- 1Department of Botany, The University of British Columbia, Vancouver, BC, Canada
- 2Agriculture and Agri-Food Canada, Summerland Research & Development Center, Summerland, BC, Canada
The Flavin Monooxygenase (FMO) gene superfamily in plants is involved in various processes most widely documented for its involvement in auxin biosynthesis, specialized metabolite biosynthesis, and plant microbial defense signaling. The roles of FMOs in defense signaling and disease resistance have recently come into focus as they may present opportunities to increase immune responses in plants including leading to systemic acquired resistance, but are not well characterized. We present a comprehensive catalogue of FMOs found in genomes across vascular plants and explore, in depth, 170 wheat TaFMO genes for sequence architecture, cis-acting regulatory elements, and changes due to Transposable Element insertions. A molecular phylogeny separates TaFMOs into three clades (A, B, and C) for which we further report gene duplication patterns, and differential rates of homoeologue expansion and retention among TaFMO subclades. We discuss Clade B TaFMOs where gene expansion is similarly seen in other cereal genomes. Transcriptome data from various studies point towards involvement of subclade B2 TaFMOs in disease responses against both biotrophic and necrotrophic pathogens, substantiated by promoter element analysis. We hypothesize that certain TaFMOs are responsive to both abiotic and biotic stresses, providing potential targets for enhancing disease resistance, plant yield and other important agronomic traits. Altogether, FMOs in wheat and other crop plants present an untapped resource to be exploited for improving the quality of crops.
1 Introduction
Flavin-containing monooxygenases (FMOs; EC 1.14.13.8) constitute a ubiquitous class of ancient, highly-conserved enzymes that have been well-characterized in bacteria, fungi, and insects, catalyzing the transformations of many molecules across all domains of life (Eswaramoorthy et al., 2006; Huijbers et al., 2014; Mascotti et al., 2015). FMOs are dimeric in nature, with Rossman folds in a bi-lateral distribution containing FAD- and NAD(P)H-binding domains (Schlaich, 2007; Rossner et al., 2017; Thodberg and Jakobsen Neilson, 2020). Typically, FMOs act on small, nucleophilic substrates that contain either a sulfur (S) or nitrogen (N) atom, resulting in the oxygenation of a wide range of compounds; this process often aides in the detoxification of xenobiotic compounds in many organisms (Eswaramoorthy et al., 2006). While FMOs are well characterized in bacteria, fungi, and insects, less is known of many of the FMOs in vascular plants, despite abundant putative plant FMOs having been observed (Schlaich, 2007).
Multiple studies from the last two decades indicate that a distinct subset of plant FMOs have evolved to participate in biosynthesis of auxin, a crucial hormone for plant growth (Kendrew, 2001; Schlaich, 2007; Yamamoto et al., 2007; Zhao, 2014). This specialized group of plant FMOs has been coined ‘YUCCA’, named after the phenotype of a dominant Arabidopsis thaliana fmo deletion mutant resembling the yucca plant (Zhao et al., 2001) and many of which have been functionally characterized in this model plant (Cao et al., 2019; Qin et al., 2020). Due to the significance auxin biosynthesis has on plant growth, development, and yield in crop plants, many studies since then have focused on identifying and characterizing the orthologous ‘YUCCA’ FMOs across a range of commercially valuable plant species, such as apple, peach, soybean, and rice (Yamamoto et al., 2007; Qin et al., 2020; Song et al., 2020; Luo et al., 2022; Zhang et al., 2022). Additionally, a subset of FMOs in A. thaliana have been characterized for their activity in the S-oxygenation of sidechains in various glucosinolate (GSL) compounds (Li et al., 2011). GSLs are potent, specialized defense metabolites found exclusively in the order Brassicales known to play a major role against insect herbivory (Kong et al., 2016; Cang et al., 2018). Grouped as “FMO-GSOXs”, these FMOs, which participate in processing metabolites, play a significant defensive role in plants that produce GSLs. Of the 29 total identified FMOs in A. thaliana, eleven are annotated to be YUCCAs, twelve are purportedly FMO-GS-OXs, one has been reported to be crucial for systemic acquired resistance (SAR) against microbial pathogens (Hartmann et al., 2018; Czarnocka et al., 2020), and five do not presently have designated functions. A recent review (Mitchell and Weng, 2019) provided a brief synopsis of these three groups of plant FMOs. Together these studies reveal that FMOs play diverse biological responses in plants, much of which have yet to be characterized.
Outside A. thaliana, much less is known about non-YUCCA FMOs, particularly in cereal crops such as Triticum aestivum (common bread wheat), an essential food source for over 50% of the world’s population (Curtis and Halford, 2014). Yang et al. (Yang et al., 2021) focused on ‘YUCCA’ genes in wheat and identified 63 TaYUCCA FMO genes which they assigned to six subclades in a gene genealogy comparison; for a subset of these genes, they analyzed transcriptional activity using public transcriptomic data in an attempt to reveal function. Gaba et al. (2023) recently surveyed the genomes of ten different wild and cultivated rice species for FMO genes and revealed how little is presently known about FMOs outside the YUCCA group. Their study emphasized the need for more in-depth studies of other FMOs in crop plants.
Here, we address the extent to which FMOs have expanded across vascular plants by surveying comprehensively FMO-encoding genes across a broad range of plant taxa. We identified 170 likely wheat FMOs (TaFMOs) in the cultivar ‘Chinese Spring’ (the fully-sequenced reference genome, RefSeq v2.1), and sought to expand knowledge of the evolution, distribution, gene expansions, and potential functional diversity for different groups of TaFMO genes by using thorough phylogenetic, gene genealogy and transcriptome analyses using data obtained from publicly available studies. In addition, we take into account existing literature pertaining to TaYUCCAs (Li et al., 2014; Yang et al., 2021) and present a more cohesive picture of the biological roles that TaFMOs may play across a broad range of conditions, with a particular focus on various pathogen defense responses.
2 Materials and methods
2.1 Genome-wide identification of FMO genes
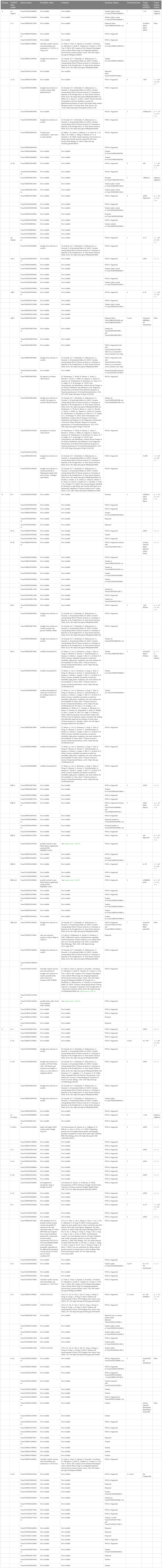
We obtained a protein-family hidden Markov model (HMM) profile for the FMO-like family (PF00743 v22) from the Protein Family (Pfam) database (https://pfam.xfam.org/), and conducted a local HMM search (HMMER v3.3.2; http://hmmer.org/) with significance parameters of E-value < 1e-2 against the International Wheat Genome Sequencing Consortium (IWGSC) fully-sequenced reference genomes for version RefSeqv2.1, to identify all putative wheat flavin-containing monooxygenase genes (TaFMOs) for the cultivar ‘Chinese Spring’. Results from HMMER were further consolidated with the use of query-based searches in both the online database Wheat Proteome (Duncan et al., 2017) with the search term ‘flavin-containing monooxygenase’ and the search term ‘flavin monooxygenase’ in the UniProt database (https://www.uniprot.org/). Similarly, all FMO candidate genes from other plant species spanning a vast range of green plants and algae, were found through a HMM-based search. Table 1 lists the total number of FMOs per plant species. We used the summary of relationships of major angiosperm lineages available at NCBI Common Tree builder (https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi) to display the distribution of FMOs in vascular plants (Figure 1; for sources of sequence and protein data and gene search details, see Supplementary Tables S1, S2; Appendix S1, Supplementary Figure S1).
