- 1Shandong Provincial Key Laboratory of Plant Stress Research, College of Life Science, Shandong Normal University, Jinan, China
- 2Research Team of Plant Pathogen Microbiology and Immunology, College of Life Science, Shandong Normal University, Jinan, China
Salt cress (Eutrema salsugineum), an Arabidopsis-related halophyte, can naturally adapt to various harsh climates and soil conditions; thus, it is considered a desirable model plant for deciphering mechanisms of salt and other abiotic stresses. Accumulating evidence has revealed that compared with Arabidopsis, salt cress possesses stomata that close more tightly and more succulent leaves during extreme salt stress, a noticeably higher level of proline, inositols, sugars, and organic acids, as well as stress-associated transcripts in unstressed plants, and they are induced rapidly under stress. In this review, we systematically summarize the research on the morphology, physiology, genome, gene expression and regulation, and protein and metabolite profile of salt cress under salt stress. We emphasize the latest advances in research on the genome adaptive evolution encountering saline environments, and epigenetic regulation, and discuss the mechanisms underlying salt tolerance in salt cress. Finally, we discuss the existing questions and opportunities for future research in halophytic Eutrema. Together, the review fosters a better understanding of the mechanism of plant salt tolerance and provides a reference for the research and utilization of Eutrema as a model extremophile in the future. Furthermore, the prospects for salt cress applied to explore the mechanism of salt tolerance provide a theoretical basis to develop new strategies for agricultural biotechnology.
Introduction
Soil salinity is one of the major environmental stress factors that restrict the functioning of plants in natural ecosystems, and greatly reduce crop yields which subsequently leads to a threat to food security (Zhu, 2002; Vera-Estrella et al., 2005; Zhao et al., 2020). As we know, the enormous value will come from a better understanding of the mechanisms through which plant tolerance of abiotic stresses is achieved (Zhao et al., 2020). Most studies on salt stress response mechanisms of plants have been conducted using the glycophyte model Arabidopsis thaliana and some important clues were gained (Wu et al., 2012). However, Arabidopsis has a relatively low capacity to survive salt stress, thus it may lack the protective mechanisms required for growing in harsh environments (Lee et al., 2013). Therefore, searching for the novel stress tolerance determinants including novel genes and novel stress tolerance mechanisms has led to increasing interest in the plants native to growing in extreme environments called “extremophytes” (John and Spangenberg, 2005; Amtmann, 2009; Gechev et al., 2012; Bressan et al., 2013; Oh et al., 2013; Cheeseman, 2015; Flowers et al., 2015; Eshel et al., 2017).
Halophytes can naturally thrive under high salinity conditions, such as in marshlands, swamps, and intertidal estuarine areas, and their tolerance to salt stress may occur through various evolutionary and molecular mechanisms, which can provide crucial insights into the underlying mechanisms that endow tolerance of plants to salt stress (Flowers et al., 1986; Bressan et al., 2001; Zhu, 2002; Amtmann, 2009; Rajalakshmi and Parida, 2012; Wu et al., 2012). The crucifer Eutrema salsugineum (salt cress) formerly classified as Thellungiella salsuginea or Thellungiella halophila is a close relative of Arabidopsis. Eutrema salsugineum can naturally grow well in extreme environments and displays exceptionally high resistance to salt as well as cold, drought, and oxidative stresses, but its genetic make-up, morphology, and development are similar to the glycophyte Arabidopsis (Bressan et al., 2001; Inan et al., 2004; Taji et al., 2004; Volkov et al., 2004; Wang et al., 2004; Gong et al., 2005; Wong et al., 2006; Amtmann, 2009; Orsini et al., 2010; Wu et al., 2012; Koch and German, 2013; Figure 1). Furthermore, salt cress is an ideal model organism due to its short life cycle, self-fertility, and being genetically transformable, and has a relatively small genome (241 Mb) with approximately twice the genome size of Arabidopsis (Inan et al., 2004; Wu et al., 2012). Importantly, salt cress has the availability of extensive ecotypes exhibiting a range of stress responses to adapt to different environmental stresses (Inan et al., 2004; Wong et al., 2005; Li et al., 2006; Amtmann, 2009; Hou and Bartels, 2015). These advantages characteristics make the halophytic Eutrema species amenable to the functional genomics approaches designed to identify the novel genes and new molecular mechanisms involved in stress resistance (Lugan et al., 2010).
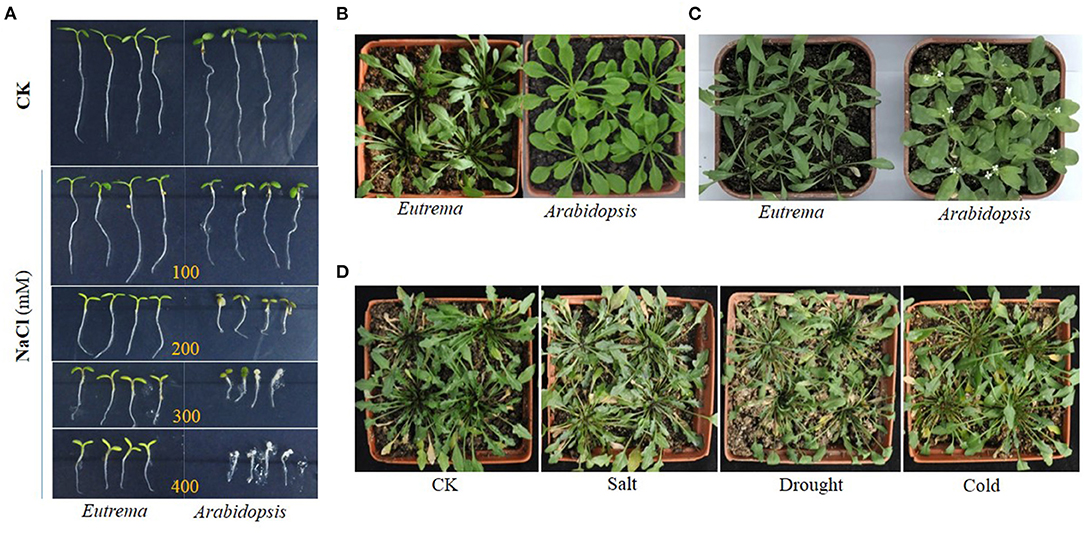
Figure 1. Growth comparison of Eutrema salsugineum (Shandong) and Arabidopsis thaliana plants in the control (CK) and stress treatment conditions. (A) Growth comparison of Eutrema and Arabidopsis plants in the control and NaCl treatment (100, 200, 300, and 400 mM NaCl) conditions. Eutrema seeds germinated 10 days, and Arabidopsis seeds germinated for 7 days in 1/2 MS medium, then the seedlings of uniform growth moved to 1/2 MS medium containing 0 (CK), 100, 200, 300, and 400 mM NaCl for growing for about 5 days. (B) Growth comparison of the 50-day-old Eutrema and 30-day-old Arabidopsis plants that grow in a mixture of vermiculite, perlite and peat moss (1:1:1), with a photoperiod of 16 h light/22°C and 8 h dark/18°C. (C) Growth comparison of the 60-day-old Eutrema and 40-day-old Arabidopsis plants. The surface sterilized Eutrema seeds planted in 1/2 MS medium, was firstly stratified treatment (4°C for 13 days) to synchronize the germination, then moved into the growth chamber for 7 days; the seedlings were transplanted in nutrient soil for 34 days, followed by 6 day-salt treatment using 200 mM NaCl. The surface sterilized Arabidopsis seeds planted in 1/2 MS medium, was firstly stratified treatment in 4°C for 3 days, then moved into a growth chamber for 7 days; the seedlings were transplanted in nutrient soil containing vermiculite, perlite, and peat moss (1:1:1) for 24 days, followed by 6-day salt treatment using 200 mM NaCl. (D) Phenotype comparison of Eutrema plants under salt, drought, and cold stresses. Eutrema seeds were planted in a mixture of vermiculite, perlite, and peat moss (1:1:1) in a greenhouse for 60 days with a photoperiod of 16 h light/22°C and 8 h dark/18°C, followed by stress treatments using 300 mM NaCl (Salt), no watering (Drought) and low temperature of 4–6°C (Cold) for 2 weeks, respectively, or kept in normal growth condition (CK).
Over the past few years, this salt cress species has come to the fore in the research into abiotic stress resistance and is considered a plant model of stress resistance well comparable with the Arabidopsis genome (Bressan et al., 2001; Inan et al., 2004; Amtmann et al., 2005; Amtmann, 2009). Firstly, the genome of salt cress provides a resource for identifying naturally occurring genetic alterations which could contribute to the adaptation of the halophyte to soil salinity, and that might be bioengineered in related crop species (Yang et al., 2013). The characterization of transcripts with unknown functions will possibly reveal the novel defense mechanisms in this halophyte (Rajalakshmi and Parida, 2012). Secondly, comparative analyses of the genome structures, protein-coding genes, microRNAs, stress-related pathways, and proteomic and metabolic profiles between Eutrema and Arabidopsis suggest that adaptation of the halophyte to environmental salt stress may occur via a global network adjustment of multiple regulatory mechanisms (Yang et al., 2013). Furthermore, the research of Eutrema is supported by growing technical resources including the physiological and molecular protocols, ecotype collections, and comparison of genome, transcriptome, and metabolome. Hence, recent scientific advances make exploration of the mechanisms of environmental adaptation in plants more feasible which could result in a larger implication and gains in genetic improvement of crops.
In this review, we systematically summarize the recent research advances in understanding morphology, physiology and biochemistry, expression and expression regulation of genes, and evaluating its usefulness as a model for research into plant stress tolerance, as well as emphasizing the possible molecular mechanisms underlying adaptation of the halophytic Eutrema to saline environments. This can provide a comprehensive insight to understand the mechanisms conferring a high level of salt stress tolerance in the halophyte.
Genome Evolution Adapted to Saline Environment
High levels of stress tolerance in plants are the result of evolutionary adaptation, thus the genotypic selection of plants for adaptation to various environments plays an important role in agriculture and ecology (Boyer, 1982; Janz et al., 2010). Due to the natural adaptations of E. salsugineum to various harsh climates and soil conditions, it is considered as an important model for deciphering the mechanisms of salt and other abiotic stress tolerance of plants (Gong et al., 2005; Griffith et al., 2007; Lamdan et al., 2012; Wang et al., 2021).
Sixteen accessions of halophytic E. salsugineum species including previously called T. salsuginea, T. halophila, and T. botschantzevii were used to investigate their natural variation in salinity tolerance and found that these accessions could survive up to 700 mM NaCl in hydroponic culture, however, their relative salt tolerance has a considerable variation (Lee et al., 2016). Using 90 resequencing whole genomes data from 90 individuals in 21 natural populations, the analysis of Eutrema species across central Asia to North America revealed that the selection signals for genes were related to the adaptation of salt and other abiotic tolerance, at the species level (Wang et al., 2021).
Gene duplication, a major source of genetic diversity that drives the adaptive evolution of plants (Ohno, 1970; Kondrashov, 2012; Monihan et al., 2019). Wu et al. (2012) demonstrated that, although the gene spaces display an extensive colinearity between E. salsugineum and A. thaliana, the salt stress-related gene families expanded in E. salsugineum genome (Wu et al., 2012). For instance, 21 transcription factor (TF) families expanded in the E. salsugineum genome, compared to A. thaliana. Among these, the AtNFXL1 gene encoding NF-X1 type zinc finger proteins was suggested to play a regulatory role in protecting photosynthesis, and be required for the growth of Arabidopsis plants under salt stress. The members of NF-X1, HSF, Trihelix, and GRAS TF families involved in abiotic stress response, were also expanded in numbers in salt cress (Table 1). The expansions of these gene family members are suggested to be associated with the adaptation of salt cress to harsh environments, due to the fact that their orthologous gene members in Arabidopsis and other plants have been documented to be associated with stress resistance (Lisso et al., 2006; Ogawa et al., 2007; Xie et al., 2009; Wu et al., 2012). These expanded family members possibly endow E. salsugineum with more flexibility in response to salinity stress (Wu et al., 2012). Additionally, the RAV gene family reported to respond to high salt and cold stresses, had been expanded from six members in A. thaliana to nine in E. salsugineum genome (Fowler et al., 2005; Sohn et al., 2006; Wu et al., 2012; Table 1). However, the AtRAV1 gene overexpressed plants showed strong growth retardation. And comparing to the wild type, a more inhibition of seed germination in salt conditions, and the transgenic plants exhibited higher transpirational water loss in drought conditions (Fu et al., 2014). Given the expressions of RAVs in Arabidopsis were reduced by salt and dryness, it is inferred that adaption of reduced AtRAV1 expression is involved in the adaption of salinity environment in Arabidopsis (Fu et al., 2014). Moreover, the transcripts of EsRAVs in the group A (A-EsRAVs) from E. salsugineum seedling exhibited a moderate decline gradually by salt treatment, and stronger inhibition of seed germination and seedling root elongation occurred in the 35S:A-EsRAV transgenic plants in presence of NaCl, suggesting the roles of 35S:A-EsRAVs in negatively controlling plant growth (Yang et al., 2016; Table 1).
