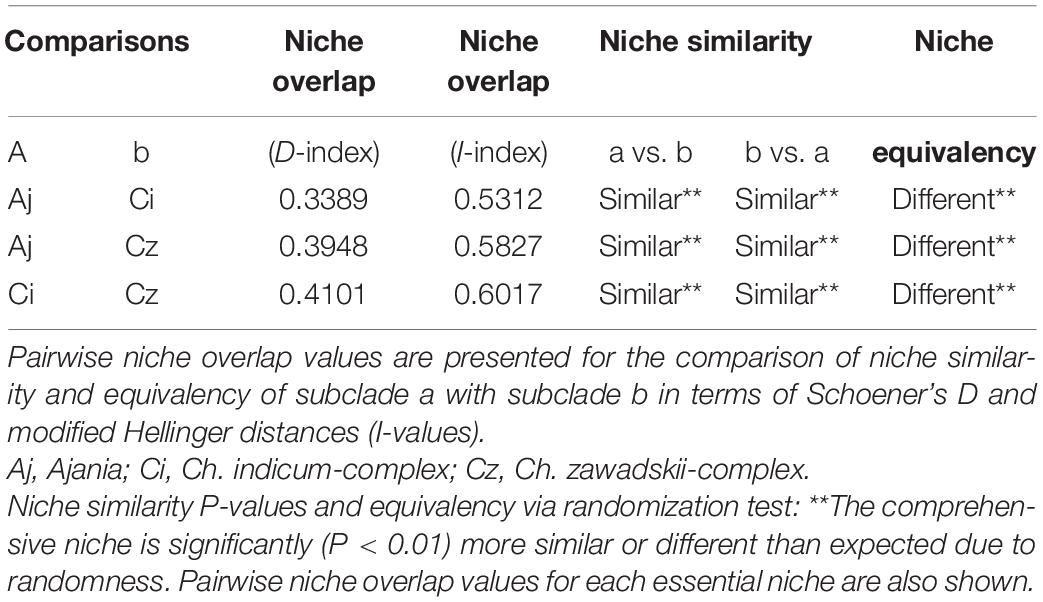
Considering the predicted distribution of the modeled lineages (Figure 4), the comprehensive pairwise D-values among lineages ranged from 0.34 to 0.4 (Table 5), suggesting ecological differentiation (Figure 4 and Table 5). The background niche similarity tests indicated that the observed overlaps were greater than 95% of the simulated values, suggesting that lineages occupy an ecological space that is more similar to each other than expected by chance (Table 5). However, the null hypothesis of niche equivalency was rejected for all comparisons between subclades. The profiling niche occupancy (PNO) of each ecological factor was computed for the detailed adaptation patterns (Supplementary Table 4). As a result, Ajania had the lowest niche overlap with two subclades of Chrysanthemum in precipitation in the driest quarter and isothermality (D-values of 0.4 and 0.46) (Figure 5 and Supplementary Table 4). Two Chrysanthemum subclades greatly diverged in temperature seasonality and mean temperature in the driest quarter (Figure 5 and Supplementary Table 4, D-value of ca. 0.46).
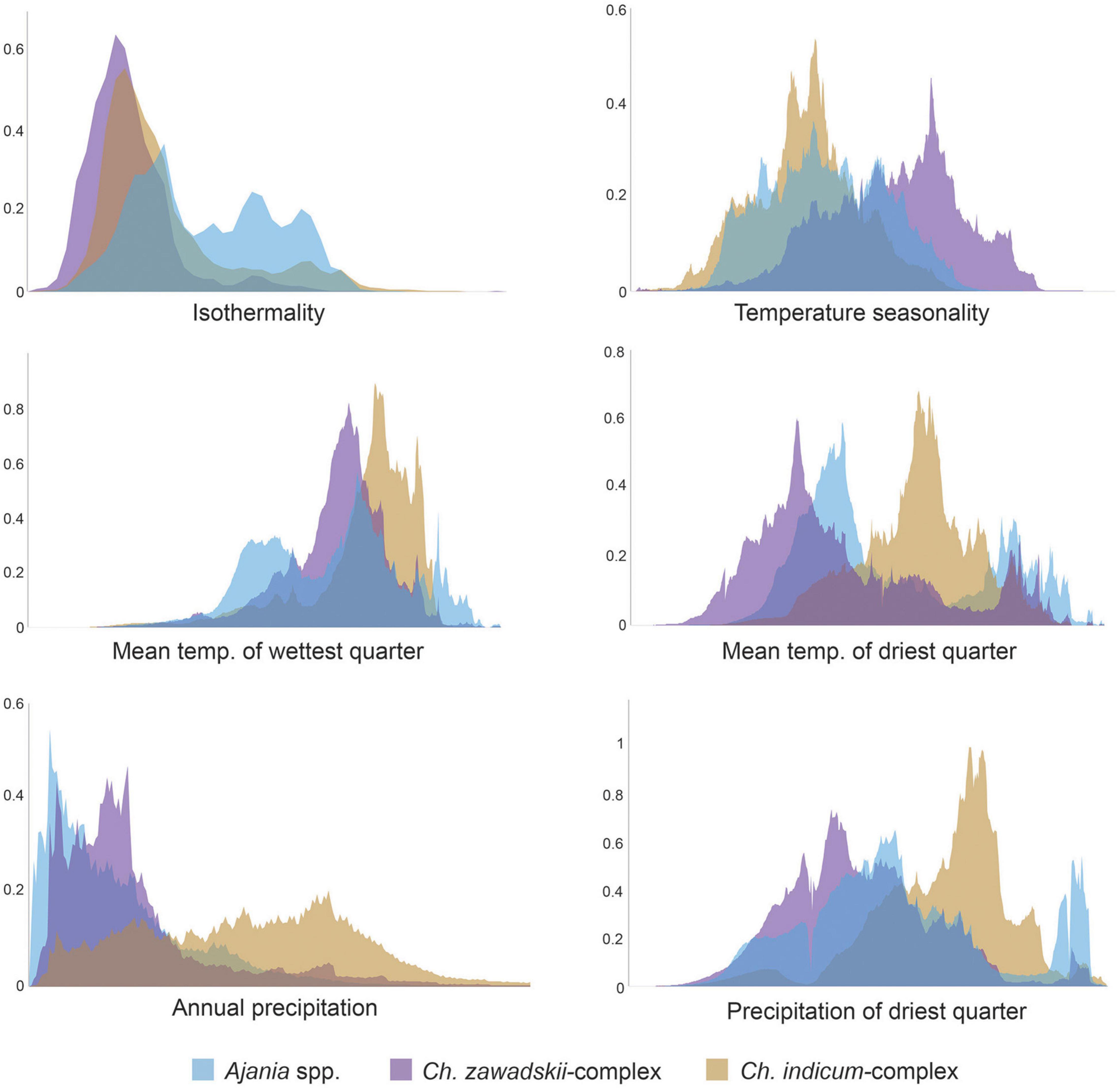
Figure 5. Diagrams displaying niche overlaps among three major lineages. Suitability diagrams were converted from potential distribution and ecological data according to the strategy reported by Evans et al. (2009). The x-axis indicates the adaptive range of such ecological factors, and the y-axis indicates suitability. Different colors correspond to different lineages, as shown in the legend at the bottom.