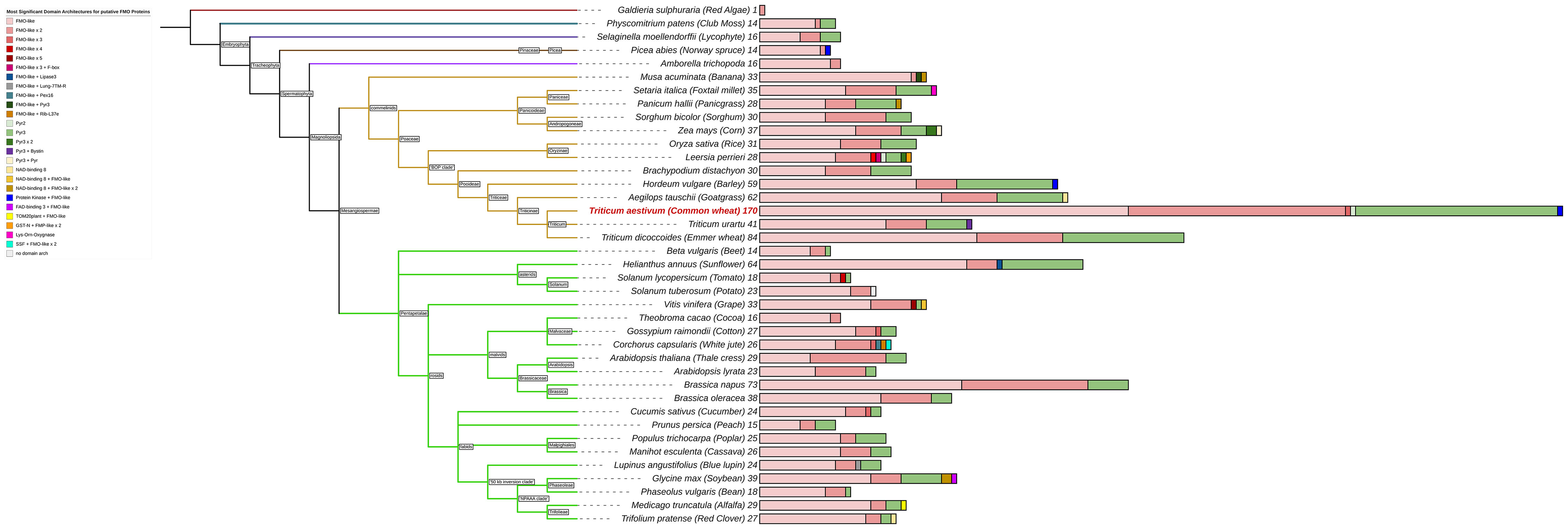
Figure 1 Total number of predicted FMOs and their main domain architectures as reported from Hidden Markov Modelling (HMM). Branch colors indicate major subdivisions amongst vascular plants. Total numbers of FMOs are reported as a number at the tips of each bar, and multivariate bars shows colors corresponding to the proportion of FMOs for each of the various kinds of domain architectures seen in the legend.
2.2 Characterization of the TaFMO family genes
The full wheat cDNA, CDS, and peptide sequences of candidate TaFMO genes identified were retrieved from IWGSC (The International Wheat Genome Sequencing Consortium (IWGSC) et al., 2018). Gene Ontology (GO) terms were retrieved from the Uniprot database (https://www.uniprot.org/; accessed January 2023). The structure of exons and introns were determined using the CDS and gene sequences of TaFMOs analyzed with the Gene Structure Display Server (GSDS) version 2.0 (gsds.cbi.pku.edu.cn; Hu et al., 2015). A small subset of putative TaFMOs were represented by splice variants and included for downstream analyses, on the basis that each variant could encode for fully functioning TaFMO proteins acting in different tissues, conditions, and developmental stages.
To assess whether the enzymatic activity of each candidate TaFMO could be carried out, the presence of three previously well-described FMO motifs (Eswaramoorthy et al., 2006; Mishina and Zeier, 2006; Schlaich, 2007; Thodberg and Jakobsen Neilson, 2020) known to be crucial for cofactor binding and required for proper FMO function—the FAD- and NAD(P)H-binding motifs crucial for oxygenation activity, the FMO-identifying motif facilitating in NAD(P)H-binding at the core pocket of the enzyme, and the ATG-containing motif thought to govern hydroxylation activity—were assessed with multiple sequence alignments (MSA) using MAFFT with the E-INS-I setting; see Appendix S2 (Katoh et al., 2019). Candidates that did not possess one or all motifs were retained in downstream phylogenetic analyses but flagged as ‘non-canonical’ (nc) FMOs with the assumption that oxygenation activity may be compromised, as we used conservation of all motifs as a proxy for predicting FMO function. We predicted the putative protein structure of all TaFMOs using the Protein Homology/Analogy Recognition Engine V 2.0 (Phyre2; Kelley et al., 2015) and then visualized them with PyMOL (Version 2.0 Schrödinger and DeLano, 2020). We used the Simple Modular Architecture Research Tool (SMART; https://smart.embl.de/; Letunic et al., 2021) program to gather protein domains for display (Figure 2). Possible transmembrane domains and signal peptides for all TaFMO were predicted using the programs DeepTMHMM and SignalP - 6.0 (Hallgren et al., 2022; Teufel et al., 2022).
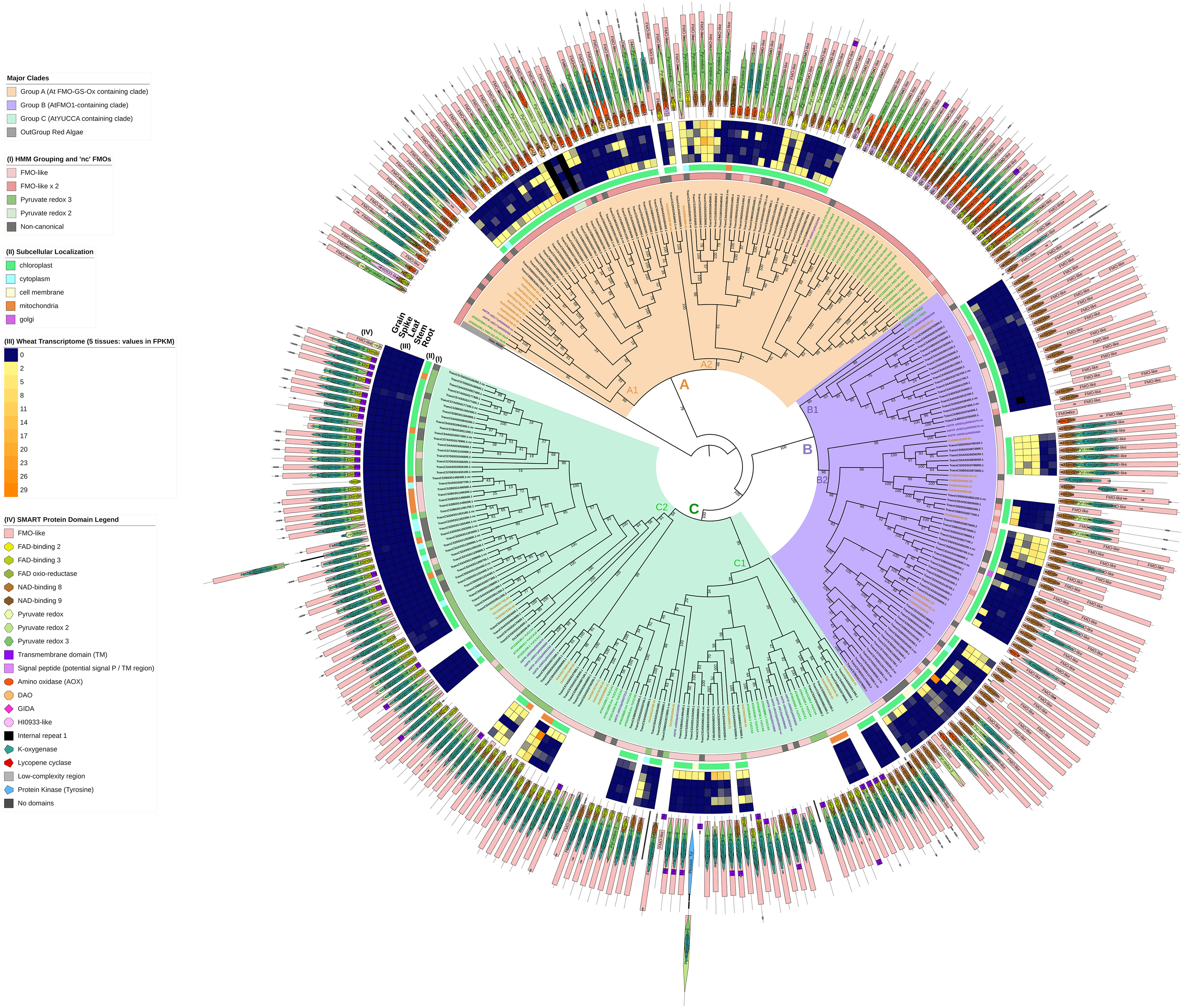
Figure 2 A maximum-likelihood (ML) consensus genealogy tree of total protein sequences of candidate TaFMOs (black gene IDs), OsjFMOs (yellow), AtFMOs (green), AmtriFMOs (purple), and GasuFMOs (grey). One GasuFMO (red microalgae) was used as outgroup. ML geneology was constructed from IQ-TREE (1000 standard bootstrap replicates, JTT+I+G4 substitution model). Three major subclades A (orange shading), B (purple) and C (green) were confidently supported across vascular plant FMOs. Outer circles: (I) shows the domain architecture the candidate FMO belongs to (from HMMER), as annotated in Figure 1; non-canonical ‘nc’ and truncated FMO protein candidates are marked with a dark grey box. (II) indicates predicted subcellular localization of TaFMO proteins. (III) represent transcript values (in FPKM) of each TaFMO candidate gene expressed under 5 tissue types (root, stem, leaves, spike, grain) from RNAseq experiments compiled on the Wheat Proteome database (Duncan et al., 2017). (IV) indicates SMART-predicted protein domains inclusive in each candidate FMO. Group A contains the least number of TaFMOs, totaling 55 splice variants pertaining to 39 genes (see S1 notes for explanation of exclusion of one TaFMO in the analysis), nine of which are categorized as ‘nc’. Group B contains a total of 63 splice variants pertaining to 57 genes, 14 of which are ‘nc’. Lastly, Group C contains 80 splice variants pertaining to 74 genes, 16 of which are ‘nc’.