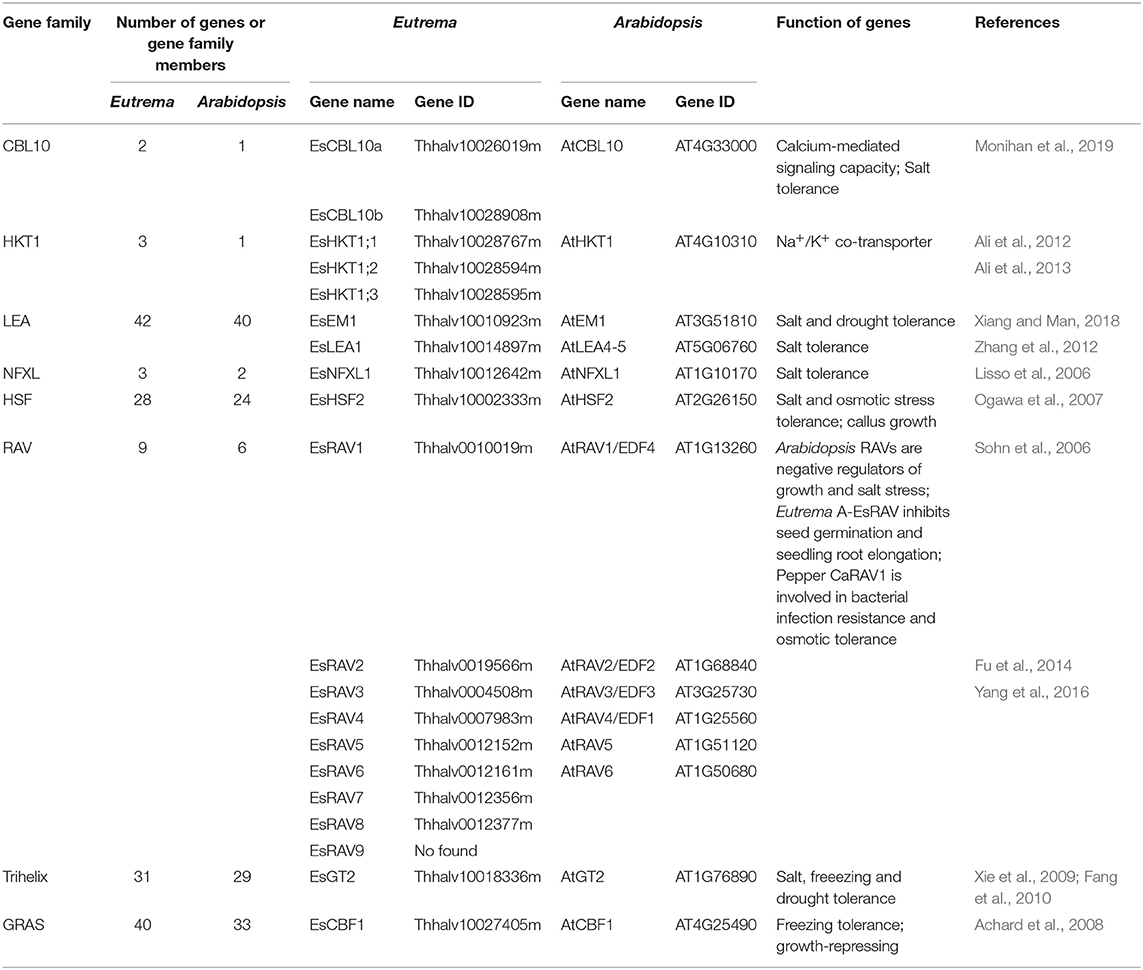
Table 1. The expanded genes or gene family members involved in response to salt stress in Eutrema compared with Arabidopsis.
LEA proteins can be involved in the “molecular shield function” to protect enzymes from induced aggregation when plants encounter stress in their environments (Goyal et al., 2005). In E. salsugineum, LEA genes were found to be higher copy numbers than their orthologous genes in the Arabidopsis genome, which is in agreement with a higher frequency of gene duplications observed in the E. salsugineum and E. parvula genomes (Table 1). This indicated that, since the last whole genome duplication event or after tandem duplications leading to neofunctionalization, the two Eutrema species (E. salsugineum and E. parvula) retained a higher fraction of duplicated genes, which is regarded as an important adaptive strategy for salt cress to survive in habitats with more extreme environmental conditions than Arabidopsis (Blanc and Wolfe, 2004a,b; Maere et al., 2005; Dassanayake et al., 2011; Wu et al., 2012; Lee et al., 2013; Oh et al., 2013).
The salt overly sensitive (SOS) pathway functions in preventing the toxic accumulation of sodium in the cytosol when A. thaliana are grown in salt-affected soils. In this pathway, Arabidopsis AtCBL10 calcium sensor interacts with the AtSOS2 kinase to activate the AtSOS1 plasma membrane sodium/proton exchanger, which initiates the transport of sodium out of the cell (Shi et al., 2000; Qiu et al., 2002, 2003; Quintero et al., 2002). In E. salsugineum, the CBL10 gene had been duplicated into EsCBL10a and EsCBL10b (Table 1), and the down-regulation of either of the two EsCBL10 genes decreased the growth of plants in the presence of salt, indicating that both genes function in the response of plants to salinity stress (Monihan et al., 2019). Further, EsCBL10b was demonstrated to have an enhanced ability to activate the SOS pathway, while the role of EsCBL10a is in an alternative pathway with different functions from AtCBL10 or EsCBL10b. Taken together, the duplication of Eutrema EsCBL10 obviously increased calcium-mediated signaling capacity and conferred an enhanced salt tolerance when compared with salt-sensitive Arabidopsis (Monihan et al., 2019).
Altogether, the maintenance of duplicated gene pairs in the genome of plants is indicative of an adaptive benefit conferred by the paralogous genes (Hahn, 2009; Kondrashov, 2012; Monihan et al., 2019). Undoubtedly, genetic variation and natural selection are fostering stress-related genes and gene interaction networks in the whole genome, which will prompt the local adaptation and differentiation of Eutrema plants to extreme salt stress in a non-random way.
Morphological Characteristics and Physiological Responses
The exposure of plants to high salt stress leads to the damage of cell structure, and disorder of many physiological functions (Sairam and Tyagi, 2004). As a consequence, this will eventually result in photosynthesis inhibition and metabolic impairment to further impaired growth and fertility, and early senescence of plants (Larcher, 2003).
Salt cress can natively grow in extreme salinity environments, and reproduce after exposure to 500 mM NaCl (Inan et al., 2004). Under treatments of 100 and 200 mM NaCl, an increased growth was observed in E. salsugineum accompanied by almost unchanged fresh and dry weights (Figures 1, 2), whereas a significant growth reduction in Arabidopsis. In a moderate salinity of 300 mM NaCl, E. salsugineum plants grew rapidly accompanied by almost unaffected chlorophyll content, a small decline of stomatal conductance and CO2 assimilation in leaves, indicating the photosynthetic apparatus was little influenced by this salt concentration (Sui and Han, 2014; Figure 2). However, the growth of Arabidopsis was remarkably decreased by the 100 and 200 mM NaCl treatments, which was associated with strong suppression in both leaf initiation and leaf expansion (M'rah et al., 2007; Sui and Han, 2014). Compared to Arabidopsis, E. salsugineum leaves are more succulent-like, have a second layer of palisade mesophyll cells, and frequently fall off during extreme salt stress, while root tissues develop an extra endodermis and cortex cell layer (Inan et al., 2004; Figure 2). Meanwhile, the stomata of E. salsugineum leaf surface are present at a higher density and less open than that in Arabidopsis, and exhibit a more tightly closing when responding to salt stress (Inan et al., 2004; Figure 2). Therefore, when compared with Arabidopsis, E. salsugineum can maintain a high water uptake and ion transport (Volkov et al., 2004; M'rah et al., 2006, 2007; Amtmann, 2009).
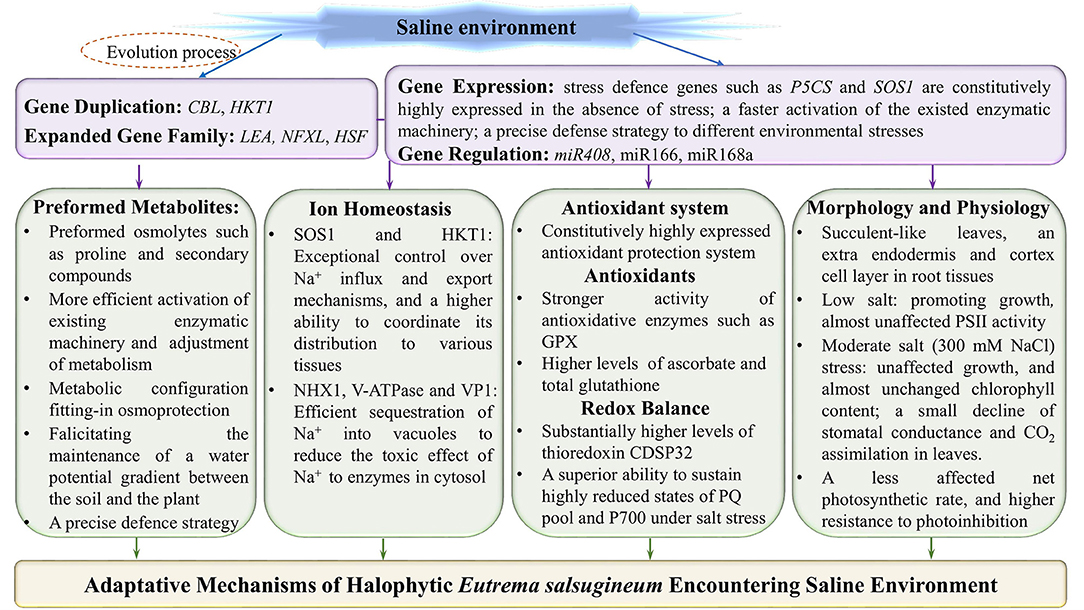
Figure 2. Adaptative mechanisms of halophytic Eutrema salsugineum encountering saline environment. The adaptative mechanisms of Eutrema plants to saline stress are mainly including: (1) The adaptive evolution endowing E. salsugineum plants with the expanded defense related genes and/or gene family members to facilitate plants more flexibility in response to salinity stress; (2) The constitutive higher expression of stress defense genes such as P5CS and SOS1 in the absence of salt stress; (3) Anticipatory prepared osmolytes such as proline and secondary compounds, and a higher efficience to activate the existed enzymatic machinery as well as the adjustment of metabolism; (4) The halophytic E. salsugineum possessing exceptional control over Na+ influx and export mechanisms, the superior ability to coordinate its distribution to various tissues, and the efficient sequestration of Na+ into vacuoles; (5) A constitutively upregulated antioxidative protection system, and the effective regulation of redox status in halophytic Eutrema; (6) The succulent-like leaves, an extra endodermis and cortex cell layer in root tissues, and higher density of stomata exhibitting a more tightly closing when responding to salt stress, as well as an almost unaffected net photosynthetic rate, in the lower or even moderate concentration of salinity. SOS1, salt overly sensitive 1 (plasma membrane Na+/H+ antiporter); NHX, vacuolar Na+/H+ antiporter; V-ATPase, vacuolar H+-ATPase; VP1 (V-Ppase), vacuolar H+-translocating inorganic pyrophosphatase (vacuolar H+-pyrophosphatase); HKT1, group 1 high-affinity K+ transporter; GPX, glutathione peroxidases; PSII, photosystem II; PQ, plastoquinone.
Photosynthesis is a primary process that is influenced by salinity stress, that mainly attributed to a decreased CO2 availability caused by diffusion limitations via the stomata and the mesophyll, or a generated secondary oxidative stress, which will seriously affect leaf photosynthetic machinery (Lawlor and Cornic, 2002; Flexas et al., 2004, 2007; Munns et al., 2006; Chaves et al., 2009). In leaves of Arabidopsis, the photosynthetic activity was suppressed after 15 days of salt treatment with higher than 50 mM NaCl, which could be explained by the stomata closure and dramatic shrinkage of the RubisCO pool. By contrast, E. salsugineum leaves still remained at 50% of the stomatal conductance of the control, and a negligible influence of RubisCO quantity, when exposed to the same salt-treated conditions (M'rah et al., 2007).
Chlorophyll (Chl) and PSII play a key role in the response of leaf photosynthesis to environmental stresses (Baker, 1991). When plants are subjected to 100 or 200 mM NaCl treatments, there are no remarkable changes in the chlorophyll content and Chl a/b ratio, as well as almost do not affect the PSII activity in E. salsugineum leaf (Figure 2), whereas a progressive decline exists in Arabidopsis leaf, which suggests this halophyte has effective mechanisms to protect photosystem against salinity (Sui and Han, 2014). In addition, E. salsugineum was found to have an increased chlorophyll a/b ratio accompanied by a higher effective photochemical quantum yield (YII) value and lower non-photochemical quenching (NPQ) value, and a more active photosystem 1 (PS1) (Wiciarz et al., 2015).
All in all, these studies clearly demonstrate that, compared to A. thaliana, the E. salsugineum displays a relatively stable stomatal conductance and CO2 assimilation, higher chlorophyll content, Chl a/b ratio, and higher resistance to photoinhibition, as well as almost unaffected net photosynthetic rate and Fv/Fm (maximal photochemical efficiency of PSII; Sui and Han, 2014; Wiciarz et al., 2015; Figure 2), thus protecting photosystem activities to maintain normal growth of salt cress, in the lower or even moderate concentration of salinity.
Maintain ION Homeostasis Under Saline Environment
To survive salt stress, plants must maintain a balance between sodium (Na+) and potassium (K+) ions, thus the homeostasis of Na+ and K+ is considered the most important mechanism to reducing NaCl stress in higher plants (Zhao et al., 2020). Halophytes can activate the protective mechanisms to prevent high Na+ accumulation in the cytosol and to maintain photosynthesis (Niewiadomska and Wiciarz, 2015; Bose et al., 2017; Kazachkova et al., 2018; Grigore, 2019).