Discussion
Clarifying Phylogenetic Positions of Some Taxa in Subtribe Artemisiinae
Despite a series of cladistical studies based on morphological and molecular information, there remain quite a number of taxa whose systematic positions are unclear in subtribe Artemisiinae (Bremer and Humphries, 1993; Torrell et al., 1999; Martin et al., 2001; Vallès and McArthur, 2001; Vallès et al., 2003; Oberprieler et al., 2007; Sanz et al., 2008; Zhao et al., 2010a, b). Phaeostima is one such taxon. It was separated from Ajania as an independent genus by Muldashev (1981). However, neither morphological data nor molecular phylogenies could resolve its relationships with Ajania, Artemisia, and Chrysanthemum (see Huang et al., 2017 and references therein). By integrating morphological analyses of 20 characters, and palynological and molecular data, Huang et al. (2017) circumscribed a monophyletic Phaeostigma by including three species formerly named under Ajania, which was in line with the proposal of Bremer and Humphries (1993) that the circumscription of Phaeostigma should be extended to some members of Ajania. Moreover, Huang et al. (2017) supposed that Phaeostigma might be phylogenetically closer to Artemisia than to Ajania (Huang et al., 2017; Chen et al., 2020), which was strongly supported by the present analysis, that is, Phaeostigma was nested in the clade of Artemisia-group (Figure 1). Nevertheless, the similarity between Ajania and Phaeostigma in terms of characteristics such as synflorescence and capitulum, as well as in diversification ages and distribution patterns, implies that they may have experienced convergent evolution under similar habitats (Figures 1, 2 and Supplementary Figure 2). More detailed studies are required to determine the adaptive significance of these traits.
Opisthopappus is another taxon worth discussion. It was placed in subtribe Tanacetinae by Bremer and Humphries (1993) mainly due to the presence of pappi or coronas on achenes but was different from other members of Tanacetinae according to its myxogenic achenes. Its systematic position in subtribe Tanacetinae was called into question by the present (Figure 2) and previous (Zhao et al., 2010a, b) molecular data that suggested a closer relationship to Chrysanthemum and Ajania. Besides, several studies reported crossability between Opisthopappus and species of Chrysanthemum-group, e.g., Ch. lavandulifolium, Ch. dichrum, A. pallasiana, and Elachanthemum intricatum and A. pacifica x Ch. vestitum (Hu and Zhao, 2008; Yang et al., 2010; Zheng et al., 2013). Combining all the evidence, we suggest a taxonomic treatment to incorporate Opisthopappus into Chrysanthemum-group. Nevertheless, to clarify its phylogenetic position, more data of morphological and ecological traits, reproductive biology and phylogenomics are required.
The systematic positions of Hippolytia, Nipponanthemum, Leucanthemella, Brachanthemum, and Tanacetum tatsienense have also been the subjects of much debate. Hippolytia and Tanacetum were formerly described in subtribe Tanacetinae, while Nipponanthemum and Leucanthemella were classified into subtribe Leucantheminae, but all were later shown to belong to subtribe Artemisiinae according to molecular phylogenetic data (Vallès and McArthur, 2001; Watson et al., 2002; Vallès et al., 2003; Oberprieler, 2005; Oberprieler et al., 2007, 2009; Masuda et al., 2009). Using multiple nuclear loci with a coalescent analytical method, we found that they belonged to Artemisiinae, which was supported by our analysis. Meanwhile, our analysis showed that Hippolytia, Nipponanthemum, Leucanthemella, Tanacetum tatsienense, and Brachanthemum were nested into different branches of Artemisia-group (Figure 1) rather than being basal grades of Artemisiinae, as suggested by previous ITS-ETS data (Watson et al., 2002; Sanz et al., 2008; Zhao et al., 2010a).
Evolution of the Capitulum Architecture in Subtribe Artemisiinae
Subtribe Artemsiinae is one of the youngest lineages in the daisy family, and it appears as an assemblage of taxa of plesiomorphic features, especially with regard to pollination syndrome, including capitulum architectures and affiliated features (Harris, 1999; Martin et al., 2001; Sanz et al., 2008; Pellicer et al., 2010; Huang et al., 2017). In terms of capitulum architecture, three types can be observed in wild Artemisiinae species: radiate, disciform and discoid. The radiate capitula are composed of many central disk florets and peripheral conspicuous ray flowers, whereas, the discoid consist of bisexual disk flowers only, and the disciform are discoid-like but with a few tubular female marginal florets). The evolutionary orientation of these capitulum types has remained a key question in teasing apart historical patterns of lineage divergence and/or hybridization within Artemisiinae. Bremer and Humphries (1993) regarded the radiate capitulum as the plesiomorphy of Artemisiinae and the discoid as a derivative of the disciform, rather than evolving directly from the radiate. However, ancestral state reconstruction based on the ITS-ETS tree suggested that the discoid was the plesiomorphy of Artemisiinae (Sanz et al., 2008). In the present study, using a similar strategy, we tried to detect the direction of capitulum evolution in Artemisiinae. The results also showed discoid or disciform flowerheads as ancestral states (Supplementary Figure 6). However, model-based ancestral character reconstruction of a particular trait may not provide really accurate information without mechanism evidences, because missing data of extinct taxa or bias in models for state transition might influence the accuracy of the inference (Omland, 1999; Griffith et al., 2015). A clear answer to the problem should rely on evidence from evolutionary developmental genetic studies. Trow (1912), as well as Ford and Gottlieb (1990) examined the inheritance of rayed and rayless heads in Senecio and Layia, respectively, and suggested that the switch between the presence and absence of ray flowers may be governed by simple genetic rules. Later molecular genetic studies indicated that the CYC2-mediated module participates in inflorescence repatterning (Broholm et al., 2008; Chapman et al., 2008, 2012; Kim et al., 2008; Tähtiharju et al., 2012; Chen et al., 2018), and the rise of rayless capitula is always linked to a reduction in the expression or gene loss of one or more CYC2 members (Chen et al., 2018). Our recent analyses demonstrated that dysfunction of CYC2g led to a shift from the radiate flowerhead to the disciform head in Chrysanthemum-group (Shen et al., 2021). Therefore, the radiate should be the ancestral state of the capitulum in subtribe Artemisiinae, despite Stilpnolepis centiflora at its basal position possessing discoid flowerheads (probably an autoapomorphic state). The shifts from radiate to disciform capitula may have happened repeatedly during the diversification of this group. Early on, in 1961, Tzvelev considered that Ajania and Artemisia were convergent lineages evolved independently from Chrysanthemum-like ancestors. Similarly, our phylogenetic tree suggested that convergent capitula evolution may also have occurred within the Artemisia-group (Figure 1). For example, the ‘disciform’ capitula in Artemisia look similar with, but may have different developmental underpinnings from that of Ajania. Therefore, the contradiction between the reconstructed ancestral state of capitulum of subtribe Artemisiinae (Sanz et al., 2008) and the evo-devo insights is probably due to the morphological convergence.