2.3 Phylogenetic analyses of the TaFMO gene family and sub-families
We inferred a maximum likelihood (ML) phylogeny to define relationships between total validated TaFMOs and validated FMOs from the model plant A. thaliana, the cereal crop Oryza sativa subsp. japonica, and sister group of all other angiosperms, Amborella trichopoda. We selected a red algae having a single FMO as an outgroup. We first aligned full-length FMO peptide sequences using MAFFT with the E-INS-i setting (Katoh et al., 2019). We validated the full-length alignment of the total FMOs across plant species using the software HOMO v2.0 (Jermiin, 2017) by using it to determine whether all FMO sequences met the phylogenetic assumption of evolution under reversible, stationary, and globally homogenous conditions (Jermiin, 2017; Jermiin et al., 2020). Following this, poorly aligned regions of the MSA were masked using the AliStat program (Wong et al., 2014) to ensure downstream analysis focused on regions where amino acid residues were readily alignable across sequences (see Appendix S2) (Wong et al., 2014). We then manually inspected alignments using Mesquite v 3.7 (Maddison and Maddison, 2023) and adjusted accordingly. Lastly, we used IQ-TREE v 2.2.0 to infer the ML tree based on amino acid alignments; we inferred the optimal substitution model using ModelFinder (JTT+I+G4), considering the Bayesian Information Criterion, BIC (Kalyaanamoorthy et al., 2017), and calculated branch support from 1000 standard bootstrapping replicates (Felsenstein, 1985). The tree figures were made with iTOL (Letunic and Bork, 2021).
We constructed separate ML phylogenies between all TaFMO and FMOs from closely-related species in the grass order, Poales—specifically, H. vulgare (barley), T. urartu (red einkorn wheat) and O. sativa ssp. japonica (rice) to improve our understanding of the relatedness of TaFMO in the three major clades A, B, and C (Supplementary Figures S2-5). For detailed notes on alignment and modelling, see Appendix S2.
2.4 Gene duplication and homeolog analysis, and protein motif analysis
We retrieved chromosomal locations of all TaFMOs in wheat from the RefSeqv2.1 gff3 file. We inferred homeologs based on well-supported phylogenetic clustering; separate sub-phylogenies were inferred for the three major partitions (clades A, B, and C) and a 70% bootstrap support value was used as a threshold to acknowledge clades with moderate support. Gene duplication and syntenic gene blocks of all TaFMOs (including ‘nc’ TaFMOs) were inferred through the MCSanX algorithm (Wang et al., 2012) via TBtools software v1.116 (Chen et al., 2022) and NOTUNG (Chen et al., 2000) to resolve tandem duplications and relationships between TaFMO homoeologous groups. Genes with no discernible relatives were denoted as ‘orphan’ TaFMO (Tables 2, 3). A comprehensive synchronized map of all TaFMO homeologs (Figure 3) connected by designated group colors assigned in Supplementary Figures S2-4 was generated using the shinyCircos software in RStudio (Yu et al., 2018; R Core Team, 2021). See Appendix S3 for more details. A de novo search for putative conserved protein motifs among all TaFMOs, HvFMOs, OsjFMOs, AtFMOs, AmTriFMOs, and PpFMOs was conducted with MEME Suite (Bailey et al., 2015): see Appendix S4.
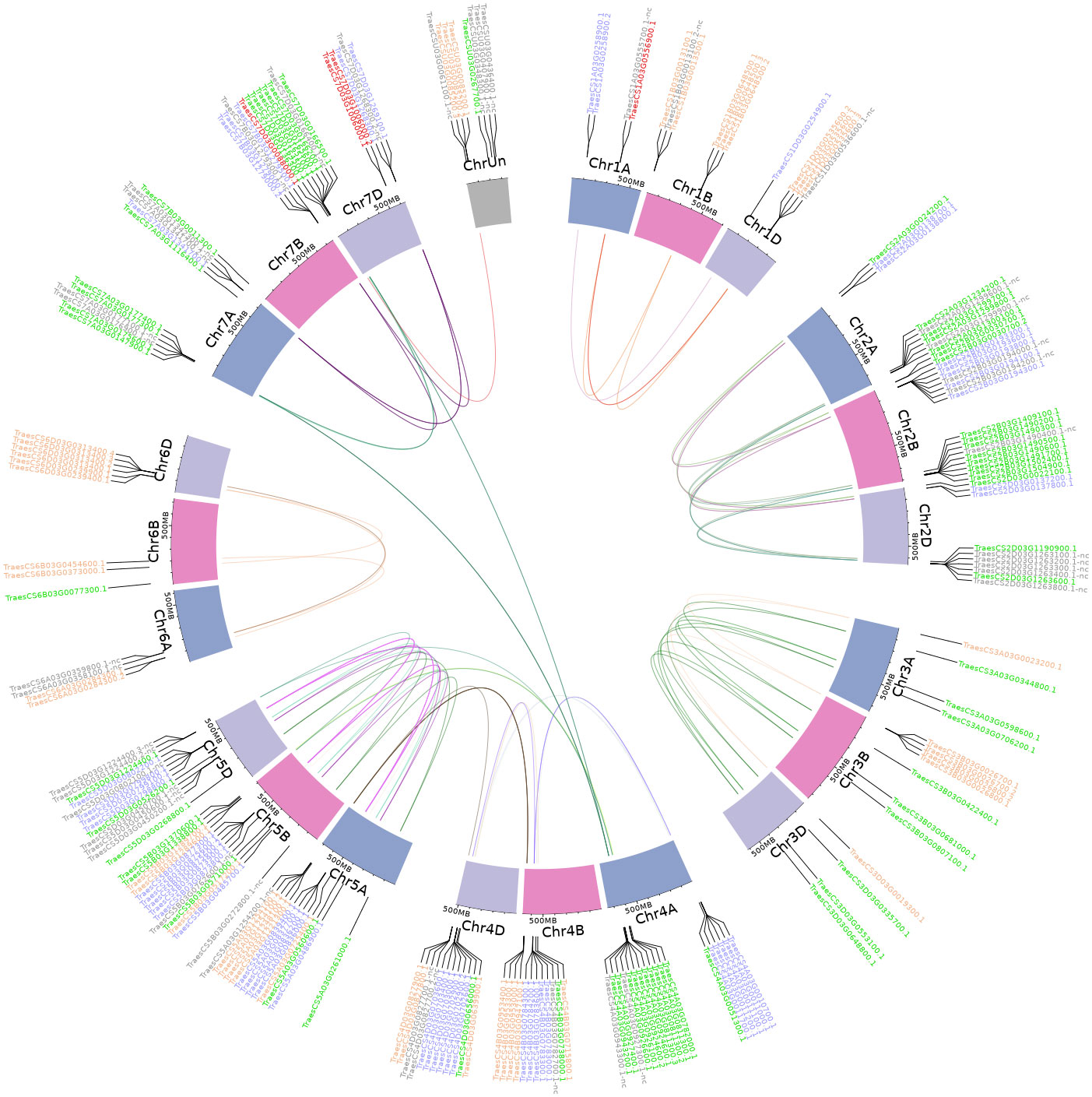
Figure 3 Genomic distribution of TaFMO groups A, B, and C candidates presented on a Circos plot (shinyCircos). Colors of gene labels correspond to the Group A (orange), B (blue) and C (green) partitions of TaFMO candidates. Red-labelled genes are orphaned/singleton TaFMOs, and grey-labelled genes are ‘nc’ TaFMOs. Cytobands exhibit chromosome tracks for chromosomes 1-7 (and unmapped chromosome, ChrUn) for each sub-genome A, B, and D in wheat. Links inside the circos plot are representative of whole-genome-duplication homeologs (syntenic triads) of wheat TaFMO genes delineated through a genome-wide synteny analysis; links are discretely colored in correspondence to annotated subclades for each of the TaFMO candidates, seen in Supplementary Figures S2-4, S2 notes.