The plasma membrane (PM) Na+/H+ antiporter (SOS1, Salt Oerly Sensitive 1) is involved in the transport of sodium ions across the plasma membrane and retrieves Na+ ions from the xylem sap, which is essential for Na+ and K+ homeostasis (Qiu et al., 2002; Shi et al., 2002). Taji et al. (2004) found, in E. salsugineum, a constitutively high expressed EsSOS1 RNA in the absence of stress, and an increased EsSOS1 expression observed in the PM of salt-treated roots and leaves (Taji et al., 2004; Vera-Estrella et al., 2005; Figures 2, 3). Likewise, Kant et al. (2006) demonstrated that E. salsugineum SOS1 transcript was strongly induced by salt in the shoots, and a constitutively high expression existed in roots in unstressed salt cress, which was consistent with a lower salt-induced Na+ concentration in xylem sap of E. salsugineum than that in Arabidopsis (Kant et al., 2006). Further, ectopic expression of EsSOS1 from E. salsugineum suppressed the salt-sensitive phenotype of Saccharomyces cerevisiae strains lacking Na+ efflux transporters (SOS1), and enhanced the salt tolerance of wild-type Arabidopsis (Oh et al., 2009). E. salsugineum SOS1-RNA interference (RNAi) significantly induced reduction of SOS1 gene, which led to faster leaf senescence and severe shoot water loss, accompanied by fragmentation of vacuoles, inhibition of endocytosis, and apoplastic Na+ influx into the stele and shoot (Oh et al., 2009). Another study also demonstrated that EsSOS1 conferred greater salt tolerance than AtSOS1 when expressed in a salt-sensitive strain of S. cerevisiae, and suggested that the salt tolerance of E. salsugineum may at least in part be derived from SOS1-mediated Na+ extrusion (Jarvis et al., 2014; Figures 2, 3). Therefore, it is suggested that EsSOS1 activity limits Na+ accumulation and affects the distribution of Na+, and acts as an important tolerance determinant in shaping the halophytic character of the E. salsugineum species (Oh et al., 2009; Figures 2, 3).
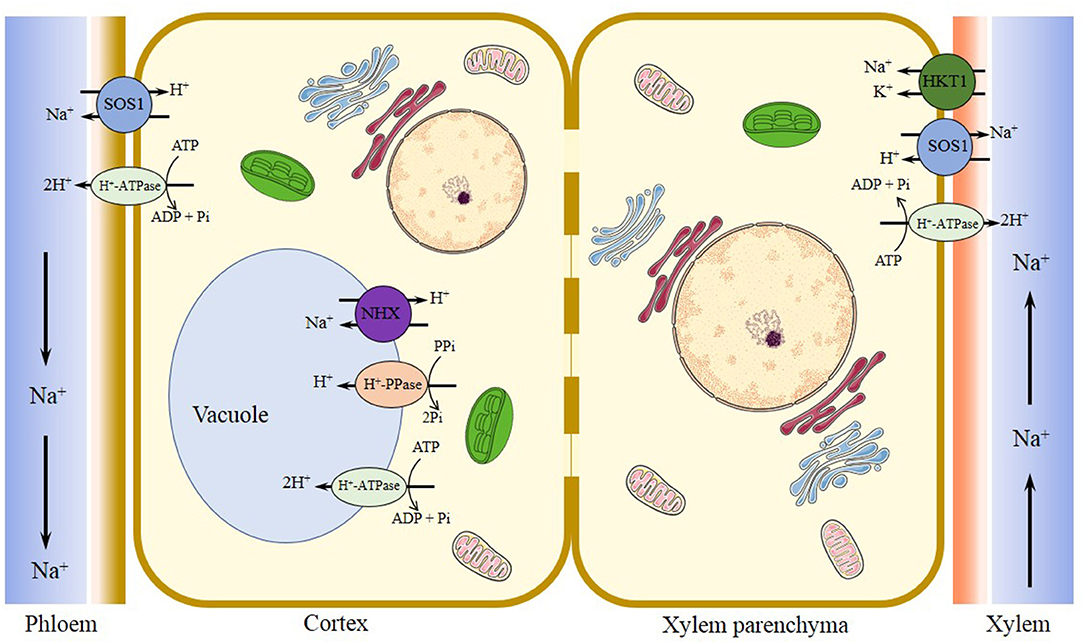
Figure 3. Major transporters regulating Na+/K+ homeostasis in Eutrema under salinized tissues. Plasma membrane Na+/H+ antiporter SOS1 functions in the regulation of ion homeostasis by extruding excessive Na+ out of cells, facilitating Na+ loading into the xylem in roots and shoots, and reducing the transfer of Na+ from roots to shoots, to counterbalance the uptake of Na+. Tonoplast-localized Na+/H+ exchanger NHX1 driven by vacuole H+-PPase (VP1) and/or H+-ATPase (V-ATPase) proton pumps is responsible for the sequestration of Na+ into the vacuole, to reduce the concentration of Na+ in the cytosol. The halophytic Eutrema HKT1;2 (EsHKT1;2 and EpHKT1;2) is considered as the Na+/K+ co-transporter, that function in Na+ retrieval from the xylem vessels under salt stress, and helps sequester ions into xylem parenchyma cells to protect photosynthetic tissues. SOS1, salt overly sensitive 1 (a plasma membrane Na+/H+ antiporter); NHX, vacuolar Na+/H+ antiporter; V-ATPase, vacuolar H+-ATPase; V-PPase, vacuolar H+-translocating inorganic pyrophosphatase (vacuolar H+-pyrophosphatase); HKT1, group 1 high-affinity K+ transporter.
The compartmentalization of Na+ into vacuole can alleviate the toxicity of Na+ and Cl− in the cytosol and enhance vacuolar osmoregulatory capacity, which could confer salt tolerance of plants (Gaxiola et al., 2001). The vacuolar ATPase (V-ATPase) and pyrophosphatase-dependent H+-pyrophosphatase (VP1) as the proton pumps function in establishing and maintaining the electrochemical proton gradient across the tonoplast (Kirsch et al., 1996; Maeshima, 2000). The Na+ sequestration process is mediated by the vacuolar Na+/H+ antiporter (NHX) that is driven by V-ATPase and VP1 (Apse et al., 2003; Bhaskaran and Savithramma, 2011). When E. salsugineum plants were treated with NaCl, a salt-induced increase in the EsNHX1 protein expression from PM fractions of roots, and a similar increased expression of the vacuolar H+-translocating ATPase E subunits (VHA-E) in leaf tissues were detected (Vera-Estrella et al., 2005; Kant et al., 2006). Taken together, salt stress enhanced H+-transport and hydrolytic activity of E. salsugineum tonoplast and plasma membrane H+-ATPases, and also greatly stimulated the activity of tonoplast Na+/H+ exchange in E. salsugineum leaves and roots (Vera-Estrella et al., 2005; Figures 2, 3).
High-affinity potassium transporters (HKTs) play a key role in reducing Na+ toxicity through K+ uptake (Rus et al., 2004). The study found that EsHKT1;2 mRNA is dramatically induced, whereas AtHKT1 is downregulated upon salt stress (Ali et al., 2013; Figures 2, 3). And EsHKT1;2-RNAi lines display a severe potassium deficiency, and are sensitive to high Na+ (Ali et al., 2013). E. salsugineum EsHKT1;2 ectopically expressed in yeast mutants lacking Na+ or K+ transporters showed a strong K+ transporter activity, and selectivity for K+ over Na+, suggesting EsHKT1;2 act as maintenance of K+ uptake under salt stress that supporting the halophytic lifestyle (Ali et al., 2012, 2013; Figures 2, 3). Further, the plants overexpressing Eutrema EsHKT1;2 in Arabidopsis athkt1 mutant was proved to be less sensitive to Na+ as well as less K+ deficiency, compared to the plants overexpressing AtHKT1 gene in athkt1 (Ali et al., 2012). Importantly, the K+ specificity of EsHKT1;2 protein is based on amino acid differences in the pore of the transporter protein relative to AtHKT1 (Ali et al., 2013). In addition, the transcript level of EpHKT1;2 from halophytic E. parvula (formerly known as Thellungiella parvula) increased rapidly in response to high salinity, and the yeast cells expressing EpHKT1;2 could tolerate high concentrations of NaCl (Ali et al., 2018). Moreover, Arabidopsis plants (Col-gl) overexpressing EpHKT1;2 gene accumulated less Na+ and more K+, and displayed significantly higher tolerance to salt stress when compared to those overexpressing EpHKT1;1 or AtHKT1. Thus, EpHKT1;2 is suggested to mediate tolerance to Na+ ion toxicity in E. parvula, and is a major contributor to its halophytic nature (Ali et al., 2018; Figure 3).
Besides, the major site of Na+ accumulation occurs in old leaves, followed by young leaves and taproots, and the least accumulation occurred in the lateral roots of E. salsugineum plants under salt stress (Vera-Estrella et al., 2005). Notably, although most genes in E. salsugineum exhibit ~90% identity with Arabidopsis genes, the size of E. salsugineum transport genes population is ~1.5 times that compared of the corresponding Arabidopsis genes (Taji et al., 2008). This suggests that the transporter encoding genes could be remarkably different from their co-orthologs in Arabidopsis, and regulate a unique ion transportation system in this halophyte (Taji et al., 2008).
Taken together, these results revealed E. salsugineum as a halophyte is able to distribute and store Na+ by very strict control of ion movement across both the tonoplast (TP) and plasma membrane (PM) (Vera-Estrella et al., 2005; Figures 2, 3). Correspondingly, Eutrema accumulated lower levels of Na+ and Cl− than Arabidopsis, and its rosette leaves displayed more efficient protective mechanisms against Na+ metabolic toxicity when plants were exposed to higher salinity stress (M'rah et al., 2007). These works of key Na+-transport mechanisms provide some detailed mechanisms analysis underlying salinity tolerance in this halophytic E. salsugineum, an Arabidopsis relative model system (Vera-Estrella et al., 2005; Figures 2, 3).
Antioxidants, Redox Balance, and Salt Stress Tolerance
A common reaction of plants to salt-triggered osmotic stress and ionic imbalance is to increase the levels of reactive oxygen species (ROS; Bose et al., 2014; Kumar et al., 2017). The ROS species of superoxide (), hydrogen peroxide (H2O2), hydroxyl radical (OH•), and singlet oxygen (1O2) are generated constantly as an unavoidable consequence of aerobic metabolism (Møller et al., 2007). The primarily negative effects of ROS molecules on the cells include lipid peroxidation, protein denaturation, and DNA damage (Møller et al., 2007). On the other hand, ROS as universal signaling metabolites has been perceived to regulate plant growth and development, as well as defense against biotic and abiotic stresses (Foyer et al., 2017; Czarnocka and Karpiński, 2018; Fichman and Mittler, 2020). Usually, low levels of H2O2 could modulate many biological and physiological roles, whereas a high concentration of H2O2 will cause damage to cellular structures resulting in severe injury to plants (Ozyigit et al., 2016). Thereby, the salt tolerance of halophytes seems to be associated with their ability to control the redox balance (Ozgur et al., 2013; Bose et al., 2014; Surówka et al., 2019; Pilarska et al., 2021; Figures 2, 4).
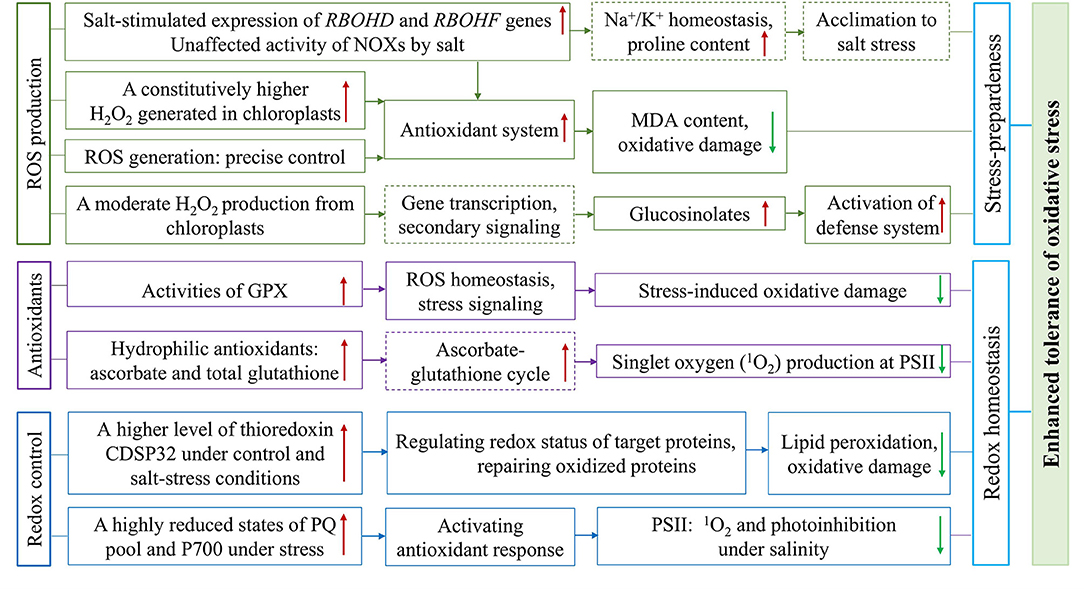
Figure 4. Overview of the constitutively higher expressed antioxidative protection system and effective regulation of redox status in halophytic Eutrema. The plasma membrane ROS produced through NADPH oxidases (NOXs), the important components of signal transduction, encoded by RBOHD and RBOHF genes, is associated with the regulation of Na+/K+ homeostasis and increased accumulation of proline involved in salt tolerance. The enhanced antioxidant tolerance of Eutrema is associated with: (1) A constitutively enhanced H2O2 generated to keep the antioxidant system up-regulated to preadapted plant to salinity stress, as well as a chloroplastic production of H2O2 inducing the accumulation of glucosinolates to activate the stress defense system, (2) Antioxidative enzymes such as GPX, and non-enzymatic antioxidants such as ascorbate and glutathione facilitating ROS homeostasis and alleviating damage of singlet oxygen (1O2) to PSII under salt stress, (3) A higher expression level of thioredoxin CDSP32 regulating redox status of target proteins and highly reduced states of PQ pool and P700 facilitating redox homeostasis and lower photoinhibition under salt stress. NOXs, NADPH oxidases; GPX, glutathione peroxidases; PSII, photosystem II; PQ, plastoquinone.