To date, there have been no data demonstrating whether there is a transition from the discoid to the disciform capitula. During our previous morphogenetic study (Ren and Guo, 2015), a uniformly acropetal developmental sequence was found on the homogamous discoid capitulum, but basipetal development probably occurred in the marginal florets of the heterogamous radiate/disciform flowerhead. This was also postulated by Harris (1995). Although functional assays of chrysanthemum ClCYC2g and sunflower HaCYC2c showed that downregulating the expression of these genes led to the appearance of dorsal petals and stamens in ray florets, we assume that the developmental process of the discoid capitulum might be completely different from that of the radiate/disciform due to lacking a module responsible for the development of zygomorphic or asymmetric marginal female flowers, namely, a marginality identity module (Pozner et al., 2012; Zhao et al., 2016; Elomaa et al., 2018; Zoulias et al., 2019; Shen et al., 2021). The finding of our previous study that the CYC2g gene was lost in the discoid species Stilpnolepis centiflora (Chen et al., 2018; Shen et al., 2021) may be a hint for this hypothesis. Certainly, to test this hypothesis, more detailed molecular developmental studies are required.
Flowerhead Morphology Associated With Climatic Niche Divergence as a Driver of Species Diversification of Chrysanthemum-Group
As mentioned above, the relationship between Chrysanthemum and Ajania and their patterns of diversification and evolution have remained highly controversial. Considering their distinct capitulum morphology and geographic distribution, with members of Ajania being more western than Chrysanthemum, and members of Ch. indicum-complex being more southeastern than Ch. zawadskii-complex (Figure 4 and Supplementary Figure 5), we postulate that the three main taxa must have experienced morphological differentiation under niche divergence.
From the environmental aspect, different landscape features and climate patterns can drive directional selection (Sobel et al., 2010; Rundle and Nosil, 2012). According to ∗BEAST analysis, separation between Chrysanthemum-group and Artemisia-group occurred around the late Miocene (ca. 9 Ma, Figure 1), and the radiation within Chrysanthemum-group was dated to ca. 2 Ma. The time frame largely corresponded to the period of the profound climatic change in Asia interior, i.e., extremely intensive aridification in Middle to Central Asia demonstrated by paleoenvironmental data on loess and clay deposits (An et al., 2001; Ma et al., 2005; Nie et al., 2014; Wang X. et al., 2016), which may be due to the synergy of recently geological activity of Northern QTP (Miao et al., 2012; Favre et al., 2015; Shi et al., 2019; Yang et al., 2019). Our DIVA analysis also indicated the origin of Chrysanthemum-group in Central Asia, followed by an eastward dispersal of the Chrysanthemum lineage, and in situ diversification or colonization of the Ajania lineage along with tectonic activities of QTP (Table 3 and Supplementary Figure 2; Chen et al., 2020). Thus, the late Miocene origin and evolution of Chrysanthemum-group was strongly influenced by the establishment and development of climatic heterogeneity in Central-East Asia.
In Asteraceae, alterations of capitulum forms are closely associated with shifts in reproductive strategies, and therefore, variations in capitulum architecture should be consequences of trade-offs between energy consumption and adaptation in response to environmental stresses (Stuessy et al., 1986; Berry and Calvo, 1989; Andersson, 1999; Nielsen et al., 2002; Chen et al., 2020). Our previous analyses have demonstrated that the radiate flowerhead with white ligules (of Chrysanthemum zawadskii-complex) represents an earlier evolutionary status, and the disciform flowerhead of Ajania evolved from the radiate at approximately 8 Ma, somewhat overlapping with the time when Chrysanthemum-group arose but occurring much earlier than the divergence time of Chrysanthemum and Ajania (Figure 1 and Supplementary Figure 7). This suggests that capitulum polymorphism had long existed in the ancestral populations of the Chrysanthemum-group. Comparing the present multilocus coalescent tree and the tree based on CYC2g sequences of Chrysanthemum-group (Shen et al., 2021), we found a high consensus of topologies, implying that the evolution of this group is linked to the alteration of flowerhead morphology (Supplementary Figure 7). Within Chrysanthemum-group, low pairwise niche overlaps were found between subclades, although they diverged rather recently (Figures 4, 5). The ecological suitability diagrams based on the current climate scenario suggested that the Ajania lineage preferred environments with lower yearly mean temperatures and precipitation levels, together with higher climatic instability, compared to the two Chrysanthemum lineages, which is consistent with the habitat shift between the plants being related to anemophily and entomophily (Culley et al., 2002) and is concordant with the good differentiation of their capitulum morphologies (Figure 5).
Shown in Figure 2, three Korea-Japan distributed Ajania species, A. pallasiana_JJ, A. pacifica, and A. shiwogiku, fell into the clade of Ch. indicum-complex. This could be due to incomplete lineage sorting of genes analyzed here. We found that the sequences of their CYC2g genes were highly similar to those of A. parviflora (Shen et al., 2021; Supplementary Figure 7) which is a member distributed in the eastern part of the range of Ajania. However, the mix-up of three Ajania species with Chrysanthemum is more likely due to secondary contacts as we found all three species are polyploids with high ploidy levels from 2n = 6x to 8x and to 10x, suggesting complicated origins probably involving parentages from Ch. indicum-complex. The present SDM analysis suggested a broader geological overlap between Ajania and Chrysanthemum during the LGM (Figure 4), implying possible introgression between the two recently diverging lineages because morphological differentiation and reproductive barriers between them might still be incomplete (Comes and Kadereit, 1998; Potts et al., 2003; Hewitt, 2004; Li et al., 2013; Chen et al., 2020).
Overall, considering the divergence time, ancestral distribution, and niche differentiation data, we propose that the divergence of three major groups in Chrysanthemum-group, Ajania, Ch. indicum-complex, and Ch. zawadskii-complex, probably occurred in a rather short time span in response to environmental heterogeneity in interior East Asia, which might be related to divergence of capitulum types.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
C-ZS conceived the study, performed the most of the experiments, and data analyses. C-JZ and JC participated in the experiments. C-ZS and Y-PG wrote the manuscript. All the authors have read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31670221), the China Postdoctoral Science Foundation (Grant No. 2019M660517), and the Support Program for Postdoctoral Innovative Talent (Grant No. BX20190043).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Prof. Hui-En Zhao at BJFU (Beijing Forestry University) for providing the living plant materials of Nipponanthemum nipponicum, Leucanthemella linearis, Brachanthemum pulvinatum, Opisthopappus longilobus, and Jian-Qiang Zhang at SNU (Shaanxi Normal University) for collecting specimens of Tanacetum tatsienense. We are grateful to Zi-Meng Wang and Qian-Yuan Liu for their technical assistance in performing R and ArcGIS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.648026/full#supplementary-material
Supplementary Figure 1 | Bayesian trees of the single-copy nuclear genes.
Supplementary Figure 2 | Historical biogeographical analyses of subtribe Artemisiinae.
Supplementary Figure 3 | Time frame comparison among four different calibration settings.
Supplementary Figure 4 | Principal component analysis (PCA) of environmental factors of the Chrysanthemum-group.
Supplementary Figure 5 | Potential suitability distribution of climate conditions of major lineages within the Chrysanthemum-group.
Supplementary Figure 6 | Ancestral state reconstruction of capitulum architectures based on the phylogenetic framework under likelihood model (right) and continuous-time Markov model (left).