2.5 TaFMO promoter regulatory elements analysis and transposable element identification
We searched promoter regions 1.5 kb upstream from the start codon of validated TaFMOs for plant cis-acting regulatory elements (PlantCARE: http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; Lescot, 2002) and visualized putative cis-acting regulatory elements (CAREs) governing the different clades of TaFMOs (Supplementary Tables S3, 4, Appendix S5). We also compiled a list of putative transposable elements (TEs) detected by the CLARI-TE program (https://github.com/jdaron/CLARI-TE) either spanning introns and exons of TaFMOs, or flanking TaFMOs within a 2 kb region on either side (Table 4), from the TE annotation file provided in IWGSC RegSeqv2.1 (International Wheat Genome Sequencing Consortium et al., 2018; Zhu et al., 2021). The TE positions flanking or inside genes were transposed into TaFMO gene structure analysis. Refer to Appendix S6 for details.
2.6 Transcriptome, proteome, and tissue localization data retrieval and display
Using public transcriptomic data across a range of conditions, we extracted candidate TaFMO gene expression information, obtaining transcript values in FPKM for five general tissues (root, stem, leaves, spike, and grain) and proteome data (peptide spectral counts) for different tissues from the developmental atlas in the Wheat Proteome database (Duncan et al., 2017). We acquired transcript data for a broad range of different conditions in values of log2 transcripts per million (tpm) for TaFMO expression under various abiotic stressors (cold, drought, heat) and biotic stressors (i.e., infection by Septoria blotch, Fusarium graminearum, or Puccinia striiformis f.sp. tritici) from the Wheat Expression Database (Ramírez-González et al., 2018) in order to survey the differential expression of TaFMO candidates, and infer their possible biological functions (see Supplementary Tables S5-8 for an overview of studies and sources used).
3 Results and discussion
3.1 FMOs are a largely expanded gene family in flowering plants
The FMO gene family is highly expanded in the vascular plants, as previously reported (Eswaramoorthy et al., 2006; Mishina and Zeier, 2006; Schlaich, 2007; Thodberg and Jakobsen Neilson, 2020). Figure 1 exhibits the general number of potential FMO protein-encoding genes among select vascular plant reference genomes. The three most commonly present types of domain architectures that represent these plant FMOs are a single FMO-like domain (FMO-like), two consecutive FMO-like domains (FMO-like x 2), and a pyruvate redox 3 (Pyr3) domain. Some FMOs are predicted as fusion proteins, such as found in both wheat (Ta) and barley (Hv) with a tyrosine protein kinase conjugated to an FMO, alluding to a possible function in environmental sensing and downstream signaling coupled with oxidation processes (Tichtinsky et al., 2003; Shumayla and Upadhyay, 2023). The phenomenon of FMO fusion proteins does not present for lycophytes, moss, or algae, where total numbers of FMOs in the genome are substantially reduced relative to flowering vascular plant species. The total number of FMOs in plant genomes also appears to be largely reflective of species ploidy levels for recently diverged taxa, as exemplified in Triticum urartu (2n, 41 putative TuFMOs), T. dicoccoides (4n, 84 putative TdFMOs) and T. aestivum (6n, 170 putative TaFMOs). However, in plants considered diploid, the number of FMOs varies substantially, such as in Arabidopsis (29 AtFMOs), rice (31 OsjFMOs), and barley (59 HvFMOs), up to 64 potential FMOs in sunflower (H. annuus), which has double the amount of most diploid species in the eudicots (lower, green clade in Figure 1). We report several additional FMO candidates for rice compared to a recent report of 28 OsjFMOs by (Gaba et al., 2023), and for barley previously reported having 41 HvFMOs (Thodberg and Jakobsen Neilson, 2020), highlighting the complexity of conducting genome-wide searches for highly expanded gene families.
An HMM search conducted against the RefSeqv2.1 wheat reference genome for cultivar ‘Chinese Spring’ yielded a total of 171 high confidence (HC) TaFMO genes, and 82 low confidence (LC) TaFMO genes. We selected 170 HC TaFMO genes including those deemed ‘unmapped’ to specific chromosomes after consolidation with other search methods (see Supplementary Figure S1, Appendix S1 for additional information). A total of 34 TaFMO candidate genes (40 including transcript splice variants) are denoted as ‘non-canonical’ (nc), based on alterations in—or the absence of—protein motifs previously reported as crucial for proper FMO function, discussed below (Table 2). The ‘nc’ TaFMOs possessing alterations in important protein motifs but were still found in synteny with other TaFMO homoeologous triads, were retained in downstream analyses; further experimental validation is required to prove their functional and biological relevance as an FMO in wheat. Other more highly truncated ‘nc’ TaFMOs unlikely functional in the canonical sense, could still highlight trends in motif diversification/loss for certain subclades when included in a phylogenetic analysis. The same logic was applied to sort for FMOs of Arabidopsis thaliana (At), Oryza sativa ssp. japonica (Osj), Hordeum vulgare (Hv), and Triticum urartu (Tu).
3.2 TaFMOs are consistently divided into three major groups across vascular plants, and span all chromosomes and sub-genomes
A maximum-likelihood phylogeny was constructed with protein sequences representing the total 170 TaFMO gene candidates (198 independent splice variants); FMOs from Amborella trichopoda (Amtri), rice Oryza sativa sp. Japonica (Osj), and A. thaliana (At) were included to probe for phylogenetic partitioning of TaFMOs among flowering plants. A red microalgae (G. sulphuraria) served as an outgroup, whose genome encodes for only one known FMO (Figure 1). We observed three major phylogenetic groupings, indicated as A, B, and C in Figure 2.
Domain architecture analysis (Figure 2, circle I) shows that Group A FMOs mostly share the same domain architecture type as the FMO from the outgroup red algae (‘FMO-like x 2’), with two TaFMOs as exceptions having the ‘pyruvate redox 2’ architecture (TraesCS3B03G0026700.1, TraesCS3B03G0026800.2). Group B (purple) and Group C (green) FMOs form a clade across vascular plants with Group B mostly composed of an ‘FMO-like’ architecture, whereas group C has a mix between the ‘FMO-like’ and ‘pyruvate redox 3’ architectures. The distribution of total FMOs across an array of vascular plants (or within one species) has been documented by Thodberg and Jakobsen Neilson (2020); Gaba et al. (2023), and Yoshimoto et al. (2015), and the phylogenies presented in these studies also find Group B (AtFMO1-containing clade) and Group C (‘YUCCA’ clade) to be more closely related to each other than to Group A (the AtFMO GSOX-containing clade).
We found that TaFMOs span all seven (and unmapped) chromosomes and sub-genomes (A, B, and D) derived from progenitor grass species Triticum urartu, Aegilops speltoides, and Aegilops tauschii, respectively (Ramírez-González et al., 2018; Zhu et al., 2021). Chromosomes 2, 4 and 7 are enriched with TaFMOs in the telomeric regions (Figure 3). Translocations of TaFMOs are inferred to have occurred most frequently between chromosomes 4A, 7A, and 7D, which are reported to be hot spots for homoeologous gene translocation events for many genes in wheat (Zhou et al., 2020) (Appendix S3). Other prominent translocations of TaFMO occurred between chromosomes 4A, 5A, and 5B in Group C TaFMO, and between chromosomes 5A, 4B, and 4D in Group A TaFMO (Figure 3, Table 2).