The H2O2 checked in A. thaliana and E. salsugineum plants treated with H2O and the salt solution containing 150 and 300 mM NaCl revealed that, in the chloroplasts of salt cress mesophyll cells, electron-dense perhydroxide cerium precipitates were detectable in control, and more pronounced visible dark spots found after salt stress (Pilarska et al., 2016). In contrast, the chloroplasts in A. thaliana leaves displayed no perhydroxide deposits in the control condition, while salt stress associated with H2O2 accumulation was mainly visible at the edges of grana in thylakoids (Pilarska et al., 2016). This result suggested that a constitutively enhanced H2O2 generated in chloroplasts may be a crucial component of stress-preparedness of this halophyte (Pilarska et al., 2016; Figures 2, 4). Similarly, increased content of H2O2 in leaves was documented in E. salsugineum than in A. thaliana. Meantime, the salinity-induced thylakoid swelling was detected only in Arabidopsis chloroplasts, while no signs of such destruction occurred in E. salsugineum chloroplasts (Pilarska et al., 2016). However, a very low level of MDA was detected, in spite of the higher ROS availability in E. salsugineum leaves, indicating a low extent of oxidative damage to membrane lipids (Pilarska et al., 2016). Thus, it is suggested that enhanced leakage of H2O2 from plastids keeps the antioxidant system up-regulated to make plants preadapted to the salinity stress (Pilarska et al., 2016; Figures 2, 4). The effect of H2O2 signaling not only depends on its type, but also on the site of its origins such as chloroplasts, mitochondria, and peroxisomes in the cell (Sewelam et al., 2014; Pilarska et al., 2021). The comparison of A. thaliana and E. salsugineum demonstrated that the latter was capable of enhancing the production of H2O2 under control and stress conditions (Wiciarz et al., 2015; Pilarska et al., 2016, 2021; Figures 2, 4). These studies indicate a different regulation of ROS formation in the two relative species, and the precise control of ROS generation may contribute to plant stress responses (Pilarska et al., 2021).
In the plasma membrane, ROS are synthesized by NADPH oxidases (NOXs), termed respiratory burst oxidase homologs (RBOHs; Kaur et al., 2018), and NOXs is considered an important component of signal transduction under salt stress (Pilarska et al., 2021). Arabidopsis NOX genes, RBOHD and RBOHF, are the main genes encoding NADPH oxidases associated with acclimation to salinity (Ma et al., 2012; Pilarska et al., 2021). After E. salsugineum was exposed to 5 days of salinity, a salt-stimulated expression in RBOHD and RBOHF genes in leaf tissues was revealed (Figures 2, 4), while their orthologous genes expression in Arabidopsis decreased (Pilarska et al., 2021). The ROS produced by RBOHD and RBOHF is associated with the regulation of Na+/K+ homeostasis under salinity (Chung et al., 2008; Ma et al., 2012), as well as the increased the accumulation of proline involved in salt tolerance (Ben Rejeb et al., 2015; Pilarska et al., 2021; Figure 4). Moreover, the different salt-induced expression patterns of RBOHD and RBOHF genes between the E. salsugineum and A. thaliana point to, in leaves of the halophytic Eutrema, the two NOX genes (RBOHD and RBOHF) be involved in the late acclimation response to ionic and osmotic stress (Pilarska et al., 2021; Figure 4). Further, the total activity of NOXs in E. salsugineum was demonstrated to be unaffected by severe salt stress (Figure 4), whereas a declined activity was seen in A. thaliana (Pilarska et al., 2021). And the prolonged mild salinity did not lead to obvious changes in the activity of NOXs in leaves, indicating that the acclimation to salinity in the halophytic E. salsugineum was in connection with the maintenance of the basal activity of NOXs (Pilarska et al., 2021; Figure 4). This opinion is in agreement with the study from Srivastava et al. (2015), that the total NOXs activity in the halophyte Sesuvium portulacastrum was unaffected, whereas a significantly decreased NOXs activity in the glycophyte Brassica juncea (Srivastava et al., 2015).
Antioxidative enzymes play an important role in scavenging toxic radicals in the different organelles of plants such as chloroplasts, cytosol, mitochondria, and peroxisomes, particularly in environmental stress such as salinity (Rajalakshmi and Parida, 2012). In E. salsugineum, the enzymatic antioxidant systems could contribute to its extremely high level of tolerance to salt, drought, cold, and oxidative stresses (Gao et al., 2014). Glutathione peroxidases (GPXs) catalyze the reduction of H2O2 and organic hydroperoxides into the water and correspondingly alcohols, using reduced glutathione (GSH). Plant GPXs play essential roles that are not only involved in ROS homeostasis, stress signaling, and protecting plants from stress-induced oxidative damage to membrane and protein, but also in plant growth and development (Gao et al., 2014; Ozyigit et al., 2016). The gene and protein expression profiles of eight GPXs identified in E. salsugineum displayed that GPX5, GPX7, and GPX8 genes were upregulated in both leaves and roots upon salt stress (Figures 2, 4), while GPX1, GPX2, and GPX3 were only upregulated in roots under salt stress; GPX1, GPX3, GPX4, GPX7 genes in leaves and almost all GPX genes in roots were up-regulated under osmotic stress (Gao et al., 2014). An enhanced GPX activity (Figures 2, 4), but decreased activity of ascorbate peroxidase (APX) were revealed in Eutrema plants (Pilarska et al., 2016). These studies documented that, the different members of the GPX gene family were modulated in specific environmental stresses and/or in different tissues, indicating the important roles of EsGPXs in salt and osmotic stress response of Eutrema plants (Gao et al., 2014).
Thioredoxin is a small oxidoreductase functioning as a hydrogen donor of target proteins and is considered a specific marker for assessment of oxidative stress (Holmgren, 1985; M'rah et al., 2007). Thioredoxin CDSP32 participating in the repair of oxidized proteins during environmental constraints is regarded as a critical component of the defense system against lipid peroxidation and oxidative damage (Rey et al., 2005; Dos Santos and Rey, 2006; M'rah et al., 2007). It was reported that potato (Solanum tuberosum) CDSP32 exhibited a substantial accumulation in chloroplast stroma and preserved chloroplastic structures against oxidative injury under drought and oxidative stresses (Broin et al., 2000). In E. salsugineum, thioredoxin CDSP32 was found to be higher in abundance than that in A. thaliana under control and salt-stress conditions (Figures 2, 4), and strongly diminished in salt-treated Arabidopsis plants (M'rah et al., 2007). Furthermore, the magnitude of electrolyte leakage and level of lipid peroxidation were modest in E. salsugineum, and very remarkable in Arabidopsis (M'rah et al., 2007).
In addition, a significantly increased hydrophilic antioxidants (ascorbate and total glutathione), but no changed lipophilic antioxidants existed in the halophytic E. salsugineum when exposed to 300 mM NaCl (Figures 2, 4). In contrast, the glycophyte Arabidopsis showed a decreased ascorbate content but increased lipophilic antioxidants (α-tocopherol, plastochromanol, and hydroxy-plastochromanol) due to 150 mM NaCl (Wiciarz et al., 2018). The redox states of plastoquinone (PQ) and P700 were differently regulated by salinity between the halophyte E. salsugineum and glycophyte A. thaliana, which was documented by effectively avoiding harmful singlet oxygen (1O2) in the PSII of Eutrema (Figures 2, 4), whereas an increased oxidation extent was observed in Arabidopsis (Wiciarz et al., 2018). Moreover, E. salsugineum has a very efficient antioxidant protection in which glucosinolates may play a specific role (Pilarska et al., 2016; Figure 4).
To sum up, these researches proposed that the salt tolerance of halophytic Eutrema species is associated with constitutively expressed protection systems against oxidative stress such as GPX and thioredoxin CDSP32, as well as the ability to sustain a highly reduced state of PQ pool and P700, to alleviate the damage of PSII under salt stress (Figures 2, 4).
Preformed Metabolites and Osmotic Adaptation
Accumulation of osmoprotectants is one of the defense mechanisms for plants to cope with abiotic stresses (Rajalakshmi and Parida, 2012; Zhao et al., 2020). Several studies have shown that plant metabolism is strongly impacted by salt stress (Flowers and Colmer, 2008; Bartels and Dinakar, 2013; Lee et al., 2016). The metabolites positively correlating with salt stress are reported to participate in the underlying mechanisms including cell-wall remodeling (hydroxyproline), osmoprotectant and storage (proline and sucrose), and photorespiration (glycine and serine; Lugan et al., 2010). The E. salsugineum had much higher levels of most metabolites than A. thaliana under the salt or osmotic stress treatment (Figure 2), despite the fact that the same metabolic pathways were regulated by salt stress in both species (Lugan et al., 2010). Importantly, significant differences observed in E. salsugineum and A. thaliana were also associated with the physicochemical properties of their metabolomes, such as water solubility and polarity (Lugan et al., 2010). Comprehensive quantification of organic and mineral solutes documented relative stability of the total solute content in the two species under various treatment conditions (Lugan et al., 2010). But E. salsugineum could cope with osmotic stress by tolerating dehydration via the metabolic configuration to lend itself to osmoprotectant rather than osmo-adjustment strategy (Lugan et al., 2010; Figure 2). Moreover, the leaves of E. salsugineum have a constitutively lower water content than in A. thaliana, and have the ability to lose more water without losing turgor, which could contribute to the maintenance of a water potential gradient between the soil and the plant (Lugan et al., 2010; Figure 2). Taken together, salt stress markedly decreased the osmotic potential of shoots, increased total solute levels to sustain a water potential gradient with the environment (Figure 2).
Furthermore, several studies supported the constitutively higher levels of proline, inositols, sugars, and organic acids in E. salsugineum, which were interpreted as metabolic stress-anticipation (Gong et al., 2005; Lee et al., 2016; Figure 2). The salt responses of most metabolites in E. salsugineum Yukon took place at 200 mM NaCl, and a few additional changes were observed between 200 and 500 mM (Lee et al., 2016).
The compatible osmolyte proline's accumulation is an important marker of salt tolerance in plants (Verbruggen and Hermans, 2008). Salt cress accumulated a markedly higher amount of proline than that of Arabidopsis under normal growth conditions, thus suggesting that the characteristic of extreme tolerance of salt cress to high salinity is due in part to the overaccumulation of proline under unstressed conditions (Taji et al., 2004; Figure 2). Likewise, Kant et al. (2006) demonstrated that E. salsugineum contained higher unstressed levels of proline than Arabidopsis, while under salt stress, Eutrema accumulated more proline mainly in shoots (Kant et al., 2006). The evaluation of salinity stress tolerance of eight different Eutrema accessions revealed that Tuva and Buriatia classified as salt susceptible ecotypes treated at 500 mM NaCl had seven-fold higher proline than that in Altai 1 and Altai 2 identified as salt tolerant ecotypes when treated with a similar concentration of NaCl (Gandour et al., 2019). Another study revealed that phenotypic and metabolic adaptive plasticity observed in E. salsugineum in different growth conditions could be an inherent trait endowing the flexibility of the halophyte that is required for an extremophile lifestyle (Guevara et al., 2012; Kazachkova et al., 2013).
In summary, these presented data support the markedly higher basal metabolite levels (proline and secondary compounds) in E. salsugineum than Arabidopsis in normal growth conditions (Figure 2), which is an important characteristic of salt tolerance of salt cress plants. The metabolic pre-adaptation of this halophyte could in part be attributed to the decreased osmotic potential in shoots and increased total solute levels, as well as the physicochemical properties of the metabolites such as water solubility and polarity (Figure 2).
Gene Expression and Regulation
Salt cress plants can grow in high salinity coastal areas, even in a 500 mM NaCl medium, but they do not have salt glands or other significant morphological alterations either before or after salt adaptation (Inan et al., 2004; Taji et al., 2004). Thus, the salt-tolerant halophyte E. salsugineum and salt-sensitive glycophyte A. thaliana are considered to use common mechanisms to cope with salinity, and the differences in salt tolerance between the two species can be mainly attributed to the changes in the regulation of a basic set of salt tolerance genes (Kant et al., 2006).
The gene expression profiles in E. salsugineum (Shandong ecotype) using an Arabidopsis full-length cDNA microarray revealed that, in contrast to Arabidopsis, only a few genes were induced by 250 mM NaCl stress, and many known stress-inducible genes such as Fe-SOD, P5CS, PDF1.2, AtNCED, P-protein, b-glucosidase, and SOS1 were expressed at high levels in salt cress even in the absence of stress (Taji et al., 2004; Figure 2). The higher expression of the P5CS gene, encoding a key enzyme of proline biosynthesis, is in accordance with a much higher level of accumulated proline in salt cress than did Arabidopsis under normal growing conditions (Taji et al., 2004). Likewise, a comparison of transcript profiles between Arabidopsis at 150 mM NaCl and Eutrema (Shandong) exposed to 250 mM NaCl, using a microarray platform and Arabidopsis homologous gene probes, demonstrated that 40% shared and 60% distinguished regulated genes existed in the halophyte and the glycophyte (Gong et al., 2005). The combined transcript and metabolite analyses notably pointed toward a stress-anticipatory preparedness in E. salsugineum (Gong et al., 2005). In comparison with a great activation of transcription detected in A. thaliana, only a slight change in gene expression was found in E. salsugineum after stress treatment (Taji et al., 2004; Wong et al., 2006). Collectively, the analysis of expression profiling suggested that the stress-inducible signaling pathways are constitutive and active in salt cress even in the absence of stress (Figure 2), which is considered an important strategy for the extreme salt tolerance of this halophyte (Taji et al., 2004; Gong et al., 2005).
Also, comparative proteomics of salt responses between A. thaliana and E. salsugineum revealed more changes in protein abundance in Arabidopsis than in Eutrema (Pang et al., 2010). This indicates that the adjustment of metabolism and activation of the already existing enzymatic machinery could serve as a faster and more efficient strategy to cope with salt stress than the synthesis of new proteins (Pilarska et al., 2016; Figure 2). Furthermore, E. salsugineum exposed to 300 mM NaCl was used to investigate global transcriptional changes, and genes of several pathways mainly involved in lignin biosynthesis and accumulations of soluble sugars, as well as genes in autophagy and peroxisome pathways, were significantly enriched in the halophyte, suggesting these upregulated genes could contribute to salt tolerance of the halophytic Eutrema (Li et al., 2021).