Supplementary Figure 7 | Comparison of the gene tree of CYC2g that regulates capitulum architectures in subtribe Artemisiinae and the present multilocus phylogeny (left, redrawn from Figures 1, 2 in the main text).
Supplementary Table 1 | Species used in the present study and the sampling information.
Supplementary Table 2 | GPS coordinates used for niche modeling in this study.
Supplementary Table 3 | Biogeographic history of subtribe Artemisiinae reconstructed with DEC and s-DEC.
Supplementary Table 4 | Pairwise niche overlap among three subclades of the Chrysanthemum-group under predicted niche occupancy profiles (PNO).
Supplementary Data Sheet 1 | Datasets of the sequence alignments.
Footnotes
- ^ www.jelitto.com
- ^ https://mafft.cbrc.jp/alignment/server/
- ^ http://www.phylo.org/
- ^ https://www.worldclim.org
- ^ https://www.gbif.org
- ^ http://www.nsii.org.cn/2017/home.php
References
Allouche, O., Tsoar, A., and Kadmon, R. (2006). Assessing the accuracy of species distribution models: prevalence, kappa, and the true skill statistic (TSS). J. Appl. Ecol. 43, 1223–1232.
An, Z. S., Kutzbach, J. E., Prell, W. L., and Porter, S. C. (2001). Evolution of Asian monsoons and phased uplift of the Himalaya-Tibetan plateau since Late Miocene times. Nature 411, 62–66. doi: 10.1038/35075035
Anacker, B. L., and Strauss, S. Y. (2014). The geography and ecology of plant speciation: range overlap and niche divergence in sister species. Proc. R. Soc. B Biol. Sci. 281:20132980. doi: 10.1098/rspb.2013.2980
Andersson, S. (1999). The cost of floral attractants in Achillea ptarmica (Asteraceae): evidence from a ray removal experiment. Plant Biol. 1, 569–572. doi: 10.1055/s-2007-978554
Annadana, S., Beekwilder, M. J., Kuipers, G., Visser, P. B., and Jongsma, M. A. (2002). Cloning of the chrysanthemum UEP1 promoter and comparative expression in florets and leaves of Dendranthema grandiflora. Transgenic Res. 11, 437–445. doi: 10.1023/A:1016313924844
Berry, P. E., and Calvo, R. N. (1989). Wind pollination, self-incompatibility, and altitudinal shifts in pollination systems in the high andean genus Espeletia (Asteraceae). Am. J. Bot. 76, 1602–1614. doi: 10.2307/2444398
Bremer, K., and Humphries, C. (1993). Generic monograph of the Asteraceae-Anthemideae. Bull. Nat. History Museum (Bot.) 23, 71–177.
Broholm, S. K., Tähtiharju, S., Laitinen, R. A., Albert, V. A., Teeri, T. H., and Elomaa, P. (2008). A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. U.S.A. 105, 9117–9122. doi: 10.1073/pnas.0801359105
Chapman, M. A., Leebens-Mack, J. H., and Burke, J. M. (2008). Positive selection and expression divergence following gene duplication in the sunflower CYCLOIDEA gene family. Mol. Biol. Evol. 25, 1260–1273. doi: 10.1093/molbev/msn001
Chapman, M. A., Tang, S., Draeger, D., Nambeesan, S., Shaffer, H., Barb, J. G., et al. (2012). Genetic analysis of floral symmetry in Van Gogh’s sunflowers reveals independent recruitment of CYCLOIDEA genes in the Asteraceae. PLoS Genet. 8:e1002628. doi: 10.1371/journal.pgen.1002628
Chen, J., Shen, C. Z., Guo, Y. P., and Rao, G. Y. (2018). Patterning the Asteraceae capitulum: duplications and differential expression of the flower symmetry CYC2-like genes. Front. Plant Sci. 9:551. doi: 10.3389/fpls.2018.00551
Chen, X., Wang, H. B., Yang, X., Jiang, J. F., Ren, G. P., Wang, Z. J., et al. (2020). Small-scale alpine topography at low latitudes and high altitudes: refuge areas of the genus Chrysanthemum and its allies. Hortic. Res. 7, 1–12. doi: 10.1038/s41438-020-00407-9
Comes, H. P., and Kadereit, J. W. (1998). The effect of Quaternary climatic changes on plant distribution and evolution. Trends Plant Sci. 3, 432–438. doi: 10.1016/S1360-1385(98)01327-2
Cowan, P. J. (2007). Geographic usage of the terms Middle Asia and Central Asia. J. Arid Environ. 69, 359–363. doi: 10.1016/j.jaridenv.2006.09.013
Culley, T. M., Weller, S. G., and Sakai, A. K. (2002). The evolution of wind pollination in angiosperms. Trends Ecol. Evol. 17, 361–369. doi: 10.1016/S0169-5347(02)02540-5
Deng, W. K., Wang, Y. B., Liu, Z. X., Cheng, H., and Xue, Y. (2014). HemI: a toolkit for illustrating heatmaps. PLoS One 9:e111988. doi: 10.1371/journal.pone.0111988
Ding, W. N., Ree, R. H., Spicer, R. A., and Xing, Y. W. (2020). Ancient orogenic and monsoon-driven assembly of the world’s richest temperate alpine flora. Science 369, 578–581. doi: 10.1126/science.abb4484
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012a). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Drummond, A. J., Xie, W., and Heled, J. (2012b). Bayesian inference of species trees from multilocus data using ∗BEAST. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/msp274
Ebersbach, J., Muellner-Riehl, A. N., Favre, A., Paule, J., Winterfeld, G., and Schnitzler, J. (2018). Driving forces behind evolutionary radiations: Saxifraga section Ciliatae (Saxifragaceae) in the region of the Qinghai-Tibet Plateau. Bot. J. Linn. Soc. 186, 304–320. doi: 10.1093/botlinnean/box100
Edwards, S. V., Xi, Z., Janke, A., Faircloth, B. C., Mccormack, J. E., Glenn, T. C., et al. (2016). Implementing and testing the multispecies coalescent model: a valuable paradigm for phylogenomics. Mol. Phylogenet. Evol. 94, 447–462. doi: 10.1016/j.ympev.2015.10.027
Elomaa, P., Zhao, Y. F., and Zhang, T. (2018). Flower heads in Asteraceae—recruitment of conserved developmental regulators to control the flower-like inflorescence architecture. Hortic. Res. 5, 1–10. doi: 10.1038/s41438-018-0056-8
Evans, M. E. K., Smith, S. A., Flynn, R. S., and Donoghue, M. J. (2009). Climate, niche evolution, and diversification of the “bird-cage” evening primroses (Oenothera, sections Anogra and Kleinia). Am. Natural. 173, 225–240. doi: 10.1086/595757
Favre, A., Päckert, M., Pauls, S. U., Jähnig, S. C., Uhl, D., Michalak, I., et al. (2015). The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. 90, 236–253. doi: 10.1111/brv.12107
Fielding, A. H., and Bell, J. F. (1997). A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 24, 38–49.