3.3 Sub-groups reveal non-uniform patterns of TaFMO gene expansion
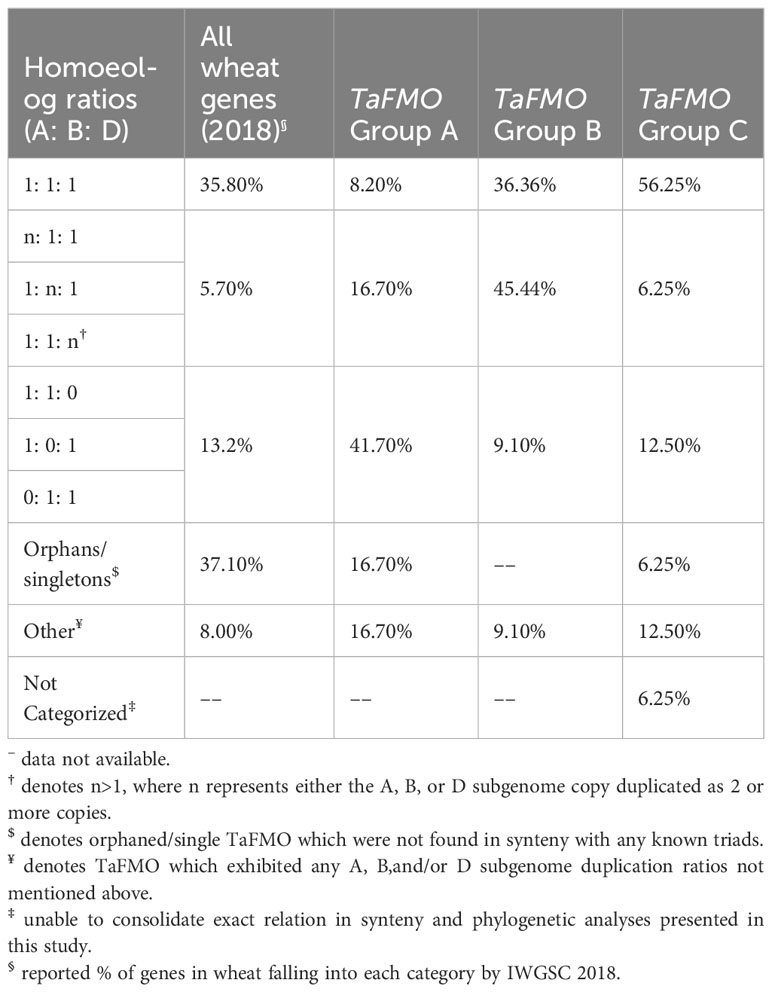
Tables 2, 3 summarize the synteny status of Groups A, B, and C TaFMO homeologs and homeolog sub-genome ratios (denoted as a TaFMO ratio of 1: 1: 1 from A, B, and D sub-genomes), respectively. Group A TaFMOs show substantially less syntenic triads with only 8.2% of triads in Group A exhibiting an even ratio of 1: 1: 1; even more, around 42% of triads in Group A were missing either an A-, B-, or D-copy, a higher frequency than the 13.2% expected for all wheat proteins in the genome (Table 3). Group C, subclade C1, showed the most conserved ‘clean’ homoeologous triads (56.3%) such that there were rarely any singleton/orphan or ‘nc’ TaFMOs, and each TaFMO triad had corresponding orthologues from other grass species (Supplementary Figure S4); the high evolutionary conservation of these TaFMO triads is perhaps due to functional constraints influencing gene retention. In contrast, TaFMOs in subclade C2 (along with barley FMOs) appear to have large, uneven homeolog expansions via tandem duplications and intra- or interchromosomal dispersion of genes (Supplementary Figure S4).
In Group B, 36.4% of TaFMOs belong to even triads which more closely resemble the anticipated percentage (35.8%) of all predicted wheat proteins belonging to even homoeologous triads (Table 3) (Ramírez-González et al., 2018). Group B appears to have many instances of sub-genome homeolog expansion and uneven tandem duplications (Table 2). Subclade B1 appears to contain three pairs of TaFMO WGD homeologs that underwent multiple duplications, with more HvFMO orthologues present than OsjFMO and TuFMO, (Supplementary Figure S3), while subclade B2 shows FMO expansions for all grass species surveyed, but none for Arabidopsis (Supplementary Figure S3).
Such divergent patterns of gene retention and expansion among homoeologous triads between the three major groups of TaFMOs might implicate differing biological roles (Birchler and Yang, 2022). make a point that genes involved in defense mechanisms are more likely to diverge after duplication events; the retention of the active sites for some of the highly duplicated Group B triads could infer a role in defense-related functions.
3.4 Variations in canonical motifs delineate the FMOs in wheat and related grasses
We detected fifteen of the most conserved protein motifs in FMOs across Arabidopsis and the grasses rice (Osj), barley (Hv), red einkorn (Tu) and wheat (Supplementary Figure S5). FMOs in plants are reported to contain three crucial protein motifs essential for ‘canonical’ FMO activity; these are the FAD-binding motif (GxGxxG), the NAD(P)H-binding motif (GxGxxG), and the FMO-identifying motif [FxGxxxHxxxY/F; (Eswaramoorthy et al., 2006; Hansen et al., 2007; Huijbers et al., 2014)]. While the FAD- and NAD(P)H-binding sites are active sites surrounding the enzyme pocket where hydroxylation takes place, the FMO-identifying motif is considered a linker region between the two active sites (Malito et al., 2004). The last and less widely talked about motif is the ‘F/LATGY’ motif, more recently referred to in some studies as the ‘ATG-containing’ or ‘TGY’ motif (Yoshimoto and Saito, 2015; Gaba et al., 2023); this motif is thought to be a crucial factor governing N-hydroxylation (Stehr et al., 1998; Fennema et al., 2016).
The FAD-binding motif at the N-terminus is very well conserved with regards to three glycine residues across all groups (Figures 4A, B). However, the residues surrounding this motif for Group A and subclade B2 from Group B is recorded to be distinct from that in subclade B1 and Group C (Figures 4A, B; Supplementary Figures S2-4); the FAD-binding site variant 1 (for Group A and B2) and the other variant are highlighted in orange in Figure 4A, B. An extra copy of the FAD-binding variant 1 also exists between the NAD(P)H-binding site and the ATG-motif in most of the examined grass species (Hv, Tu, Ta) in subclade C2, but not for rice. This may indicate a novel function for FMOs specific to Triticeae. The NAD(P)H-binding motif across all FMOs (Supplementary Figure S5) is well-conserved for the first glycine residue, but not for the other two glycine residues (Gxgxxg; Figure 4A). The FMO-identifying motif for FMOs in Group A has the first residue swapped from the canonical F (Phenylalanine) for W (Tryptophan), which is the case for most of the plant species examined (Figure 4A). Additionally, the ATG-containing motif for Group A FMOs across all flowering plants surveyed are highly conserved, represented as HCTGY or YCTGY. The differences of the Group A ATG-motif and FMO-identifying motifs relative to Groups B and C may allude to specificity for S-oxygenation of compounds and specialized metabolites yet uncharacterized in cereals, where Group A FMOs from A. thaliana (FMO-GSOX enzymes) are well-characterized for their S-oxygenation activity during synthesis of specialized metabolites (Hansen et al., 2007). The ATG-containing motif is most varied for Group B FMOs, where in addition to the canonical ‘(F/L)ATGY’ residues reported by (Schlaich, 2007), other variations are present (LATGF, FLATGF, FATGY, LATGY, FGTGF) in the B2 expanded subclade comprising other grass species (Tu, Hv, Osj). By contrast, Group C FMOs mostly present with ‘LATGY’ motifs (and some ‘MATGY’ motifs in barley and wheat) in subclade C1 and ‘FATGY’ motifs in subclade C2 (Figure 4A).