In addition, the cDNA microarray containing 3,628 stress-induced E. salsugineum (Yukon ecotype) unique sequences were used to investigate the gene expression-profiling between Yukon and Arabidopsis subjected to saline, simulated drought, and cold stress as well as recovery from water deficits. The result revealed that, among 154 differentially regulated transcripts obtained in all studied conditions, only six genes responded to all three stresses of salinity, drought, and cold (Wong et al., 2006). The low overlapping genes indicated an extensive divergence that exists in the responses of Eutrema Yukon to different stresses (Wong et al., 2006). There were relatively few transcripts responding to high salinity in this halophytic E. salsugineum, in contrast with A. thaliana (Wong et al., 2006). Intriguingly, in addition to activating the expression of some well-known stress-responsive genes, a large number of biotic stress-related genes under salinity and drought treatments were found to be down-regulated, suggesting a precise defense strategy to different environmental stresses in E. salsugineum (Figure 2), thereby maximizing its survival potential via conserving energy and resources (Wong et al., 2006).
MicroRNAs (miRNAs) are known to regulate gene expression by cleavage of complementary target mRNA, repression of translation of target mRNA, and transcriptional silencing of target mRNA (Sunkar and Zhu, 2004; Mittal et al., 2016). Solexa sequencing platform together with microarray and qRT-PCR was used to identify salt-stress responsive miRNAs in E. salsugineum. The differential response of the conserved high abundance miR166f, miR168a, miR408, and miR408-5p to salt stress suggests that they may play a key role in salt tolerance of Eutrema (Zhang et al., 2013; Figure 2). In agreement with this result, miR166, miR168a, and miR408 were all previously consistently demonstrated to be responsive to stress conditions, including salt and drought stress (Liu et al., 2008; Kong et al., 2010; Trindade et al., 2010; Zhou et al., 2010; Li et al., 2012; Figure 2). Another research on the large-scale characterization of E. salsugineum miRNAs and analysis of potential targets demonstrated that 11 conserved miRNA families including miR160, miR161, miR162, miR164, miR171, miR319, miR390, and miR827, and four novel miRNAs displayed a significant response to 300 mM NaCl stress (Wu et al., 2016). Importantly, E. salsugineum miR408 was strongly induced by 200 mM NaCl treatment, and Arabidopsis miR408 was induced by cold and mannitol, but not by salt (Liu et al., 2008; Zhang et al., 2013). The higher expression of miR408 leads to improved tolerance to salinity, cold and oxidative stress in Arabidopsis (Ma et al., 2015). Therefore, it is suggested that miR408 could play a critical role in the salt tolerance of E. salsugineum (Figure 2).
Long non-coding RNAs (lncRNAs) are implicated in the regulation of gene expression at the level of chromatin modification, transcription, and post-transcriptional processing, and are suggested to play an important role in a wide range of biological processes, such as genomic imprinting, chromatin remodeling, transcriptional activation, transcriptional interference, and cell cycle (Mercer et al., 2009; Sun et al., 2018). Recently, lncRNAs were proposed to play an important role in response to abiotic and biotic stress including a significant role in plant salt stress response (Gai et al., 2018). The leaf transcriptomes of Yukon and Shandong E. salsugineum plants subjected to a progressive, two-stage drought treatment were analyzed, and 1,007 lncRNAs were detected. Among them, 8% were only expressed in stress-treated plants, which suggested a documented association between lncRNA expression and stress (Simopoulos et al., 2020).
In conclusion, these analyses of gene expression profiles based on the microarray platform and transcriptome in E. salsugineum provide useful global insight for elucidating the underlying salt tolerance mechanisms that distinguish it from Arabidopsis and help our more in-depth analyzing the extreme stress tolerance in this halophyte species. The investigation of the patterns of miRNA accumulation and target gene analysis revealed the possible roles of miRNAs in the adaptive response of E. salsugineum to salt stress (Figure 2), which enables us to access the salt tolerance mechanism of this halophyte from the perspective of small RNA regulation. However, research on the role of lncRNAs in salt resistance of plants is in its infancy, and the function and specific mechanism of action of the lncRNAs in salt tolerance of salt cress await to be studied and explored.
Candidate Genes for Salt Tolerance
Halophytes have the potential to serve as a rich repository for genes to study abiotic stress mechanisms in plants (Rajalakshmi and Parida, 2012). Based on the various plant defense mechanisms adopted by halophytes, many groups have identified some genes functioning in tolerance to salinity via expressing them in transgenic systems (Rajalakshmi and Parida, 2012). In E. salsugineum, some important candidate genes for salt tolerance have been identified.
Aquaporins Genes AQPs
The exposure of plants to abiotic stresses challenges the plant water status and triggers highly specific hydraulic responses. Aquaporins (AQPs), a large family of channel proteins (Maurel et al., 2009; Wang et al., 2014), could selectively transport water molecules across the cell membrane that function in the plant water balance (Martre et al., 2002; Li et al., 2011, 2014; Qin et al., 2019). Plant AQP genes respond to diverse abiotic stresses such as salinity and drought (Alexandersson et al., 2005; Guo et al., 2006), and play important functions in the responses to environmental stimuli (Maurel et al., 2008; Aroca et al., 2012; Wang et al., 2014).
In Arabidopsis, the PIPs (plasma membrane intrinsic proteins) aquaporins were well-characterized to be involved in the regulation of root hydraulic conductivity (Lpr) (Javot et al., 2003; Tournaire-Roux et al., 2003). It has been reported that Arabidopsis PIP2;1 and PIP2;2 contribute to approximately 40% of the root hydraulic conductivity (Sutka et al., 2011; Péret et al., 2012), and the modification and localization of PIP aquaporins and root hydraulic conductivity could be regulated by the salt stress (Boursiac et al., 2008; Prak et al., 2008; Martinière et al., 2012; Qin et al., 2019). In E. salsugineum (T. halophila) seedlings, expression of the EsPIP1 gene in the shoot and root tissues were induced by high salinity. Overexpression of the EsPIP1 gene in rice markedly enhanced the tolerance of transgenic plants to salt stress via regulating the osmotic potential, accumulation of organic small molecules substances, and the ratio of K+/Na+ in the plant cells (Qiang et al., 2015). Moreover, EsPIP1 protein was shown to specifically interact with EsPIP2 and a non-specific lipid-transfer protein 2, suggesting that E. salsugineum EsPIP1 probably plays key roles in the response to multiple external stimuli, and participates in various physiological processes when plants subjected to salt stress (Qiang et al., 2015). Several E. salsugineum PIPs were observed to have a more stable expression in the roots compared with that in the shoots under salt stress conditions, and this specific expression pattern of PIP aquaporins is suggested to improve their ability to maintain a higher level of water transport activity in the roots, and decreasing water transpiration in the shoots (Qin et al., 2019).
The tonoplast intrinsic protein gene EsTIP1;2 from the halophyte E. salsugineum was found to have water channel activity when expressed in Xenopus oocytes. And this gene was also able to conduct H2O2 into yeast cells in response to oxidative stress, and the inhibited growth of EsTIP1;2 transgenic yeast cells were suggested as a result of H2O2 influx (Wang et al., 2014). In plants, ectopic overexpression of E. salsugineum EsTIP1;2 in A. thaliana plants revealed an enhanced oxidative stress tolerance of plants after methyl viologen (MV) treatment, accompanied by a decrease of H2O2 level in the leaf cells (Wang et al., 2014). Meanwhile, EsTIP1;2 could facilitate the entry of Na+ ions into plant cell vacuoles under high salinity conditions, through an indirect process (Wang et al., 2014). In all, the tonoplast AQP gene EsTIP1;2, was induced by multiple external stimuli, and a significantly elevated tolerance to salt, drought, and oxidative stress was observed in EsTIP1;2 transgenic Arabidopsis plants (Wang et al., 2014). This suggested that EsTIP1;2 could act as a multifunctional contributor to the survival of Eutrema plants in highly stressful habitats via mediating the conduction of both H2O and H2O2 across membranes.
To sum up, the E. salsugineum aquaporin gene EsPIP1;4 (ThPIP1) or EsTIP1;2 (TsTIP1;2) overexpressed in heterologous species displayed an enhanced plant abiotic stress tolerance in transgenic plants (Wang et al., 2014; Qiang et al., 2015). These data enhance our understanding of the water balance regulation mechanisms involved in plant water status and root hydraulic conductivity, as well as the responses of aquaporin families to salt stress, in this halophyte.
Late Embryogenesis Abundant Genes (LEAs)
Late embryogenesis abundant (LEA) proteins described as multi-functional stress proteins that play active and significant roles in the stress resistance of plants via their function in defencing against dehydration caused by drought and salt (Close, 1996; Tunnacliffe and Wise, 2007). When plants are exposed to salinity, drought, or cold stresses, LEA2 protein members typically accumulated in dehydrating plant seeds and/or in tissues (Veeranagamallaiah et al., 2011), while dehydrins (DHNs) referred to as RAB proteins could be induced in vegetative tissues (Hundertmark and Hincha, 2008; Zhang et al., 2020). The expression of HVA1, a LEA3 protein from barley (Hordeum vulgare L.) conferred tolerance of transgenic rice plants to salt stress (Xu et al., 1996). Lee et al. (2013) found that, in E. salsugineum, 52 LEA protein sequences represented a high sequence similarity with the orthologs in Arabidopsis (Lee et al., 2013). Furthermore, 64 E. salsugineum LEA proteins were identified to be classified into eight families, based on transcriptome sequencing, as well as the domain structures, numbers, and composition of conserved repeat motifs in LEAs. The remarkably induced expression of EsLEA1, EsLEA4, EsLEA6, and EsDehydrin genes was revealed in leaves and/or roots, which implies that the expression of LEA proteins could be one of the important strategies for E. salsugineum to deal with salt stress (Li et al., 2021).
Eutrema salsugineum (Eutrema halophilum) EsEm1 (EhEm1) is a hydrophilic protein and contains three different 20-mer conserved motifs of N-domain, M-domain, and C-domain. EsEm1gene was revealed to be categorized into group 1 LEA proteins, and its expression was dramatically induced by salt, dehydration, abscisic acid (ABA), and cold stresses in young seedlings (Xiang and Man, 2018; Table 1). Overexpression of the EhEm1 gene in rice demonstrated an enhanced tolerance to salt and drought stresses in transgenic plants, which was based on the better germination performances, higher survival rates, more accumulation of proline and increased peroxidase activity, as well as the reduced chlorophyll damage and less accumulation of malondialdehyde (Xiang and Man, 2018). And the EsEm1 gene overexpressed in rice led to an upregulated transcript levels of several stress-responsive genes of OsCDPK6, OsCDPK9, OsCDPK13, and rab16a under both salt and drought conditions. In addition, EsEm1 protein could effectively prevent the lactate dehydrogenase (LDH) enzyme from inactivation caused by salt and desiccation (Xiang and Man, 2018). Therefore, it is suggested that the EsEm1 gene was associated with an enhanced salt and drought tolerance in transgenic rice, possibly through its contribution to the stabilization of LDH activity, and the up-regulation of OsCDPK genes (Xiang and Man, 2018). In addition, the LEA protein gene EsLEA1 (TsLEA1, EU365627) was characterized as a salt induced gene, and it could improve the survival of salt-sensitive yeast cells, as well as the salt tolerance of transgenic Arabidopsis plants when ectopically expressed (Zhang et al., 2012; Table 1). Taken together, it was suggested that the increased expression of LEA protein families acts as protects plants from dehydrating damage, which probably is one of the outstanding mechanisms for adaptation of salt cress to the salt environment.
Conclusions and Perspectives
Salt cress (E. salsugineum or E. parvula), a close relative of the model plant A. thaliana, occurs and thrives in harsh environments, thus a number of factors contribute to its salt stress tolerance. First, the duplicated gene pairs, particularly the expansion of important resistance genes such as ion transport-related genes in the genome of Eutrema may be considered an important adaptive benefit; the generated sub-functionalization and neo-functionalization of duplicated genes could be factors for salt tolerance of salt cress (Hahn, 2009; Kondrashov, 2012; Monihan et al., 2019; Figure 2). Second, the transcriptional and metabolic comparisons of salt tolerant Eutrema and susceptible Arabidopsis pointed to a stress-anticipatory preparedness with constitutively high expression of the defense genes and relatively few salt-responsive genes detected, accompanied by preformed osmolyte of proline and secondary compounds, and lending metabolite configuration to fit osmoprotectant. And a precise defense strategy facilitates to conservation of energy that makes this halophyte to adapt various extreme environments (Figure 2). Third, E. salsugineum emerging as the halophyte, a major advantage appears to be their exceptional control over Na+ influx together with export mechanisms, the ability to coordinate its distribution to various tissues, and efficient sequestration of Na+ into vacuoles (Figure 2). Fourthly, the combined numerous studies revealed that the key components of underlying mechanisms in salt tolerance of the halophytic Eutrema, which is involved in a stronger antioxidative enzymatic system and substantially higher levels of thioredoxin, as well as the superior ability to sustain a highly reduced states of plastoquinone (PQ) pool and P700 under salt stress; the constitutively expressed protection systems against oxidative stress will facilitate to maintain a relatively stable stomatal conductance and CO2 assimilation, and enhanced chlorophyll content to keep almost unaffected net photosynthetic rate (Figure 2). Finally, the stress-related genes such as EsSOS1, EsEm1, and EsTIP1;2 that are expressed in transgenic systems led to enhanced tolerance to salinity stress, which indicates they play critical roles in the salt tolerance of E. salsugineum.
Tremendous genetic diversity that exists in the halophytic Eutrema can be exploited (Koornneef et al., 2004; Gong et al., 2005; Bohnert et al., 2006). And the comparative and functional genomics from this halophyte species have provided important lessons on stress tolerance. Also, overexpression of some abiotic stress-responsive genes from this halophyte into the transgenic model plants and crops has been achieved limited success. However, due to the fact that abiotic stress tolerance cannot be manipulated by a single quantitative trait locus (QTL), and genes could be a part of several pathways at different weights, thus the analysis of a vast of data is needed to explore the complicated regulating network in plants (Rajalakshmi and Parida, 2012). Considering the multigenic nature of abiotic stress tolerance, researchers are working toward regulating networks with more than one kind of gene implicated in stress tolerance, to improve the efficacies of transgenic systems (Rajalakshmi and Parida, 2012).