Ford, V., and Gottlieb, L. (1990). Genetic studies of floral evolution in Layia. Heredity 64, 29–44. doi: 10.1038/hdy.1990.5
Ge, J. Y., Dai, Y., Zhang, Z. S., Zhao, D. A., Li, Q., Zhang, Y., et al. (2013). Major changes in East Asian climate in the mid-Pliocene: triggered by the uplift of the Tibetan Plateau or global cooling? J. Asian Earth Sci. 69, 48–59. doi: 10.1016/j.jseaes.2012.10.009
Guo, Z. T., Peng, S. Z., Hao, Q. Z., Biscaye, P. E., An, Z. S., and Liu, T. S. (2004). Late Miocene–Pliocene development of Asian aridification as recorded in the Red-Earth Formation in northern China. Global Planet. Change 41, 135–145. doi: 10.1016/j.gloplacha.2004.01.002
Griffith, O. W., Blackburn, D. G., Brandley, M. C., Van Dyke, J. U., Whittington, C. M., and Thompson, M. B. (2015). Ancestral state reconstructions require biological evidence to test evolutionary hypotheses: a case study examining the evolution of reproductive mode in squamate reptiles. J. Exp. Zool. 324, 493–503. doi: 10.1002/jez.b.22614
Hansen, J., Sato, M., Russell, G., and Kharecha, P. (2013). Climate sensitivity, sea level and atmospheric carbon dioxide. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 371:20120294. doi: 10.1098/rsta.2012.0294
Harris, E. M. (1995). Inflorescence and floral ontogeny in Asteraceae: a synthesis of historical and current concepts. Bot. Rev. 61, 93–278. doi: 10.1007/BF02887192
Harris, E. M. (1999). Capitula in the Asteridae: a widespread and varied phenomenon. Bot. Rev. 65, 348–369. doi: 10.1007/BF02857754
He, K., Li, Y. J., Brandley, M. C., Lin, L. K., and Jiang, X. L. (2010). A multi-locus phylogeny of Nectogalini shrews and influences of the paleoclimate on speciation and evolution. Mol. Phylogenet. Evol. 56, 734–746. doi: 10.1016/j.ympev.2010.03.039
Heibl, C., and Calenge, C. (2013). Phyloclim: Integrating Phylogenetics and Climatic Niche Modelling. R Package Version 0.9, 4.
Hewitt, G. M. (2004). Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. B Biol. Sci. 359, 183–195. doi: 10.1098/rstb.2003.1388
Hijmans, R., Cruz, M., Rojas, E., and Guarino, L. (2001). DIVA-GIS version 1.4: A Geographic Information System for the Analysis of Biodiversity Data—Manual. Lima: International Potato Center, 73.
Hu, X., and Zhao, H. E. (2008). A preliminary study on distant hybridization of Opisthopappus with Chrysanthemum and Ajania. Modern Agric. Sci. 15, 13–14.
Huang, Y., An, Y. M., Meng, S. Y., Guo, Y. P., and Rao, G. Y. (2017). Taxonomic status and phylogenetic position of Phaeostigma in the subtribe Artemisiinae (Asteraceae). J. Syst. Evol. 55, 426–436. doi: 10.1111/jse.12257
Kim, M., Cui, M. L., Cubas, P., Gillies, A., Lee, K., Chapman, M. A., et al. (2008). Regulatory genes control a key morphological and ecological trait transferred between species. Science 322, 1116–1119. doi: 10.1126/science.1164371
Kim, S. J., Lee, C. H., Kim, J., and Kim, K. S. (2014). Phylogenetic analysis of Korean native Chrysanthemum species based on morphological characteristics. Sci. Hortic. 175, 278–289. doi: 10.1016/j.scienta.2014.06.018
Kishimoto, S., Aida, R., and Shibata, M. (2003). Identification of chloroplast DNA variations by PCR-RFLP analysis in Dendranthema. J. Jpn. Soc. Hortic. Sci. 72, 197–204. doi: 10.2503/jjshs.72.197
Kitamura, S. (1978). Dendranthema et Nipponanthemum. Acta Phytotaxon. Geobot. 29, 165–170. doi: 10.18942/bunruichiri.KJ00001078257
Knope, M. L., Funk, V. A., Johnson, M. A., Wagner, W. L., Datlof, E. M., Johnson, G., et al. (2020). Dispersal and adaptive radiation of Bidens (Compositae) across the remote archipelagoes of Polynesia. J. Syst. Evol. 58, 805–822. doi: 10.1111/jse.12704
Kondo, K., Abd El Twab, M., Idesawa, R., Kimura, S., and Tanaka, R. (2003). Genome phylogenetics in Chrysanthemum sensu lato. Plant Genome Biodiv. Evol. 1, 117–200.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, Y. N. (1969). A cytotaxonomic study on Chrysanthemum zawadskii complex in Korea polyploidy. J. Plant Biol. 12, 35–48.
Landis, J. R., and Koch, G. G. (1977). The measurement of observer agreement for categorical data. Biometrics 33, 159–174.
Li, J., Wan, Q., Abbott, R. J., and Rao, G. Y. (2013). Geographical distribution of cytotypes in the Chrysanthemum indicum complex as evidenced by ploidy level and genome-size variation. J. Syst. Evol. 51, 196–204. doi: 10.1111/j.1759-6831.2012.00241.x
Li, J., Wan, Q., Guo, Y. P., Abbott, R. J., and Rao, G. Y. (2014). Should I stay or should I go: biogeographic and evolutionary history of a polyploid complex (Chrysanthemum indicum complex) in response to Pleistocene climate change in China. New Phytol. 201, 1031–1044. doi: 10.1111/nph.12585
Li, J. J., Zhou, S. Z., Zhou, Z. J., and Zhang, J. (2015). The Qingzang Movement: the major uplift of the Qinghai-Tibetan Plateau. Sci. China Earth Sci. 58, 2113–2122.
Liu, C., Berry, P. M., Dawson, T. P., and Pearson, R. G. (2005). Selecting thresholds of occurrence in the prediction of species distributions. Ecography 28, 385–393.
Liu, L., Wu, S. Y., and Yu, L. L. (2015). Coalescent methods for estimating species trees from phylogenomic data. J. Syst. Evol. 53, 380–390. doi: 10.1111/jse.12160
Liu, P. L., Wan, J. N., Guo, Y. P., Ge, S., and Rao, G. Y. (2012a). Adaptive evolution of the chrysanthemyl diphosphate synthase gene involved in irregular monoterpene metabolism. BMC Evol. Biol. 12:214. doi: 10.1186/1471-2148-12-214
Liu, P. L., Wan, Q., Guo, Y. P., Yang, J., and Rao, G. Y. (2012b). Phylogeny of the genus Chrysanthemum L.: evidence from single-copy nuclear gene and chloroplast DNA sequences. PLoS One 7:e48970. doi: 10.1371/journal.pone.0048970
Lu, L. M., Mao, L. F., Yang, T., Ye, J. F., Liu, B., Li, H. L., et al. (2018). Evolutionary history of the angiosperm flora of China. Nature 554, 234–238. doi: 10.1038/nature25485
Ma, Y. P., Chen, M. M., Wei, J. X., Zhao, L., Liu, P. L., Dai, S. L., et al. (2016). Origin of Chrysanthemum cultivars — Evidence from nuclear low-copy LFY gene sequences. Biochem. Syst. Ecol. 65, 129–136. doi: 10.1016/j.bse.2016.02.010
Ma, Y. Z., Wu, F. L., Fan, X. M., Li, J. J., An, Z. S., and Wang, W. (2005). Pollen record from red clay sequence in the central Loess Plateau between 8.1 and 2.6 Ma. Chin. Sci. Bull. 50, 2234–2243.