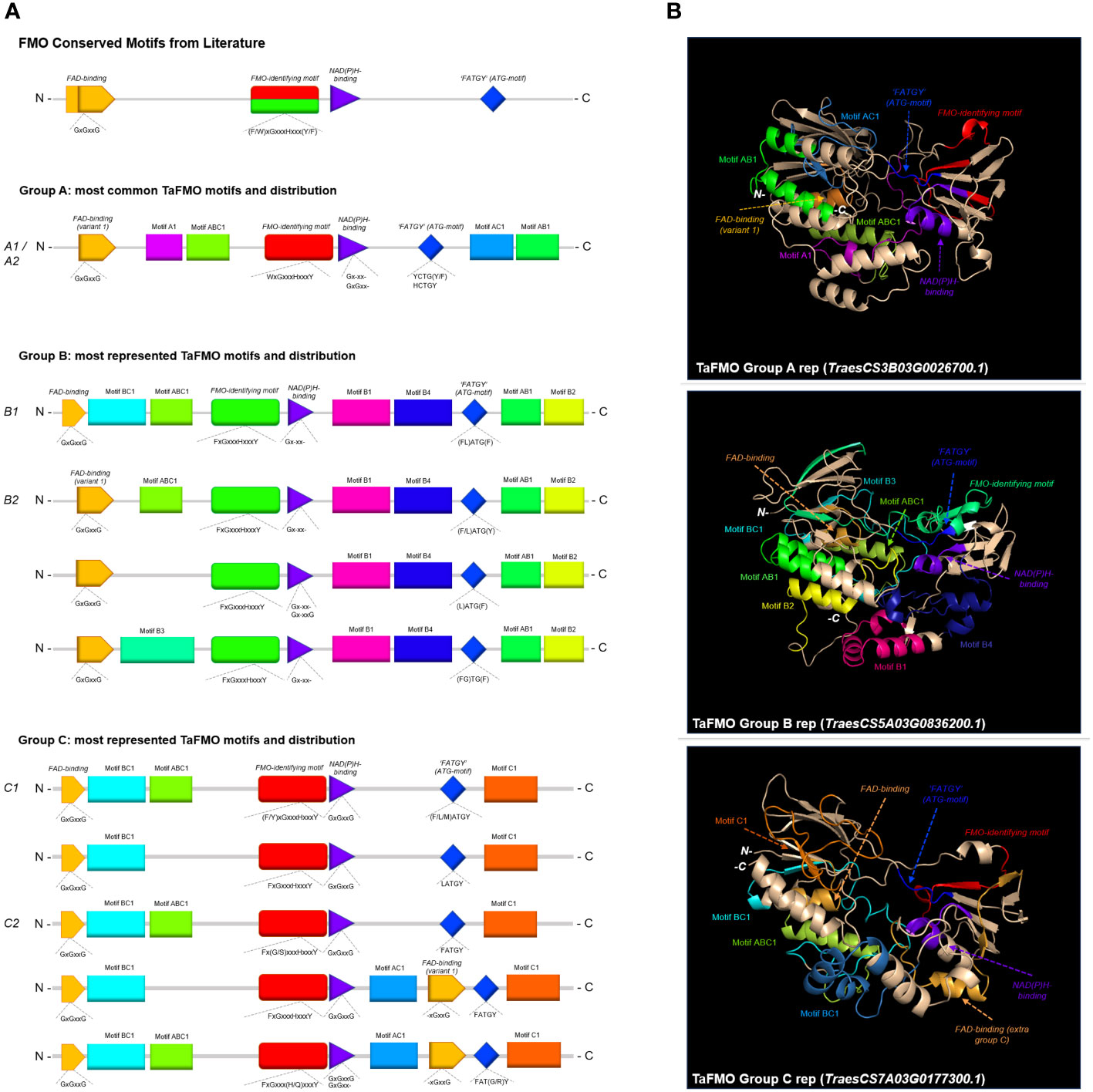
Figure 4 (A) Consensus MEME motifs for conserved and unique motifs across Groups A, B, and C TaFMO proteins. (B) Predicted 3D protein structures for Groups A, B, and C TaFMO with highlighted motifs.
Beyond the documented protein motifs discussed above, one novel motif is reported for each Group A and Group C FMOs (referred to here as motif-A1 and motif-C1, respectively; see Supplementary Figure S5 for these and other motifs referred to in this section). Meanwhile, Group B FMOs have four clade-specific motifs (motif-B1-4) (Figure 4). Groups A, B, and C FMOs all share one novel motif each with each other (motif-AB1, motif-AC1, motif-BC1). Additionally, there is a novel motif common to almost all FMOs located at the N-terminus following the FAD-binding site for most FMOs, reported here as motif-ABC1 (Figure 4A). A look at predicted protein models highlights that many of these novel motifs may present as-yet-unclassified linker regions contributing to the integrity of the enzyme, or motifs that govern substrate specificity and binding affinity (Figure 4B). We hypothesize that differences in tertiary protein structures, including these clade-specific motifs, likely play a crucial role in substrate specificity (Li et al., 2008; Zhu et al., 2019). While no plant FMOs have been crystalized to date, the use of 3D protein homology modelling and ligand-docking simulations—in corroboration with functional analyses—may shed some light on which compounds TaFMO, and other cereal FMOs, may be acting upon.
3.5 Possible roles of group A and C TaFMO: from pathogen defense to grain development
Group A FMOs contain all the Arabidopsis FMO-GSOX (glucosinolate oxidase) genes. These FMOs have been experimentally validated to participate in glucosinolate (GSL) biosynthesis by catalyzing the S-hydroxylation of methylthioalkyls to methylsulfinylalkyls, where GSLs are a class of specialized sulphur-containing metabolites involved in plant-herbivory defenses (Hansen et al., 2007; Li et al., 2011; Kong et al., 2016; Cang et al., 2018; Schall et al., 2020). GSLs are predominantly found in the mustard seed family (Brassicaceae), although there is evidence that 500 neighboring eudicot species outside this family may also contain one or more of the 120 documented GSLs (Flamini, 2012). The possible metabolites that these Group A FMOs help modify in cereals are not well-understood, as cereals are not reported to produce GSLs (Hansen et al., 2007; Thodberg and Jakobsen Neilson, 2020). More recently, other A. thaliana FMOs have been documented to synthesize trimethylamine N-oxide (TMAO) (Fennema et al., 2016), the levels of which are increased in plants under low temperatures, drought, and high salt stress conditions (Catalá et al., 2021). Group A FMOs segregate into two major subclades, A1 and A2 (Figures 2, Supplementary Figure S2). While little is known for Group A FMOs in barley and red einkorn, it is reported that five rice FMOs from subclade A1 (Os01t0368000-nc, Os02t0580600-nc, Os07t0111700, Os07t0111900, Os07t0112000) are co-expressed with WRKY13 (a transcription factor involved in pathogen defense) upon attack by rice sheath-infecting fungi (John Lilly and Subramanian, 2019). For TaFMOs in subclade A1, the literature supports involvement in root drought tolerance, tiller growth, and nutrient accumulation in the grain (Table 2; Supplementary Figure S6). The rice FMO Os10t0553800 in subclade A2 may play an important role in rice germination and seedling establishment during floods, via epigenetic methylation responses to anaerobic conditions (Castano-Duque et al., 2021). By contrast, several TaFMOs in subclade A2 are implicated in disease resistance against both biotrophic (Navathe et al., 2022) and necrotrophic pathogens (Nussbaumer et al., 2015), in addition to root drought tolerance (Supplementary Figure S6). The amino oxidase (AOX) protein domain which is seen in almost all FMOs in Group A at the N-terminus but rarely for Group B (and which is missing from Group C), is larger in subclade A2 AtFMOs (~150 amino acid residues), but not for wheat (~60 residues) or other plant FMOs (Figure 2). The AOX domain in Group A TaFMOs appears to encompass the motif-A1 mentioned previously unique to Group A and may participate in substrate binding specificity for novel compounds in wheat.
Group C contains the “YUCCA” (YUC) genes that are known to participate in auxin (idole-3-acetic acid, IAA) biosynthesis (Kendrew, 2001; Cao et al., 2019), where auxin is involved in many plant development processes (Schlaich, 2007; Chandler, 2015). We identified 74 TaFMOs in Group C, which can be split into two main subclades, C1 and C2. Several TaFMOs in subclade C1 are implicated in the response to drought stress (Table 2; Supplementary Figure S7), found to be upregulated during root drought treatment (Grzesiak et al., 2019), or associated with an increased reactive oxygen species (ROS, H2O2) burst in response to root drought conditions (Kamruzzaman et al., 2022). In subclade C2, several TaFMOs are implicated in plant development, including in grain (Mangini et al., 2021) and seed development (Li et al., 2014), and tiller number increase (Yu et al., 2021). To date, only three homoeologous genes involved in seed development have been functionally characterized via gene cloning (Li et al., 2014). A catalogue of 63 ‘TaYUC’ gene candidates involved in auxin biosynthesis by Yang et al. (2021) included several TaFMO genes outside Group C (dispersed throughout subclade B2; Supplementary Table S4), hinting towards a flexibility in TaFMO involvement in auxin biosynthesis-regulated plant development. In the absence of a full gene family survey and experimental characterization, adopting a more wheat-specific nomenclature for the FMO gene family via the guidelines put forward by the Wheat Initiative community (Boden et al., 2023), may minimize confusion on gene names in wheat to streamline future research efforts.