Going forward, these genetic variations of E. salsugineum are providing valuable resources for further deciphering mechanisms underlying the stress tolerance and local adaptations of this halophytic species (Wang et al., 2021). Moreover, Eutrema plants undoubtedly have the potential to serve as a rich repository for genes, and the QTL-based approaches in halophytes may be utilized in breeding for abiotic stress tolerance (Rajalakshmi and Parida, 2012). Herein, we provide the recent scientific advance underlying the halophytic Eutrema that makes exploration of these fundamental adaptive mechanisms of salt stress more feasible, and to gain opportunities for improving the production of crops in unfavorable salt environments.
Author Contributions
QZ conceived the manuscript. QZ and YanZ wrote the manuscript with contributions from CL, CD, HZ, YaoZ, and ZM. CL and CD performed the collection of documents and arrangement of data. All authors have read and approved the final manuscript.
Funding
This work was supported by the Key Technology Research and Development Program of Shandong (2019GSF107089), the Key Program of Nature Science Foundation of Shandong Province (ZR2020KC018), the Granary Science and Technology Demonstration project in the Bohai Sea (2019BHLC001), and the National Major Science and Technology Project of China (2018ZX08009-10B-004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Achard, P., Gong, F., Cheminant, S., Alioua, M., Hedden, P., and Genschik, P. (2008). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20, 2117–2129. doi: 10.1105/tpc.108.058941
Alexandersson, E., Fraysse, L., Sjövall-Larsen, S., Gustavsson, S., Fellert, M., Karlsson, M., et al. (2005). Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol. 59, 469–484. doi: 10.1007/s11103-005-0352-1
Ali, A., Cheol Park, H., Aman, R., Ali, Z., and Yun, D.-J. (2013). Role of HKT1 in Thellungiella salsuginea, a model extremophile plant. Plant Signal. Behav. 8:e25196. doi: 10.4161/psb.25196
Ali, A., Khan, I. U., Jan, M., Khan, H. A., Hussain, S., Nisar, M., et al. (2018). The high-affinity potassium transporter EpHKT1; 2 from the extremophile Eutrema parvula mediates salt tolerance. Front. Plant Sci. 9:1108. doi: 10.3389/fpls.2018.01108
Ali, Z., Park, H. C., Ali, A., Oh, D.-H., Aman, R., Kropornicka, A., et al. (2012). TsHKT1; 2, a HKT1 homolog from the extremophile Arabidopsis relative Thellungiella salsuginea, shows K+ specificity in the presence of NaCl. Plant Physiol. 158, 1463–1474. doi: 10.1104/pp.111.193110
Amtmann, A. (2009). Learning from evolution: thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol. Plant. 2, 3–12. doi: 10.1093/mp/ssn094
Amtmann, A., Bohnert, H. J., and Bressan, R. A. (2005). Abiotic stress and plant genome evolution. Search for new models. Plant Physiol. 138, 127–130. doi: 10.1104/pp.105.059972
Apse, M. P., Sottosanto, J. B., and Blumwald, E. (2003). Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 36, 229–239. doi: 10.1046/j.1365-313X.2003.01871.x
Aroca, R., Porcel, R., and Ruiz-Lozano, J. M. (2012). Regulation of root water uptake under abiotic stress conditions. J. Exp. Bot. 63, 43–57. doi: 10.1093/jxb/err266
Baker, N. R. (1991). A possible role for photosystem II in environmental perturbations of photosynthesis. Physiol. Plant. 81, 563–570. doi: 10.1111/j.1399-3054.1991.tb05101.x
Bartels, D., and Dinakar, C. (2013). Balancing salinity stress responses in halophytes and non-halophytes: a comparison between Thellungiella and Arabidopsis thaliana. Funct. Plant Biol. 40, 819–831. doi: 10.1071/FP12299
Ben Rejeb, K., Lefebvre-De Vos, D., Le Disquet, I., Leprince, A. S., Bordenave, M., Maldiney, R., et al. (2015). Hydrogen peroxide produced by NADPH oxidases increases proline accumulation during salt or mannitol stress in Arabidopsis thaliana. New Phytol. 208, 1138–1148. doi: 10.1111/nph.13550
Bhaskaran, S., and Savithramma, D. (2011). Co-expression of Pennisetum glaucum vacuolar Na+/H+ antiporter and Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic tomato. J. Exp. Bot. 62, 5561–5570. doi: 10.1093/jxb/err237
Blanc, G., and Wolfe, K. H. (2004a). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16, 1679–1691. doi: 10.1105/tpc.021410
Blanc, G., and Wolfe, K. H. (2004b). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16, 1667–1678. doi: 10.1105/tpc.021345
Bohnert, H. J., Gong, Q., Li, P., and Ma, S. (2006). Unraveling abiotic stress tolerance mechanisms-getting genomics going. Curr. Opin. Plant Biol. 9, 180–188. doi: 10.1016/j.pbi.2006.01.003
Bose, J., Munns, R., Shabala, S., Gilliham, M., Pogson, B., and Tyerman, S. D. (2017). Chloroplast function and ion regulation in plants growing on saline soils: lessons from halophytes. J. Exp. Bot. 68, 3129–3143. doi: 10.1093/jxb/erx142
Bose, J., Rodrigo-Moreno, A., and Shabala, S. (2014). ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 65, 1241–1257. doi: 10.1093/jxb/ert430
Boursiac, Y., Boudet, J., Postaire, O., Luu, D. T., Tournaire-Roux, C., and Maurel, C. (2008). Stimulus-induced downregulation of root water transport involves reactive oxygen species-activated cell signalling and plasma membrane intrinsic protein internalization. Plant J. 56, 207–218. doi: 10.1111/j.1365-313X.2008.03594.x
Boyer, J. S. (1982). Plant productivity and environment. Science 218, 443–448. doi: 10.1126/science.218.4571.443
Bressan, R. A., Park, H. C., Orsini, F., Oh, D.-,h., Dassanayake, M., Inan, G., et al. (2013). Biotechnology for mechanisms that counteract salt stress in extremophile species: a genome-based view. Plant Biotechnol. Rev. 7, 27–37. doi: 10.1007/s11816-012-0249-9
Bressan, R. A., Zhang, C., Zhang, H., Hasegawa, P. M., Bohnert, H. J., and Zhu, J.-K. (2001). Learning from the Arabidopsis experience. The next gene search paradigm. Plant Physiol. 127, 1354–1360. doi: 10.1104/pp.010752
Broin, M., Cuiné, S., Peltier, G., and Rey, P. (2000). Involvement of CDSP 32, a drought-induced thioredoxin, in the response to oxidative stress in potato plants. FEBS Lett. 467, 245–248. doi: 10.1016/S0014-5793(00)01165-0
Chaves, M. M., Flexas, J., and Pinheiro, C. (2009). Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann. Bot. 103, 551–560. doi: 10.1093/aob/mcn125
Cheeseman, J. M. (2015). The evolution of halophytes, glycophytes and crops, and its implications for food security under saline conditions. New Phytol. 206, 557–570. doi: 10.1111/nph.13217
Chung, J. S., Zhu, J. K., Bressan, R. A., Hasegawa, P. M., and Shi, H. (2008). Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis. Plant J. 53, 554–565. doi: 10.1111/j.1365-313X.2007.03364.x
Close, T. J. (1996). Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol. Plant 97, 795–803. doi: 10.1111/j.1399-3054.1996.tb00546.x
Czarnocka, W., and Karpiński, S. (2018). Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 122, 4–20. doi: 10.1016/j.freeradbiomed.2018.01.011
Dassanayake, M., Oh, D.-H., Haas, J. S., Hernandez, A., Hong, H., Ali, S., et al. (2011). The genome of the extremophile crucifer Thellungiella parvula. Nat. Genet. 43, 913–918. doi: 10.1038/ng.889
Dos Santos, C. V., and Rey, P. (2006). Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci. 11, 329–334. doi: 10.1016/j.tplants.2006.05.005
Eshel, G., Shaked, R., Kazachkova, Y., Khan, A., Eppel, A., Cisneros, A., et al. (2017). Anastatica hierochuntica, an Arabidopsis desert relative, is tolerant to multiple abiotic stresses and exhibits species-specific and common stress tolerance strategies with its halophytic relative, Eutrema (Thellungiella) salsugineum. Front. Plant Sci. 7:1992. doi: 10.3389/fpls.2016.01992
Fang, Y., Xie, K., Hou, X., Hu, H., and Xiong, L. (2010). Systematic analysis of GT factor family of rice reveals a novel subfamily involved in stress responses. Mol. Genet. Genomics. 283, 157–169. doi: 10.1007/s00438-009-0507-x
Fichman, Y., and Mittler, R. (2020). Rapid systemic signaling during abiotic and biotic stresses: is the ROS wave master of all trades? Plant J. 102, 887–896. doi: 10.1111/tpj.14685
Flexas, J., Bota, J., Loreto, F., Cornic, G., and Sharkey, T. (2004). Diffusive and metabolic limitations to photosynthesis under drought and salinity in C3 plants. Plant Biol. 6, 269–279. doi: 10.1055/s-2004-820867
Flexas, J., DIAZ-ESPEJO, A., Galmes, J., Kaldenhoff, R., Medrano, H., and Ribas-Carbo, M. (2007). Rapid variations of mesophyll conductance in response to changes in CO2 concentration around leaves. Plant Cell Environ. 30, 1284–1298. doi: 10.1111/j.1365-3040.2007.01700.x
Flowers, T., Hajibagheri, M., and Clipson, N. (1986). Halophytes. Q. Rev. Biol. 61, 313–337. doi: 10.1086/415032
Flowers, T. J., and Colmer, T. D. (2008). Salinity tolerance in halophytes. New Phytol. 945–963. doi: 10.1111/j.1469-8137.2008.02531.x
Flowers, T. J., Munns, R., and Colmer, T. D. (2015). Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann. Bot. 115, 419–431. doi: 10.1093/aob/mcu217
Fowler, S. G., Cook, D., and Thomashow, M. F. (2005). Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 137, 961–968. doi: 10.1104/pp.104.058354
Foyer, C. H., Ruban, A. V., and Noctor, G. (2017). Viewing oxidative stress through the lens of oxidative signalling rather than damage. Biochem. J. 474, 877–883. doi: 10.1042/BCJ20160814
Fu, M., Kang, H. K., Son, S.-H., Kim, S.-K., and Nam, K. H. (2014). A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA. Plant Cell Physiol. 55, 1892–1904. doi: 10.1093/pcp/pcu118
Gai, Y.-P., Yuan, S.-S., Zhao, Y.-N., Zhao, H.-N., Zhang, H.-L., and Ji, X.-L. (2018). A novel LncRNA, MuLnc1, associated with environmental stress in Mulberry (Morus multicaulis). Front. Plant Sci. 9:669. doi: 10.3389/fpls.2018.00669
Gandour, M., Gharred, J., Taamalli, W., and Abdelly, C. (2019). Comparison of salinity tolerance in geographically diverse collections of Thellungiella accessions. Russ. J. Ecol. 50, 249–255. doi: 10.1134/S1067413619030020
Gao, F., Chen, J., Ma, T., Li, H., Wang, N., Li, Z., et al. (2014). The glutathione peroxidase gene family in Thellungiella salsuginea: genome-wide identification, classification, and gene and protein expression analysis under stress conditions.Int. J. Mol. Sci. 15, 3319–3335. doi: 10.3390/ijms15023319
Gaxiola, R. A., Li, J., Undurraga, S., Dang, L. M., Allen, G. J., Alper, S. L., et al. (2001). Drought-and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc. Natl. Acad. Sci. U.S.A. 98, 11444–11449. doi: 10.1073/pnas.191389398
Gechev, T. S., Dinakar, C., Benina, M., Toneva, V., and Bartels, D. (2012). Molecular mechanisms of desiccation tolerance in resurrection plants. Cell. Mol. Life Sci. 69, 3175–3186. doi: 10.1007/s00018-012-1088-0
Gong, Q., Li, P., Ma, S., Indu Rupassara, S., and Bohnert, H. J. (2005). Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J. 44, 826–839. doi: 10.1111/j.1365-313X.2005.02587.x
Goyal, K., Walton, L. J., and Tunnacliffe, A. (2005). LEA proteins prevent protein aggregation due to water stress. Biochem. J. 388, 151–157. doi: 10.1042/BJ20041931
Griffith, M., Timonin, M., Wong, A. C., Gray, G. R., Akhter, S. R., Saldanha, M., et al. (2007). Thellungiella: an Arabidopsis-related model plant adapted to cold temperatures. Plant Cell Environ. 30, 529–538. doi: 10.1111/j.1365-3040.2007.01653.x
Grigore, M.-N. (2019). “Defining halophytes-A conceptual and historical approach in an ecological frame,” in Halophytes and Climate Change: Adaptive Mechanisms and Potential Uses, eds M. Hasanuzzaman, S. Shabala, and M. Fujita (Wallingford: CABI), 3–18. doi: 10.1079/9781786394330.0003
Guevara, D. R., Champigny, M. J., Tattersall, A., Dedrick, J., Wong, C. E., Li, Y., et al. (2012). Transcriptomic and metabolomic analysis of Yukon Thellungiella plants grown in cabinets and their natural habitat show phenotypic plasticity. BMC Plant Biol. 12:175. doi: 10.1186/1471-2229-12-175
Guo, L., Wang, Z. Y., Lin, H., Cui, W. E., Chen, J., Liu, M., et al. (2006). Expression and functional analysis of the rice plasma-membrane intrinsic protein gene family. Cell Res. 16, 277–286. doi: 10.1038/sj.cr.7310035