Maddison, W. P., and Maddison, D. R. V. (2009). MESQUITE: a modular system for evolutionary analysis. Evolution 11, 1103–1118.
Martin, J., Torrell, M., and Valles, J. (2001). Palynological features as a systematic marker in Artemisia L. and related genera (Asteraceae, Anthemideae). Plant Biol. 3, 372–378.
Masuda, Y., Yukawa, T., and Kondo, K. (2009). Molecular phylogenetic analysis of members of Chrysanthemum and its related genera in the tribe Anthemideae, the Asteraceae in East Asia on the basis of the internal transcribed spacer (ITS) region and the external transcribed spacer (ETS) region of nrDNA. Chrom. Bot. 4, 25–36. doi: 10.3199/iscb.4.25
Meng, S. Y., Wu, L., and Shen, C. Z. (2020). Chrysanthemum bizarre, a new species of Chrysanthemum from Hunan, China. Phytotaxa 442, 215–224. doi: 10.11646/phytotaxa.442.3.7
Miao, Y., Herrmann, M., Wu, F., Yan, X., and Yang, S. (2012). What controlled mid–late miocene long-term aridification in Central Asia? — Global cooling or Tibetan Plateau uplift: a review. Earth Sci. Rev. 112, 155–172. doi: 10.1016/j.earscirev.2012.02.003
Miao, Y., Meng, Q., Fang, X., Yan, X., Wu, F., and Song, C. (2011). Origin and development of Artemisia (Asteraceae) in Asia and its implications for the uplift history of the Tibetan plateau: a review. Quat. Int. 236, 3–12.
Muldashev, A. (1981). Novyi rod Phaeostigma (Asteraceae) iz vostochnoi Azii. Botanischeskii Zhurnal 66, 584–588.
Muldashev, A. (1983). A critical review of the genus Ajania (Asteraceae-Anthemideae). Botanischeskii Zhurnal 68, 207–214.
Nie, J., Peng, W., Andreas, M., Song, Y., Stockli, D. F., Stevens, T., et al. (2014). Provenance of the upper miocene–pliocene red clay deposits of the chinese loess plateau. Earth Planet. Sci. Lett. 407, 35–47.
Nielsen, L. R., Philipp, M., and Siegismund, H. R. (2002). Selective advantage of ray florets in Scalesia affinis and S. pedunculata (Asteraceae), two endemic species from the Galápagos. Evol. Ecol. 16, 139–153. doi: 10.1023/A:1016301027929
Oberprieler, C. (2005). Temporal and spatial diversification of circum-Mediterranean Compositae-Anthemideae. Taxon 54, 951–966.
Oberprieler, C., Himmelreich, S., Källersjö, M., Vallès, J., Watson, L. E., and Vogt, R. (2009). “Tribe Anthemideae Cass,” in Systematics, Evolution, and Biogeography of Compositae, eds V. A. Funk, A. Susanna, T. F. Stuessy, and R. J. Bayer (Washington, DC: International Association for Plant Taxonomy), 631–666. doi: 10.1006/mpev.1999.0635
Oberprieler, C., Himmelreich, S., and Vogt, R. (2007). A new subtribal classification of the tribe Anthemideae (Compositae). Willdenowia 37, 89–114. doi: 10.3372/wi.37.37104
Oberprieler, C., Vogt, R., and Watson, L. E. (2006). “Tribe Anthemideae Cass. (1819),” in The Families and Genera of Vascular Plants 8, eds J. W. Kadereit and C. Jeffrey (Berlin: Springer), 342–374.
Ohashi, H., and Yonekura, K. (2004). New combinations in Chrysanthemum (Compositae-Anthemideae) of Asia with a list of Japanese species. J. Japanese Bot. 79, 186–195.
Omland, K. E. (1999). The assumptions and challenges of ancestral state reconstructions. Syst. Biol. 3, 604–611. doi: 10.1080/106351599260175
Ortiz-Rodriguez, A. E., Ornelas, J. F., and Ruiz-Sanchez, E. (2018). A jungle tale: molecular phylogeny and divergence time estimates of the Desmopsis-Stenanona clade (Annonaceae) in Mesoamerica. Mol. Phylogenet. Evol. 122, 80–94. doi: 10.1016/j.ympev.2018.01.021
Papeş, M., and Gaubert, P. (2007). Modelling ecological niches from low numbers of occurrences: assessment of the conservation status of poorly known viverrids (Mammalia, Carnivora) across two continents. Divers. Distrib. 13, 890–902. doi: 10.1111/j.1472-4642.2007.00392.x
Paradis, E., and Schliep, K. (2019). ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528. doi: 10.1093/bioinformatics/bty633
Pellicer, J., Hidalgo, O., Garcia, S., Garnatje, T., Korobkov, A. A., Vallcs, J., et al. (2010). Palynological study of Ajania and related genera (Asteraceae, Anthemideae). Bot. J. Linn. Soc. 161, 171–189. doi: 10.1111/j.1095-8339.2009.00998.x
Pennell, M. W., Eastman, J. M., Slater, G. J., Brown, J. W., Uyeda, J. C., Fitzjohn, R. G., et al. (2014). geiger v2.0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 30, 2216–2218. doi: 10.1093/bioinformatics/btu181
Phillips, S. J., Anderson, R. P., and Schapire, R. E. (2006). Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259. doi: 10.1016/j.ecolmodel.2005.03.026
Phillips, S. J., and Dudík, M. (2008). Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31, 161–175. doi: 10.1111/j.0906-7590.2008.5203.x
Posada, D. (2008). jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Potts, B., Barbour, R., Hingston, A., and Vaillancourt, R. (2003). Genetic pollution of native eucalypt gene pools—identifying the risks. Aust. J. Bot. 51, 1–25. doi: 10.1071/bt02035
Pozner, R., Zanotti, C., and Johnson, L. A. (2012). Evolutionary origin of the Asteraceae capitulum: insights from Calyceraceae. Am. J. Bot. 99, 1–13. doi: 10.3732/ajb.1100256
Qin, A. L., Wang, M. M., Cun, Y. Z., Yang, F. S., Wang, S. S., Ran, J. H., et al. (2013). Phylogeographic evidence for a link of species divergence of Ephedra in the Qinghai-Tibetan Plateau and adjacent regions to the Miocene Asian aridification. PLoS One 8:e56243.