3.6 Group B TaFMOs: major players in broad-spectrum disease resistance
Group B FMOs can be split into two distinct subclades, B1 and B2 (Figure 2). This group contains the Arabidopsis AtFMO1 and AtFMO2 genes. AtFMO1 hydroxylates the N atom of an L-lysine catabolic product to form N-hydroxylated pipecolic acid (NHP), crucial for establishing systemic acquired resistance (SAR) to combat microbial pathogen invasions (Conrath, 2006; Mishina and Zeier, 2006; Chen et al., 2018; Hartmann et al., 2018), and a lack of NHP (due to defects in its biosynthesis) results in immunocompromised Arabidopsis plants. Various studies have recently contributed to better understanding the mechanism of NHP-mediated SAR regulation in other plants, including cereal crops, by searching for functional orthologs to AtFMO1 and their possible involvement in disease resistance (Holmes et al., 2019; Lenk et al., 2019; Schnake et al., 2020; Vlot et al., 2021; Zhang et al., 2021).
In subclade B1, AtFMO1 and AtFMO2, the only Group B FMOs in Arabidopsis, form a clade with 18 TaFMOs, where only three candidates are ‘nc’ (Table 2, Figure 4B). All FMOs from subclade B1 possess the most minimal number of known protein domains via SMART analysis, whereas multiple protein domains are present for TaFMO from all other clades (Figure 2). Of note, AtFMO1 harbors an additional K-oxygenase domain (IPR025700) in the first half of its protein sequence, which is not seen in any of the B1 TaFMOs or AtFMO2 but is present in most B2 TaFMOs and TaFMOs in Groups A and C. The K-oxygenase domain is involved in siderophore biosynthesis, and in ornithine and lysine hydroxylation (Visca et al., 1994; Krithika et al., 2006); therefore, presence of this K-oxygenase may play a critical role in hydroxylation of L-lysine and/or L-ornithine derived compounds.
While subclade B1 TaFMOs show little to no transcriptional activity in the conditions surveyed, certain subclade B2 TaFMOs exhibit higher transcriptional activity (Figures 2, 5 – III/IV). The TaFMO TraesCS5D03G0799800.1 in subclade B2 (B2β-3b) was detected as the most transcriptionally upregulated “FMO1-like” TaFMO in response to treatment of wheat coleoptiles with NHP (Zhang et al., 2021). Two homoeologous triads in B2α-2 (Table 2) show transcriptional upregulation in root, stem, and leaf tissues (Figure 2), and peptide abundance of these triads was also detected in a wide variety of wheat tissues (Figure 5 – II). These TaFMOs in B2α-2 undergo transcriptional activation in wheat seedlings during infection by the fungal pathogens Zymoseptoria tritici (Zt) (Yang et al., 2013; Rudd et al., 2015) and Puccinia striiformis f.sp. tritici (Pst) (Cantu et al., 2013; Dobon et al., 2016), but have very little to no transcriptional activation during older developmental stages of wheat, such as seen in controls during infections by Fusarium graminearum (Fg) in the wheat grain head (Schweiger et al., 2016) and Magnaporthe oryza (Mo) at the grain-filling stage (Islam et al., 2016) (Figure 5 - III).
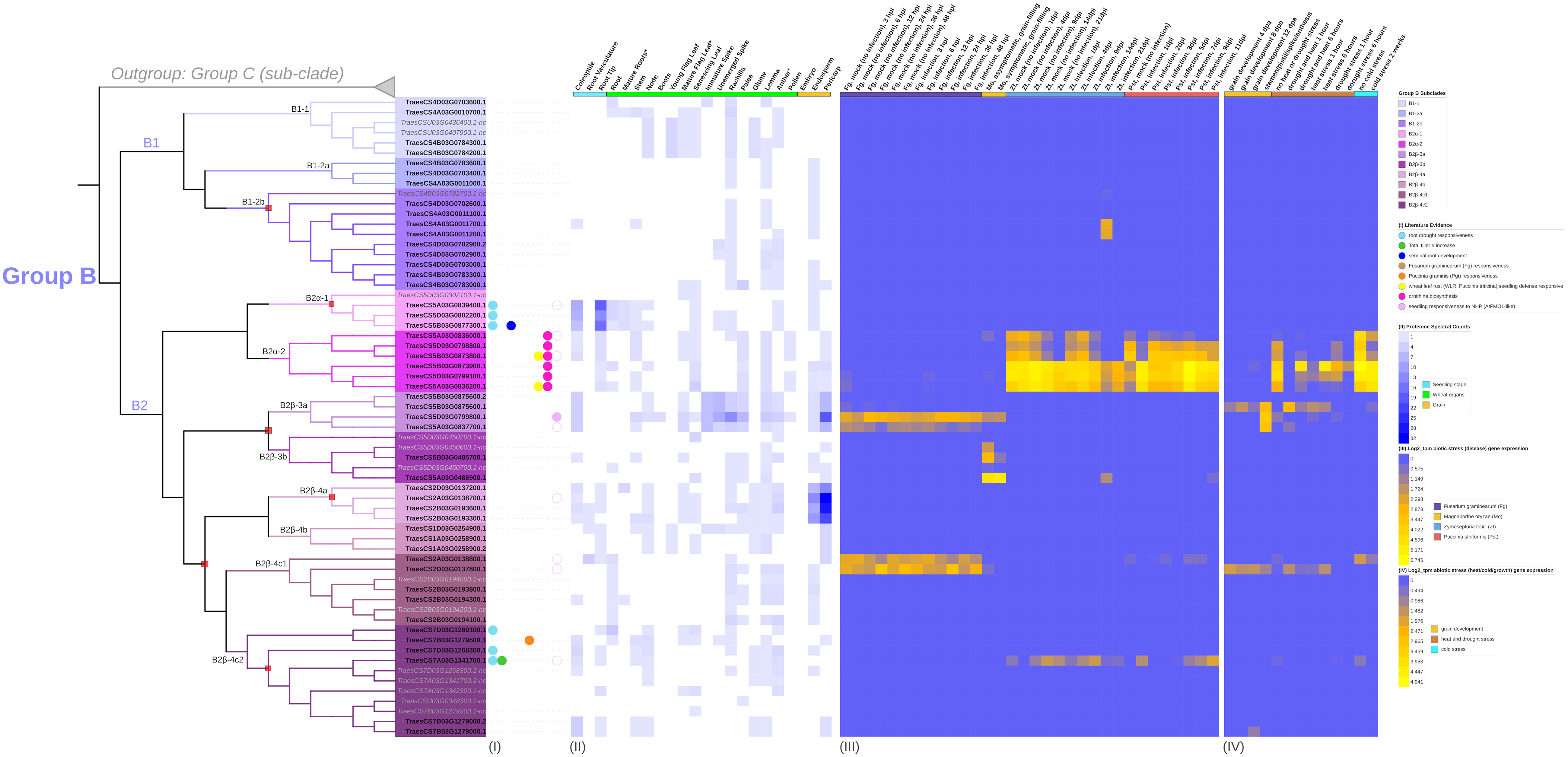
Figure 5 Transcription of group B TaFMOs under various conditions explored from literature data. (Columns I) reports of previous evidence of transcript detection or cloning under various conditions described in the legend. (Columns II) Protein expression (spectral peptide counts) for each candidate in various tissues from https://wheatproteome.org/ (Duncan et al., 2017). (Columns III) Heatmap of TaFMO transcripts in response to four types of biotic stresses; response to Fusarium graminearum infection(Fg) (Schweiger et al., 2016), mock inoculation versus pathogen infection in a time-course of 3, 6, 12, 24, 36, and 48 hours post infection (hpi); Magnaporthe oryza (Mo) (Islam et al., 2016), asymptomatic (control) versus symptomatic (infection) conditions at grain-filling stage; Zymoseptoria tritici (Zt) (Yang et al., 2013; Rudd et al., 2015), mock inoculation versus pathogen infection in a time-course of 1, 4, 9, 14, 21 days post infection (dpi); Puccinia striiformis f.sp. tritici (Pst) (Cantu et al., 2013; Dobon et al., 2016), uninfected control versus pathogen infection in a time-course of 1, 2, 3, 5, 7, 9, 11 dpi. (Columns IV) Heatmap of TaFMO transcript response to grain development over a time-course (4, 8 12 days post anthesis) (Li et al., 2013; Yang et al., 2015), heat and drought stress (1 and 6 hours) (Liu et al., 2015), and cold stress (control versus 2-week cold stress) (Li et al., 2015). Heatmap data is displayed as Log2 transcripts per million (tpm), with blue being 0 and higher-fold expression approaching bright yellow (see legends).