Hahn, M. W. (2009). Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 100, 605–617. doi: 10.1093/jhered/esp047
Holmgren, A. (1985). Thioredoxin. Annu. Rev. Plant Biol. 54, 237–271. doi: 10.1146/annurev.bi.54.070185.001321
Hou, Q., and Bartels, D. (2015). Comparative study of the aldehyde dehydrogenase (ALDH) gene superfamily in the glycophyte Arabidopsis thaliana and Eutrema halophytes. Ann. Bot. 115, 465–479. doi: 10.1093/aob/mcu152
Hundertmark, M., and Hincha, D. K. (2008). LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9:188. doi: 10.1186/1471-2164-9-118
Inan, G., Zhang, Q., Li, P., Wang, Z., Cao, Z., Zhang, H., et al. (2004). Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol. 135, 1718–1737. doi: 10.1104/pp.104.041723
Janz, D., Behnke, K., Schnitzler, J.-P., Kanawati, B., Schmitt-Kopplin, P., and Polle, A. (2010). Pathway analysis of the transcriptome and metabolome of salt sensitive and tolerant poplar species reveals evolutionary adaption of stress tolerance mechanisms. BMC Plant Biol. 10:150. doi: 10.1186/1471-2229-10-150
Jarvis, D. E., Ryu, C.-H., Beilstein, M. A., and Schumaker, K. S. (2014). Distinct roles for SOS1 in the convergent evolution of salt tolerance in Eutrema salsugineum and Schrenkiella parvula. Mol. Biol. Evol. 31, 2094–2107. doi: 10.1093/molbev/msu152
Javot, H., Lauvergeat, V., Santoni, V., Martin-Laurent, F., Güçlü, J., Vinh, J., et al. (2003). Role of a single aquaporin isoform in root water uptake. Plant Cell. 15, 509–522. doi: 10.1105/tpc.008888
John, U. P., and Spangenberg, G. C. (2005). Xenogenomics: genomic bioprospecting in indigenous and exotic plants through EST discovery, cDNA microarray-based expression profiling and functional genomics. Comp. Funct. Genomics 6, 230–235. doi: 10.1002/cfg.475
Kant, S., Kant, P., Raveh, E., and Barak, S. (2006). Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T. halophila. Plant Cell Environ. 29, 1220–1234. doi: 10.1111/j.1365-3040.2006.01502.x
Kaur, G., Guruprasad, K., Temple, B. R., Shirvanyants, D. G., Dokholyan, N. V., and Pati, P. K. (2018). Structural complexity and functional diversity of plant NADPH oxidases. Amino Acids 50, 79–94. doi: 10.1007/s00726-017-2491-5
Kazachkova, Y., Batushansky, A., Cisneros, A., Tel-Zur, N., Fait, A., and Barak, S. (2013). Growth platform-dependent and-independent phenotypic and metabolic responses of Arabidopsis and its halophytic relative, Eutrema salsugineum, to salt stress. Plant Physiol. 162, 1583–1598. doi: 10.1104/pp.113.217844
Kazachkova, Y., Eshel, G., Pantha, P., Cheeseman, J. M., Dassanayake, M., and Barak, S. (2018). Halophytism: what have we learnt from Arabidopsis thaliana relative model systems? Plant Physiol. 178, 972–988. doi: 10.1104/pp.18.00863
Kirsch, M., Zhigang, A., Viereck, R., Löw, R., and Rausch, T. (1996). Salt stress induces an increased expression of V-type H+-ATPase in mature sugar beet leaves. Plant Mol. Biol. 32, 543–547. doi: 10.1007/BF00019107
Koch, M. A., and German, D. A. (2013). Taxonomy and systematics are key to biological information: Arabidopsis, Eutrema (Thellungiella), Noccaea and Schrenkiella (Brassicaceae) as examples. Front. Plant Sci. 4:267. doi: 10.3389/fpls.2013.00267
Kondrashov, F. A. (2012). Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. R. Soc. B 279, 5048–5057. doi: 10.1098/rspb.2012.1108
Kong, Y., Elling, A. A., Chen, B., and Deng, X. (2010). Differential expression of microRNAs in maize inbred and hybrid lines during salt and drought stress. Am. J. Plant Physiol. 1:69. doi: 10.4236/ajps.2010.12009
Koornneef, M., Alonso-Blanco, C., and Vreugdenhil, D. (2004). Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55, 141–172. doi: 10.1146/annurev.arplant.55.031903.141605
Kumar, V., Khare, T., Sharma, M., and Wani, S. H. (2017). “ROS-induced signaling and gene expression in crops under salinity stress,” in Reactive Oxygen Species and Antioxidant Systems in Plants: Role and Regulation Under Abiotic Stress, eds M. I. R. Khan and N. A. Khan (Singapore: Springer), 159–184. doi: 10.1007/978-981-10-5254-5_7
Lamdan, N. L., Attia, Z., Moran, N., and Moshelion, M. (2012). The Arabidopsis-related halophyte Thellungiella halophila: boron tolerance via boron complexation with metabolites? Plant Cell Environ. 35, 735–746. doi: 10.1111/j.1365-3040.2011.02447.x
Larcher, W. (2003). Physiological Plant Ecology: Ecophysiology and Stress Physiology of Functional Groups, 4th Edn. New York, NY: Springer.
Lawlor, D. W., and Cornic, G. (2002). Photosynthetic carbon assimilation and associated metabolism in relation to water deficits in higher plants. Plant Cell Environ. 25, 275–294. doi: 10.1046/j.0016-8025.2001.00814.x
Lee, Y. P., Funk, C., Erban, A., Kopka, J., Köhl, K. I., Zuther, E., et al. (2016). Salt stress responses in a geographically diverse collection of Eutrema/Thellungiella spp. accessions. Funct. Plant Biol. 43, 590–606. doi: 10.1071/FP15285
Lee, Y. P., Giorgi, F. M., Lohse, M., Kvederaviciute, K., Klages, S., Usadel, B., et al. (2013). Transcriptome sequencing and microarray design for functional genomics in the extremophile Arabidopsis relative Thellungiella salsuginea (Eutrema salsugineum). BMC Genomics 14:793. doi: 10.1186/1471-2164-14-793
Li, C., Qi, Y., Zhao, C., Wang, X., and Zhang, Q. (2021). Transcriptome profiling of the salt stress response in the leaves and roots of halophytic Eutrema salsugineum. Front. Genet. 12:770742. doi: 10.3389/fgene.2021.770742
Li, G., Santoni, V., and Maurel, C. (2014). Plant aquaporins: roles in plant physiology. Biochim. Biophys. Acta Gen. Subj. 1840, 1574–1582. doi: 10.1016/j.bbagen.2013.11.004
Li, P., Mane, S. P., Sioson, A. A., Robinet, C. V., Heath, L. S., Bohnert, H. J., et al. (2006). Effects of chronic ozone exposure on gene expression in Arabidopsis thaliana ecotypes and in Thellungiella halophila. Plant Cell Environ. 29, 854–868. doi: 10.1111/j.1365-3040.2005.01465.x
Li, W., Cui, X., Meng, Z., Huang, X., Xie, Q., Wu, H., et al. (2012). Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 158, 1279–1292. doi: 10.1104/pp.111.188789
Li, X., Wang, X., Yang, Y., Li, R., He, Q., Fang, X., et al. (2011). Single-molecule analysis of PIP2; 1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell 23, 3780–3797. doi: 10.1105/tpc.111.091454
Lisso, J., Altmann, T., and Müssig, C. (2006). The AtNFXL1 gene encodes a NF-X1 type zinc finger protein required for growth under salt stress. FEBS Lett. 580, 4851–4856. doi: 10.1016/j.febslet.2006.07.079
Liu, H.-H., Tian, X., Li, Y.-J., Wu, C.-A., and Zheng, C.-C. (2008). Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14, 836–843. doi: 10.1261/rna.895308
Lugan, R., Niogret, M. F., Leport, L., Guégan, J. P., Larher, F. R., Savouré, A., et al. (2010). Metabolome and water homeostasis analysis of Thellungiella salsuginea suggests that dehydration tolerance is a key response to osmotic stress in this halophyte. Plant J. 64, 215–229. doi: 10.1111/j.1365-313X.2010.04323.x
Ma, C., Burd, S., and Lers, A. (2015). miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 84, 169–187. doi: 10.1111/tpj.12999
Ma, L., Zhang, H., Sun, L., Jiao, Y., Zhang, G., Miao, C., et al. (2012). NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. J. Exp. Bot. 63, 305–317. doi: 10.1093/jxb/err280
Maere, S., De Bodt, S., Raes, J., Casneuf, T., Van Montagu, M., Kuiper, M., et al. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 5454–5459. doi: 10.1073/pnas.0501102102
Maeshima, M. (2000). Vacuolar H+-pyrophosphatase. Biochim. Biophys. Acta Biomembr. 1465, 37–51. doi: 10.1016/S0005-2736(00)00130-9
Martinière, A., Li, X., Runions, J., Lin, J., Maurel, C., and Luu, D.-T. (2012). Salt stress triggers enhanced cycling of Arabidopsis root plasma-membrane aquaporins. Plant Signal. Behav. 7, 529–532. doi: 10.4161/psb.19350
Martre, P., Morillon, R., Barrieu, F., North, G. B., Nobel, P. S., and Chrispeels, M. J. (2002). Plasma membrane aquaporins play a significant role during recovery from water deficit. Plant Physiol. 130, 2101–2110. doi: 10.1104/pp.009019
Maurel, C., Santoni, V., Luu, D.-T., Wudick, M. M., and Verdoucq, L. (2009). The cellular dynamics of plant aquaporin expression and functions. Curr. Opin. Plant Biol. 12, 690–698. doi: 10.1016/j.pbi.2009.09.002
Maurel, C., Verdoucq, L., Luu, D.-T., and Santoni, V. (2008). Plant aquaporins: membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 59, 595–624. doi: 10.1146/annurev.arplant.59.032607.092734
Mercer, T. R., Dinger, M. E., and Mattick, J. S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159. doi: 10.1038/nrg2521
Mittal, D., Sharma, N., Sharma, V., Sopory, S., and Sanan-Mishra, N. (2016). Role of microRNAs in rice plant under salt stress. Ann. Appl. Biol. 168, 2–18. doi: 10.1111/aab.12241
Møller, I. M., Jensen, P. E., and Hansson, A. (2007). Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 58, 459–481. doi: 10.1146/annurev.arplant.58.032806.103946
Monihan, S. M., Ryu, C.-H., Magness, C. A., and Schumaker, K. S. (2019). Linking duplication of a calcium sensor to salt tolerance in Eutrema salsugineum. Plant Physiol. 179, 1176–1192. doi: 10.1104/pp.18.01400
M'rah, S., Ouerghi, Z., Berthomieu, C., Havaux, M., Jungas, C., Hajji, M., et al. (2006). Effects of NaCl on the growth, ion accumulation and photosynthetic parameters of Thellungiella halophila. J. Plant Physiol. 163, 1022–1031. doi: 10.1016/j.jplph.2005.07.015
M'rah, S., Ouerghi, Z., Eymery, F., Rey, P., Hajji, M., Grignon, C., et al. (2007). Efficiency of biochemical protection against toxic effects of accumulated salt differentiates Thellungiella halophila from Arabidopsis thaliana. J. Plant Physiol. 164, 375–384. doi: 10.1016/j.jplph.2006.07.013
Munns, R., James, R. A., and Läuchli, A. (2006). Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 57, 1025–1043. doi: 10.1093/jxb/erj100
Niewiadomska, E., and Wiciarz, M. (2015). “Adaptations of chloroplastic metabolism in halophytic plants,” in Progress in Botany, Vol. 76, eds U. Lüttge and W. Beyschlag (Cham: Springer International Publishing), 177–193. doi: 10.1007/978-3-319-08807-5_7
Ogawa, D., Yamaguchi, K., and Nishiuchi, T. (2007). High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J. Exp. Bot. 58, 3373–3383. doi: 10.1093/jxb/erm184
Oh, D.-H., Dassanayake, M., Bohnert, H. J., and Cheeseman, J. M. (2013). Life at the extreme: lessons from the genome. Genome Biol. 13, 1–9. doi: 10.1186/gb-2012-13-3-241
Oh, D.-H., Leidi, E., Zhang, Q., Hwang, S.-M., Li, Y., Quintero, F. J., et al. (2009). Loss of halophytism by interference with SOS1 expression. Plant Physiol. 151, 210–222. doi: 10.1104/pp.109.137802
Orsini, F., D'Urzo, M. P., Inan, G., Serra, S., Oh, D.-H., Mickelbart, M. V., et al. (2010). A comparative study of salt tolerance parameters in 11 wild relatives of Arabidopsis thaliana. J. Exp. Bot. 61, 3787–3798. doi: 10.1093/jxb/erq188
Ozgur, R., Uzilday, B., Sekmen, A. H., and Turkan, I. (2013). Reactive oxygen species regulation and antioxidant defence in halophytes. Funct. Plant Biol. 40, 832–847. doi: 10.1071/FP12389
Ozyigit, I. I., Filiz, E., Vatansever, R., Kurtoglu, K. Y., Koc, I., Öztürk, M.X., et al. (2016). Identification and comparative analysis of H2O2-scavenging enzymes (ascorbate peroxidase and glutathione peroxidase) in selected plants employing bioinformatics approaches. Front. Plant Sci. 7:301. doi: 10.3389/fpls.2016.00301
Pang, Q., Chen, S., Dai, S., Chen, Y., Wang, Y., and Yan, X. (2010). Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J. Proteome Res. 9, 2584–2599. doi: 10.1021/pr100034f
Péret, B., Li, G., Zhao, J., Band, L. R., Vo,ß, U., Postaire, O., et al. (2012). Auxin regulates aquaporin function to facilitate lateral root emergence. Nat. Cell Biol. 14, 991–998. doi: 10.1038/ncb2573
Pilarska, M., Bartels, D., and Niewiadomska, E. (2021). Differential regulation of NAPDH oxidases in salt-tolerant Eutrema salsugineum and salt-sensitive Arabidopsis thaliana. Int. J. Mol. Sci. 22, 10341. doi: 10.3390/ijms221910341