R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67:901. doi: 10.1093/sysbio/syy032
Ren, J. B., and Guo, Y. P. (2015). Behind the diversity: ontogenies of radiate, disciform, and discoid capitula of Chrysanthemum and its allies. J. Syst. Evol. 53, 520–528. doi: 10.1111/jse.12154
Revell, L. J. (2012). phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. doi: 10.1111/j.2041-210X.2011.00169.x
Rivera, S. B., Swedlund, B. D., King, G. J., Bell, R. N., Hussey, C. E., Shattuck-Eidens, D. M., et al. (2001). Chrysanthemyl diphosphate synthase: isolation of the gene and characterization of the recombinant non-head-to-tail monoterpene synthase from Chrysanthemum cinerariaefolium. Proc. Natl. Acad. Sci. U.S.A. 98, 4373–4378. doi: 10.1073/pnas.071543598
Rundle, H. D., and Nosil, P. (2010). Ecological speciation. Ecol. Lett. 8, 336–352. doi: 10.1111/j.1461-0248.2004.00715.x
Sanz, M., Vilatersana, R., Hidalgo, O., Garcia-Jacas, N., Susanna, A., Schneeweiss, G. M., et al. (2008). Molecular phylogeny and evolution of floral characters of Artemisia and allies (Anthemideae, Asteraceae): evidence from nrDNA ETS and ITS sequences. Taxon 57, 66–78.
Schoener, T. W. (1970). Nonsynchronous spatial overlap of lizards in patchy habitats. Ecology 51, 408–418. doi: 10.2307/1935376
Sha, S., Chen, D., Liu, M., Li, K. L., Jiang, C. K., Wang, D. H., et al. (2017). To be serrate or pinnate: diverse leaf forms of yarrows (Achillea) are linked to differential expression patterns of NAM genes. Ann. Bot. 121, 255–266. doi: 10.1093/aob/mcx152
Shen, C. Z., Chen, J., Zhang, C. J., Rao, G. Y., and Guo, Y. P. (2021). Dysfunction of CYC2g is responsible for evolutionary shift from radiate to disciform flowerhead in the Chrysanthemum group (Asteraceae: Anthemideae). Plant J. doi: 10.1111/tpj.15216
Shi, Z. G., Liu, X. D., and Sha, Y. Y. (2015). Did northern Tibetan Plateau uplift during Pliocene? A modeling test. J. Earth Environ. 6, 67–80. doi: 10.7515/JEE201502001
Shi, Z., Sha, Y., Liu, X., Xie, X., and Li, X. (2019). Effect of marginal topography around the Tibetan Plateau on the evolution of Central Asian arid climate: Yunnan–Guizhou and Mongolian Plateaux as examples. Clim. Dynamics 53, 4433–4445.
Shih, C., and Fu, G. (1983). Angiospermae, Dicotyledoneae, Compositae (3) Anthemideae, Angiospermae. Flora Republicae Popularis Sinicae 76. Beijing: Science Press.
Shimizu, T. (1961). Cytogeographical notes on Chrysanthemum zawadskii Herb. and its allies. J. Japanese Bot. 36, 176–180.
Shishkin, B. K., and Bobrov, E.G. eds (1995). Flora of the USSR Vol. XXVI., Leningrad: Akademiya Nauk.
Sobel, J. M., Chen, G. F., Watt, L. R., and Schemske, D. W. (2010). The biology of speciation. Evol. Int. J. Org. Evol. 64, 295–315. doi: 10.1111/j.1558-5646.2009.00877.x
Sonboli, A., Stroka, K., Osaloo, S. K., and Oberprieler, C. (2012). Molecular phylogeny and taxonomy of Tanacetum L. (Compositae, Anthemideae) inferred from nrDNA ITS and cpDNA trnH-psbA sequence variation. Plant Syst. Evol. 298, 431–444. doi: 10.1007/s00606-011-0556-6
Song, C., Liu, Y., Song, A., Dong, G., Zhao, H., Sun, W., et al. (2018). The Chrysanthemum nankingense genome provides insights into the evolution and diversification of chrysanthemum flowers and medicinal traits. Mol. Plant 11, 1482–1491. doi: 10.1016/j.molp.2018.10.003
Spencer, J. E., and Robinson, H. (1968). Monsoon Asia: a geographical survey. Pacific Aff. 41, 545–552. doi: 10.2307/2755008
Spicer, R. A. (2017). Tibet, the himalaya, asian monsoons and biodiversity-in what ways are they related? Plant Divers. 39, 5–16. doi: 10.1016/j.pld.2017.09.001
Stuessy, T. F., Spooner, D. M., and Evans, K. A. (1986). Adaptive significance of ray corollas in Helianthus grosseserratus (Compositae). Am. Midland Natural. 115, 191–197. doi: 10.2307/2425849
Tähtiharju, S., Rijpkema, A. S., Vetterli, A., Albert, V. A., Teeri, T. H., and Elomaa, P. (2012). Evolution and diversification of the CYC/TB1 gene family in Asteraceae—a comparative study in Gerbera (Mutisieae) and sunflower (Heliantheae). Mol. Biol. Evol. 29, 1155–1166. doi: 10.1093/molbev/msr283
Tomasello, S., Álvarez, I., Vargas, P., and Oberprieler, C. (2015). Is the extremely rare Iberian endemic plant species Castrilanthemum debeauxii (Compositae, Anthemideae) a “living fossil”? Evidence from a multi-locus species tree reconstruction. Mol. Phylogenet. Evol. 82, 118–130. doi: 10.1016/j.ympev.2014.09.007
Torrell, M., Garcia-Jacas, N., Susanna, A., and Vallès, J. (1999). Phylogeny in Artemisia (Asteraceae, Anthemideae) inferred from nuclear ribosomal DNA (ITS) sequences. Taxon 48, 721–736. doi: 10.2307/1223643
Trow, A. (1912). On the inheritance of certain characters in the common groundsel—Senecio vulgaris, Linn.—and its segregates. J. Genet. 2, 239–276. doi: 10.1007/BF01876701
Vallès, J., and McArthur, E. D. (2001). “Artemisia systematics and phylogeny: cytogenetic and molecular insights,” in Proceedings of the Shrubland Ecosystem Genetics and Biodiversity, eds E. McArthur, F. Durant, and J. Daniel (Washington, D.C: US Department of Agriculture), 67–74.
Vallès, J., Torrell, M., Garnatje, T., Garcia-Jacas, N., Vilatersana, R., and Susanna, A. (2003). The genus Artemisia and its allies: phylogeny of the subtribe Artemisiinae (Asteraceae, Anthemideae) based on nucleotide sequences of nuclear ribosomal DNA internal transcribed spacers (ITS). Plant Biol. 5, 274–284. doi: 10.1055/s-2003-40790
Wall, D. P., and Deluca, T. (2007). “Ortholog detection using the reciprocal smallest distance algorithm,” in Comparative Genomics, ed. N. H. Bergman (New Jersey: Humana Press), 95–110.
Wang, M., Le Moigne, M. A., Bertheloot, J., Crespel, L., Perez-Garcia, M. D., Ogé, L., et al. (2019). BRANCHED1: a key hub of shoot branching. Front. Plant Sci. 10:76. doi: 10.3389/fpls.2019.00076
Wang, W. M., and Zhang, D. H. (1990). Tertiary sporo-pollen assemblages from the shangdou-huade basin, inner mongoliad with discussion on the formation of steppe vegetation in China. Acta Micropalaeontol. Sin. 7, 239–252.
Wang, X., Wei, H., Taheri, M., Khormali, F., Danukalova, G., and Chen, F. (2016). Early Pleistocene climate in western arid central Asia inferred from loess-palaeosol sequences. Sci. Rep. 6:20560. doi: 10.1038/srep20560
Wang, Z. M., Meng, S. Y., and Rao, G. Y. (2019). Quaternary climate change and habitat preference shaped the genetic differentiation and phylogeography of Rhodiola sect. Prainia in the southern Qinghai-Tibetan Plateau. Ecol. Evol. 9, 8305–8319. doi: 10.1002/ece3.5406
Warren, D. L., Glor, R. E., and Turelli, M. (2008). Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution 62, 2868–2883. doi: 10.1111/j.1558-5646.2008.00482.x
Warren, D. L., Glor, R. E., and Turelli, M. (2010). ENMTools: a toolbox for comparative studies of environmental niche models. Ecography 33, 607–611. doi: 10.1111/j.1600-0587.2009.06142.x
Watson, L. E., Bates, P. L., Evans, T. M., Unwin, M. M., and Estes, J. R. (2002). Molecular phylogeny of subtribe Artemisiinae (Asteraceae), including Artemisia and its allied and segregate genera. BMC Evol. Biol. 2:17. doi: 10.1186/1471-2148-2-17
Watson, L. E., Evans, T. M., and Boluarte, T. (2000). Molecular phylogeny and biogeography of tribe Anthemideae (Asteraceae), based on chloroplast gene ndhF. Mol. Phylogenet. Evol. 15, 59–69. doi: 10.1006/mpev.1999.0714
Wiens, J. J., and Graham, C. H. (2005). Niche conservatism: integrating evolution, ecology, and conservation biology. Annu. Rev. Ecol. Evol. Syst. 36, 519–539. doi: 10.1146/annurev.ecolsys.36.102803.095431
Xia, X. H. (2017). DAMBE6: new tools for microbial genomics, phylogenetics, and molecular evolution. J. Heredity 108, 431–437. doi: 10.1093/jhered/esx033
Xie, Y., Li, D. M., and MacKinnon, J. (2002). Preliminary researches on bio-geographical divisions of China. Acta Ecol. Sin. 22, 1599–1615. doi: 10.1007/s11769-002-0041-9
Yan, Y., Yang, X., and Tang, Z. (2013). Patterns of species diversity and phylogenetic structure of vascular plants on the Qinghai-Tibetan Plateau. Ecol. Evol. 3, 4584–4595. doi: 10.1002/ece3.847
Yang, D., Hu, X., Liu, Z., and Zhao, H. E. (2010). Intergeneric hybridizations between Opisthopappus taihangensis and Chrysanthemum lavandulifolium. Sci. Hortic. 125, 718–723. doi: 10.1016/j.scienta.2010.05.002
Yang, R., Yang, Y., Fang, X., Ruan, X., Galy, A., Ye, C. C., et al. (2019). Late miocene intensified tectonic uplift and climatic aridification on the Northeastern Tibetan Plateau: evidence from clay mineralogical and geochemical records in the Xining Basin. Geochem. Geophys. Geosyst. 20, 829–851. doi: 10.1029/2018GC007917
Yu, Y., Harris, A. J., Blair, C., and He, X. (2015). RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Mol. Phylogenet. Evol. 87, 46–49. doi: 10.1016/j.ympev.2015.03.008
Zhang, H., Xia, R., Meyers, B. C., and Walbot, V. (2015). Evolution, functions, and mysteries of plant ARGONAUTE proteins. Curr. Opin. Plant Biol. 27, 84–90. doi: 10.1016/j.pbi.2015.06.011
Zhao, H. B., Chen, F. D., Chen, S. M., Wu, G. S., and Guo, W. M. (2010a). Molecular phylogeny of Chrysanthemum, Ajania and its allies (Anthemideae, Asteraceae) as inferred from nuclear ribosomal ITS and chloroplast trnL-F IGS sequences. Plant Syst. Evol. 284, 153–169. doi: 10.1007/s00606-009-0242-0
Zhao, H. B., Miao, H. B., Wu, G. S., Chen, F. D., and Guo, W. M. (2010b). Intergeneric phylogenetic relationship of Dendranthema (DC.) Des Moul., Ajania Poljakov and their allies based on amplified fragment length polymorphism. Sci. Agric. Sin. 43, 1302–1313.
Zhao, H. E., Liu, Z. H., Hu, X., Yin, J. L., Li, W., Rao, G. Y., et al. (2009). Chrysanthemum genetic resources and related genera of Chrysanthemum collected in China. Genet. Resour. Crop Evol. 56:937. doi: 10.1007/s10722-009-9412-8
Zhao, W. P., Zhao, J., He, L., Sun, Y. H., and Cai, H. (2013). Molecular structure and the second introns variation of gene F3’H of two medicinal Chrysanthemum morifolium populations. Biochem. Syst. Ecol. 51, 251–258. doi: 10.1016/j.bse.2013.09.012
Zhao, Y. F., Zhang, T., Broholm, S. K., Tähtiharju, S., Mouhu, K., Albert, V., et al. (2016). Co-opting floral meristem identity genes for patterning of the flower-like Asteraceae inflorescence. Plant Physiol. 172, 284–296. doi: 10.1104/pp.16.00779
Zheng, H. B., Powell, C. M., An, Z. S., Zhou, J., and Dong, G. R. (2000). Pliocene uplift of the northern Tibetan Plateau. Geology 28, 715–718.
Zheng, Y., Shen, J., An, Y. M., Zhang, J. Q., Rao, G. Y., Zhang, Q. X., et al. (2013). Intergeneric hybridization between Elachanthemum intricatum (Franch.) Ling et Ling and Opisthopappus taihangensis (Y. Ling) C. Shih. Genet. Resour. Crop Evol. 60, 473–482. doi: 10.1007/s10722-012-9850-6
Zhou, X. Y., Chen, X. L., Bi, L., Ma, N., and Zhao, L. J. (2012). Isolation and expression analysis of BRANCHED1 in Dendranthema grandiflora’Jinba’. J. China Agric. Univ. 17, 100–104.
Keywords: Chrysanthemum, Ajania, coalescence, phylogeny, niche differentiation
Citation: Shen C-Z, Zhang C-J, Chen J and Guo Y-P (2021) Clarifying Recent Adaptive Diversification of the Chrysanthemum-Group on the Basis of an Updated Multilocus Phylogeny of Subtribe Artemisiinae (Asteraceae: Anthemideae). Front. Plant Sci. 12:648026. doi: 10.3389/fpls.2021.648026
Received: 31 December 2020; Accepted: 20 April 2021;
Published: 26 May 2021.
Edited by:
Nina Rønsted, National Tropical Botanical Garden, United StatesReviewed by:
Christoph Oberprieler, University of Regensburg, GermanyMario Fernández-Mazuecos, Complutense University of Madrid, Spain
Haibin Wang, Nanjing Agricultural University, China
Copyright © 2021 Shen, Zhang, Chen and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan-Ping Guo, guoyanping@bnu.edu.cn
 Chu-Ze Shen
Chu-Ze Shen Chu-Jie Zhang2
Chu-Jie Zhang2 Jie Chen
Jie Chen