Pilarska, M., Wiciarz, M., Jaji,ć, I., Kozieradzka-Kiszkurno, M., Dobrev, P., Vankov,á, R., et al. (2016). A different pattern of production and scavenging of reactive oxygen species in halophytic Eutrema salsugineum (Thellungiella salsuginea) plants in comparison to Arabidopsis thaliana and its relation to salt stress signaling. Front. Plant Sci. 7:1179. doi: 10.3389/fpls.2016.01179
Prak, S., Hem, S., Boudet, J., Viennois, G., Sommerer, N., Rossignol, M., et al. (2008). Multiple phosphorylations in the C-terminal tail of plant plasma membrane aquaporins: role in subcellular trafficking of AtPIP2; 1 in response to salt stress. Mol. Cell Proteomics 7, 1019–1030. doi: 10.1074/mcp.M700566-MCP200
Qiang, X.-J., Yu, G. H., Jiang, L. L., Sun, L. L., Zhang, S. H., Li, W., et al. (2015). Thellungiella halophila ThPIP1 gene enhances the tolerance of the transgenic rice to salt stress. J. Integr. Agric. 14, 1911–1922. doi: 10.1016/S2095-3119(15)61045-0
Qin, S., Liu, Y., Han, Y., Xu, G., Wan, S., Cui, F., et al. (2019). Aquaporins and their function in root water transport under salt stress conditions in Eutrema salsugineum. Plant Sci. 287:110199. doi: 10.1016/j.plantsci.2019.110199
Qiu, Q.-S., Barkla, B. J., Vera-Estrella, R., Zhu, J.-K., and Schumaker, K. S. (2003). Na+/H+ exchange activity in the plasma membrane of Arabidopsis. Plant Physiol. 132, 1041–1052. doi: 10.1104/pp.102.010421
Qiu, Q.-S., Guo, Y., Dietrich, M. A., Schumaker, K. S., and Zhu, J.-K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436–8441. doi: 10.1073/pnas.122224699
Quintero, F. J., Ohta, M., Shi, H., Zhu, J.-K., and Pardo, J. M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. U.S.A. 99, 9061–9066. doi: 10.1073/pnas.132092099
Rajalakshmi, S., and Parida, A. (2012). Halophytes as a source of genes for abiotic stress tolerance. J. Plant Biochem. Biotechnol. 21, 63–67. doi: 10.1007/s13562-012-0146-x
Rey, P., Cuiné, S., Eymery, F., Garin, J., Court, M., Jacquot, J. P., et al. (2005). Analysis of the proteins targeted by CDSP32, a plastidic thioredoxin participating in oxidative stress responses. Plant J. 41, 31–42. doi: 10.1111/j.1365-313X.2004.02271.x
Rus, A., Lee, B. H., Munoz-Mayor, A., Sharkhuu, A., Miura, K., Zhu, J.-K., et al. (2004). AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol. 136, 2500–2511. doi: 10.1104/pp.104.042234
Sairam, R., and Tyagi, A. (2004). Physiology and molecular biology of salinity stress tolerance in plants. Curr. Sci. 86, 407–421. doi: 10.1007/1-4020-4225-6
Sewelam, N., Jaspert, N., Van Der Kelen, K., Tognetti, V. B., Schmitz, J., Frerigmann, H., et al. (2014). Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Mol. Plant 7, 1191–1210. doi: 10.1093/mp/ssu070
Shi, H., Ishitani, M., Kim, C., and Zhu, J.-K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97, 6896–6901. doi: 10.1073/pnas.120170197
Shi, H., Quintero, F. J., Pardo, J. M., and Zhu, J.-K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14, 465–477. doi: 10.1105/tpc.010371
Simopoulos, C., MacLeod, M. J., Irani, S., Sung, W. W., Champigny, M. J., Summers, P. S., et al. (2020). Coding and long non-coding RNAs provide evidence of distinct transcriptional reprogramming for two ecotypes of the extremophile plant Eutrema salsugineum undergoing water deficit stress. BMC Genomics 21:396. doi: 10.1186/s12864-020-06793-7
Sohn, K. H., Lee, S. C., Jung, H. W., Hong, J. K., and Hwang, B. K. (2006). Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol. Biol. 61, 897–915. doi: 10.1007/s11103-006-0057-0
Srivastava, A. K., Srivastava, S., Lokhande, V. H., D'Souza, S. F., and Suprasanna, P. (2015). Salt stress reveals differential antioxidant and energetics responses in glycophyte (Brassica juncea L.) and halophyte (Sesuvium portulacastrum L.). Front. Environ. Sci. 3:19. doi: 10.3389/fenvs.2015.00019
Sui, N., and Han, G. (2014). Salt-induced photoinhibition of PSII is alleviated in halophyte Thellungiella halophila by increases of unsaturated fatty acids in membrane lipids. Acta Physiol. Plant 36, 983–992. doi: 10.1007/s11738-013-1477-5
Sun, X., Zheng, H., and Sui, N. (2018). Regulation mechanism of long non-coding RNA in plant response to stress. Biochem. Biophys. Res. Commun. 503, 402–407. doi: 10.1016/j.bbrc.2018.07.072
Sunkar, R., and Zhu, J.-K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019. doi: 10.1105/tpc.104.022830
Surówka, E., Latowski, D., Libik-Konieczny, M., and Miszalski, Z. (2019). “ROS signalling, and antioxidant defence network in halophytes,” in Halophytes and Climate Change: Adaptive Mechanisms and Potential Uses, eds M. Hasanuzzaman, S. Shabala, and M. Fujita (Boston, MA: CABI), 179–195. doi: 10.1079/9781786394330.0179
Sutka, M., Li, G., Boudet, J., Boursiac, Y., Doumas, P., and Maurel, C. (2011). Natural variation of root hydraulics in Arabidopsis grown in normal and salt-stressed conditions. Plant Physiol. 155, 1264–1276. doi: 10.1104/pp.110.163113
Taji, T., Sakurai, T., Mochida, K., Ishiwata, A., Kurotani, A., Totoki, Y., et al. (2008). Large-scale collection and annotation of full-length enriched cDNAs from a model halophyte, Thellungiella halophila. BMC Plant Biol. 8:115. doi: 10.1186/1471-2229-8-115
Taji, T., Seki, M., Satou, M., Sakurai, T., Kobayashi, M., Ishiyama, K., et al. (2004). Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol. 135, 1697–1709. doi: 10.1104/pp.104.039909
Tournaire-Roux, C., Sutka, M., Javot, H., Gout, E., Gerbeau, P., Luu, D.-T., et al. (2003). Cytosolic pH regulates root water transport during anoxic stress through gating of aquaporins. Nature 425, 393–397. doi: 10.1038/nature01853
Trindade, I., Capitão, C., Dalmay, T., Fevereiro, M. P., and Santos, D. M. (2010). miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 231, 705–716. doi: 10.1007/s00425-009-1078-0
Tunnacliffe, A., and Wise, M. J. (2007). The continuing conundrum of the LEA proteins. Naturwissenschaften 94, 791–812. doi: 10.1007/s00114-007-0254-y
Veeranagamallaiah, G., Prasanthi, J., Reddy, K. E., Pandurangaiah, M., Babu, O. S., and Sudhakar, C. (2011). Group 1 and 2 LEA protein expression correlates with a decrease in water stress induced protein aggregation in horsegram during germination and seedling growth. J. Plant Physiol. 168, 671–677. doi: 10.1016/j.jplph.2010.09.007
Vera-Estrella, R., Barkla, B. J., García-Ramírez, L., and Pantoja, O. (2005). Salt stress in Thellungiella halophila activates Na+ transport mechanisms required for salinity tolerance. Plant Physiol. 139, 1507–1517. doi: 10.1104/pp.105.067850
Verbruggen, N., and Hermans, C. (2008). Proline accumulation in plants: a review. Amino Acids 35, 753–759. doi: 10.1007/s00726-008-0061-6
Volkov, V., Wang, B., Dominy, P., Fricke, W., and Amtmann, A. (2004). Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, possesses effective mechanisms to discriminate between potassium and sodium. Plant Cell Environ. 27, 1–14. doi: 10.1046/j.0016-8025.2003.01116.x
Wang, L.-L., Chen, A.-P., Zhong, N.-Q., Liu, N., Wu, X.-M., Wang, F., et al. (2014). The Thellungiella salsuginea tonoplast aquaporin TsTIP1; 2 functions in protection against multiple abiotic stresses. Plant Cell Physiol. 55, 148–161. doi: 10.1093/pcp/pct166
Wang, X., Rao, H., Ma, J., Chen, X., Li, G., and Zhao, G. (2021). Genomic Variation Landscape of the Model Salt Cress Eutrema salsugineum. Front. Plant Sci. 12:700161. doi: 10.3389/fpls.2021.700161
Wang, Z.-L., Li, P. H., Fredricksen, M., Gong, Z. Z., Kim, C., Zhang, C., et al. (2004). Expressed sequence tags from Thellungiella halophila, a new model to study plant salt-tolerance. Plant Sci. 166, 609–616. doi: 10.1016/j.plantsci.2003.10.030
Wiciarz, M., Gubernator, B., Kruk, J., and Niewiadomska, E. (2015). Enhanced chloroplastic generation of H2O2 in stress-resistant Thellungiella salsuginea in comparison to Arabidopsis thaliana. Physiol. Plant 153, 467–476. doi: 10.1111/ppl.12248
Wiciarz, M., Niewiadomska, E., and Kruk, J. (2018). Effects of salt stress on low molecular antioxidants and redox state of plastoquinone and P700 in Arabidopsis thaliana (glycophyte) and Eutrema salsugineum (halophyte). Photosynthetica 56, 811–819. doi: 10.1007/s11099-017-0733-0
Wong, C., Li, Y., Whitty, B., Diaz-Camino, C., Akhter, S., Brandle, J., et al. (2005). Expressed sequence tags from the Yukon ecotype of Thellungiella reveal that gene expression in response to cold, drought and salinity shows little overlap. Plant Mol. Biol. 58, 561–574. doi: 10.1007/s11103-005-6163-6
Wong, C. E., Li, Y., Labbe, A., Guevara, D., Nuin, P., Whitty, B., et al. (2006). Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol. 140, 1437–1450. doi: 10.1104/pp.105.070508
Wu, H.-J., Zhang, Z., Wang, J.-Y., Oh, D.-H., Dassanayake, M., Liu, B., et al. (2012). Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl. Acad. Sci. U.S.A. 109, 12219–12224. doi: 10.1073/pnas.1209954109
Wu, Y., Guo, J., Cai, Y., Gong, X., Xiong, X., Qi, W., et al. (2016). Genome-wide identification and characterization of Eutrema salsugineum microRNAs for salt tolerance. Physiol. Plant 157, 453–468. doi: 10.1111/ppl.12419
Xiang, D., and Man, L. (2018). EhEm1, a novel Em-like protein from Eutrema halophilum, confers tolerance to salt and drought stresses in rice. Mol. Breed. 38, 1–15. doi: 10.1007/s11032-017-0750-5
Xie, Z.-M., Zou, H.-F., Lei, G., Wei, W., Zhou, Q.-Y., Niu, C.-F., et al. (2009). Soybean Trihelix transcription factors GmGT-2A and GmGT-2B improve plant tolerance to abiotic stresses in transgenic Arabidopsis. PLoS ONE 4:e6898. doi: 10.1371/journal.pone.0006898
Xu, D., Duan, X., Wang, B., Hong, B., Ho, T.-H. D., and Wu, R. (1996). Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 110, 249–257. doi: 10.1104/pp.110.1.249
Yang, R., Jarvis, D. J., Chen, H., Beilstein, M., Grimwood, J., Jenkins, J., et al. (2013). The reference genome of the halophytic plant Eutrema salsugineum. Front. Plant Sci. 4:46. doi: 10.3389/fpls.2013.00046
Yang, S., Luo, C., Song, Y., and Wang, J. (2016). Two groups of Thellungiella salsuginea RAVs exhibit distinct responses and sensitivity to salt and ABA in transgenic Arabidopsis. PLoS ONE 11:e0153517. doi: 10.1371/journal.pone.0153517
Zhang, H. F., Liu, S. Y., Ma, J. H., Wang, X. K., Haq, S. U., Meng, Y. C., et al. (2020). CaDHN4, a salt and cold stress-responsive dehydrin gene from pepper decreases abscisic acid sensitivity in Arabidopsis. Int. J. Mol. Sci. 21:26. doi: 10.3390/ijms21010026
Zhang, Q., Zhao, C., Li, M., Sun, W., Liu, Y., Xia, H., et al. (2013). Genome-wide identification of Thellungiella salsuginea microRNAs with putative roles in the salt stress response. BMC Plant Biol. 13:180. doi: 10.1186/1471-2229-13-180
Zhang, Y., Li, Y., Lai, J., Zhang, H., Liu, Y., Liang, L., et al. (2012). Ectopic expression of a LEA protein gene TsLEA1 from Thellungiella salsuginea confers salt-tolerance in yeast and Arabidopsis. Mol. Biol. Rep. 39, 4627–4633. doi: 10.1007/s11033-011-1254-8
Zhao, C., Zhang, H., Song, C., Zhu, J.-K., and Shabala, S. (2020). Mechanisms of plant responses and adaptation to soil salinity. Innovation 1:100017. doi: 10.1016/j.xinn.2020.100017
Zhou, L., Liu, Y., Liu, Z., Kong, D., Duan, M., and Luo, L. (2010). Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J. Exp. Bot. 61, 4157–4168. doi: 10.1093/jxb/erq237
Keywords: Arabidopsis relative model system, salt cress, salt stress tolerance, saline adaptation, antioxidant system, osmo-adaptation, ion homeostasis, gene expression
Citation: Li C, Duan C, Zhang H, Zhao Y, Meng Z, Zhao Y and Zhang Q (2022) Adaptative Mechanisms of Halophytic Eutrema salsugineum Encountering Saline Environment. Front. Plant Sci. 13:909527. doi: 10.3389/fpls.2022.909527
Received: 31 March 2022; Accepted: 01 June 2022;
Published: 28 June 2022.
Edited by:
Zhi Qi, Inner Mongolia University, ChinaReviewed by:
Katalin Solymosi, Eötvös Loránd University, HungaryWen-Cheng Liu, Henan University, China
Copyright © 2022 Li, Duan, Zhang, Zhao, Meng, Zhao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanxiu Zhao, zhaoyx@sdnu.edu.cn; Quan Zhang, zhangquan@sdnu.edu.cn
†These authors have contributed equally to this work
 Chuanshun Li
Chuanshun Li Chonghao Duan1†
Chonghao Duan1† Yanxiu Zhao
Yanxiu Zhao Quan Zhang
Quan Zhang