One of the B2α-2 homoeologous triads (TraesCS5A03G0836200.1, TraesCS5B03G0873900.1, TraesCS5D03G0799100.1) shows greater upregulation of transcription after Pst infection at one day post-infection (dpi), which is sustained through 11 dpi; TraesCS5A03G0836200.1 is also reported by (Vikas et al., 2022) in a genome-wide association study (GWAS) to be likely involved in seedling resistance to the biotrophic fungal pathogen Puccinia triticina (Pt), the wheat leaf rust pathogen closely related to Pst. Another TaFMO in sub-clade B2β-4c(2), TraesCS7B03G1279500.1, might be involved in Puccinia graminis f.sp tritici (Pgt, stem rust) resistance (Sahu et al., 2021). In a QTL analysis by (Matros et al., 2017), the two triads in subclade B2α-2 were also detected as significantly enriched in SNPs associated with ornithine metabolism in wheat. Ornithine is an amino acid bearing great resemblance to lysine and has functional significance in plant abiotic stress tolerance (Kalamaki et al., 2009) through its role in osmoregulation during drought and salinity stress (Roosens et al., 1998; Xue et al., 2009). These two triads are among the only Group B TaFMOs to be transcriptionally responsive in the abiotic conditions presented, downregulated during drought stress, heat stress, and combined drought + heat stress at one hour, but bounce back to nearly control levels after six hours of treatment for certain splice variants (Figure 5 – IV) (Liu et al., 2015). Transcripts of the TaFMO triad in B2α-1 (except for the ‘nc’ 5D homeolog) have been detected in wheat roots (Figure 2, Table 2) (Grzesiak et al., 2019). reported that genes of this triad were upregulated in response to water deprivation in wheat root tissues, though no upregulation of transcription in leaf tissues was detected in the drought study by Liu et al. (2015) (Figure 5 – IV), highlighting a need for careful interpretation of tissue-specific expression patterns of TaFMOs. Furthermore, a GWAS analysis indicates that the B-copy gene of this triad, TraesCS5B03G0877300.1, influences root-growth and biomass yield-related traits (Zhao et al., 2023). Trends for other candidates of interest are found in Appendix S7.
In summary, several lines of evidence hint at roles in responsiveness to various microbial diseases for several TaFMOs in subclade B2. Additionally, certain subclade B2 TaFMOs may operate during plant development (tiller increase) and drought, heat, and salinity tolerance. Thodberg et al. (2020) showed that a novel Group B FMO in fern (FOS1) participates in both the N-hydroxylation of a novel class of metabolites not previously reported in vascular plants and in the biosynthesis of cyanogenic glycoside. Likewise, the exploration and characterization of these B2 TaFMOs may identify novel functions and metabolites towards plant development, biotic, and abiotic stressors.
3.7 TaFMO UTRs are populated with elements involved in complex regulation
Untranslated regions in transcripts (UTRs) have been reported in plants as important regulatory regions often harboring important cis-acting regulatory elements (CAREs) that influence gene expression, modulate translational efficiency, and increase the coding capacity of genes that have different splice variations (Mignone et al., 2002; Srivastava et al., 2018). We found a wide variety of CAREs in the promoter regions 1.5 kb upstream of the start codon of TaFMO genes (Figures 6; Supplementary Figures S8, S9). The presence and sizes of 5’- and 3’-UTRs among TaFMOs varied; we observed that for all TaFMOs, the 3’-UTRs were longer than the 5’-UTRs, when UTRs were present (Figures 7; Supplementary Figures S10, S11). Srivastava et al. (2018) reported elongated UTRs (particularly 3’-UTRs) in rice compared to Arabidopsis. In wheat, 3’-UTRs have also been reported to have a critical involvement in mediating drought stress (Ma et al., 2023) and in establishing resistance against stripe rust (Zhang et al., 2019), possibly through regulating mRNA stability. Of note, all the tandemly duplicated wheat TaFMOs in subclade C2 lack any UTRs (Supplementary Figure S11) and show virtually no (or low) transcript responsiveness to the biotic and abiotic conditions explored (Supplementary Figure S7 – III, IV), with minimal presence of expressed peptides detected in various tissue types.
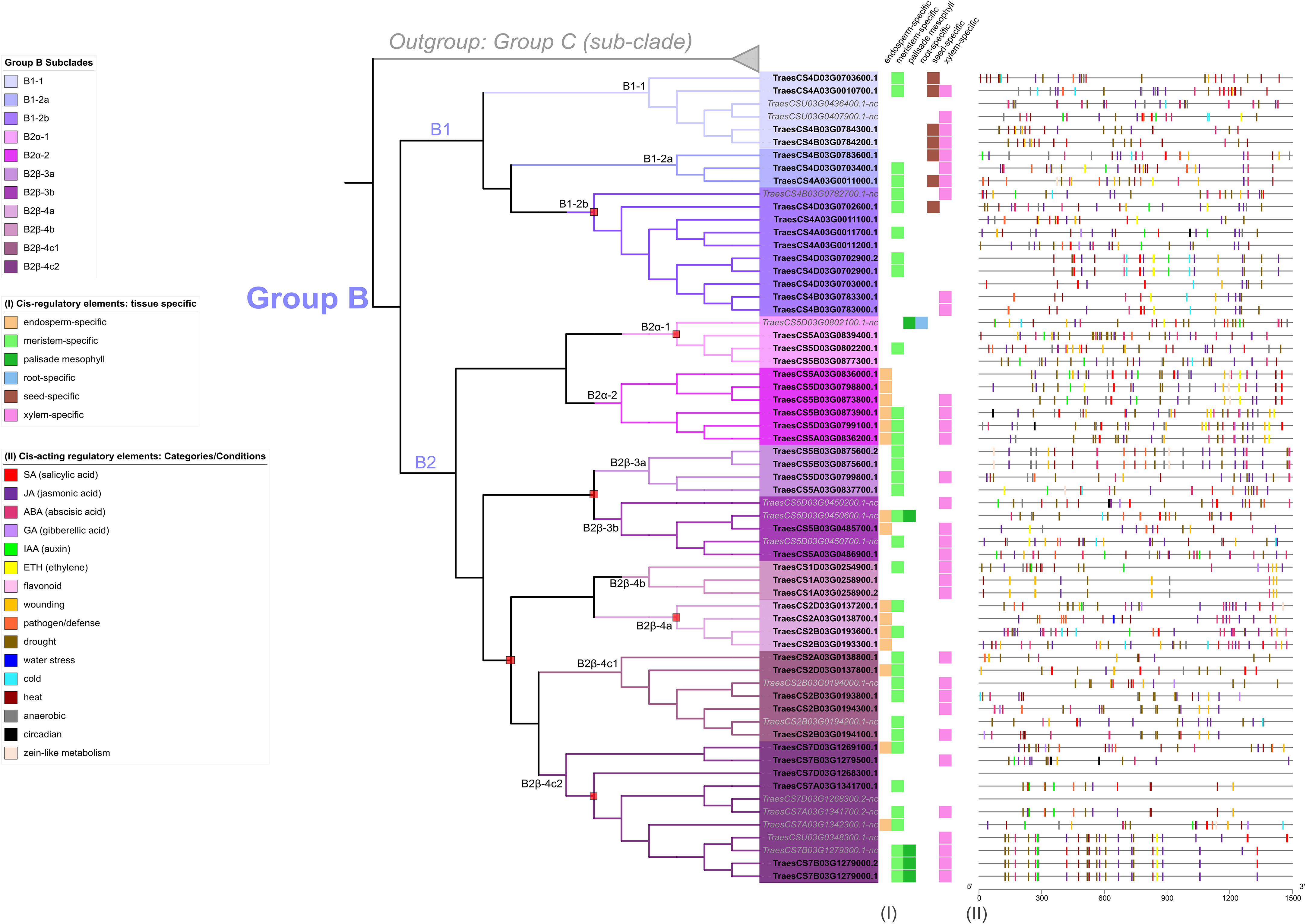
Figure 6 Plant cis-acting regulatory elements (CARE) detected in a 1.5 kb region upstream of start codons for all TaFMO in Group B X-axis denote the number of base pairs away from the start codon, where 5’ is 1,500 bp upstream ATG start codon (3’ and 0 bp is right before the ATG start codon). Polytomy regions are denoted by a red box. Presence of regulatory elements specific to different tissues are described in (I). Other responsive promoter elements are denoted in the figure legend and displayed as rectangles of corresponding color to the condition in (II).
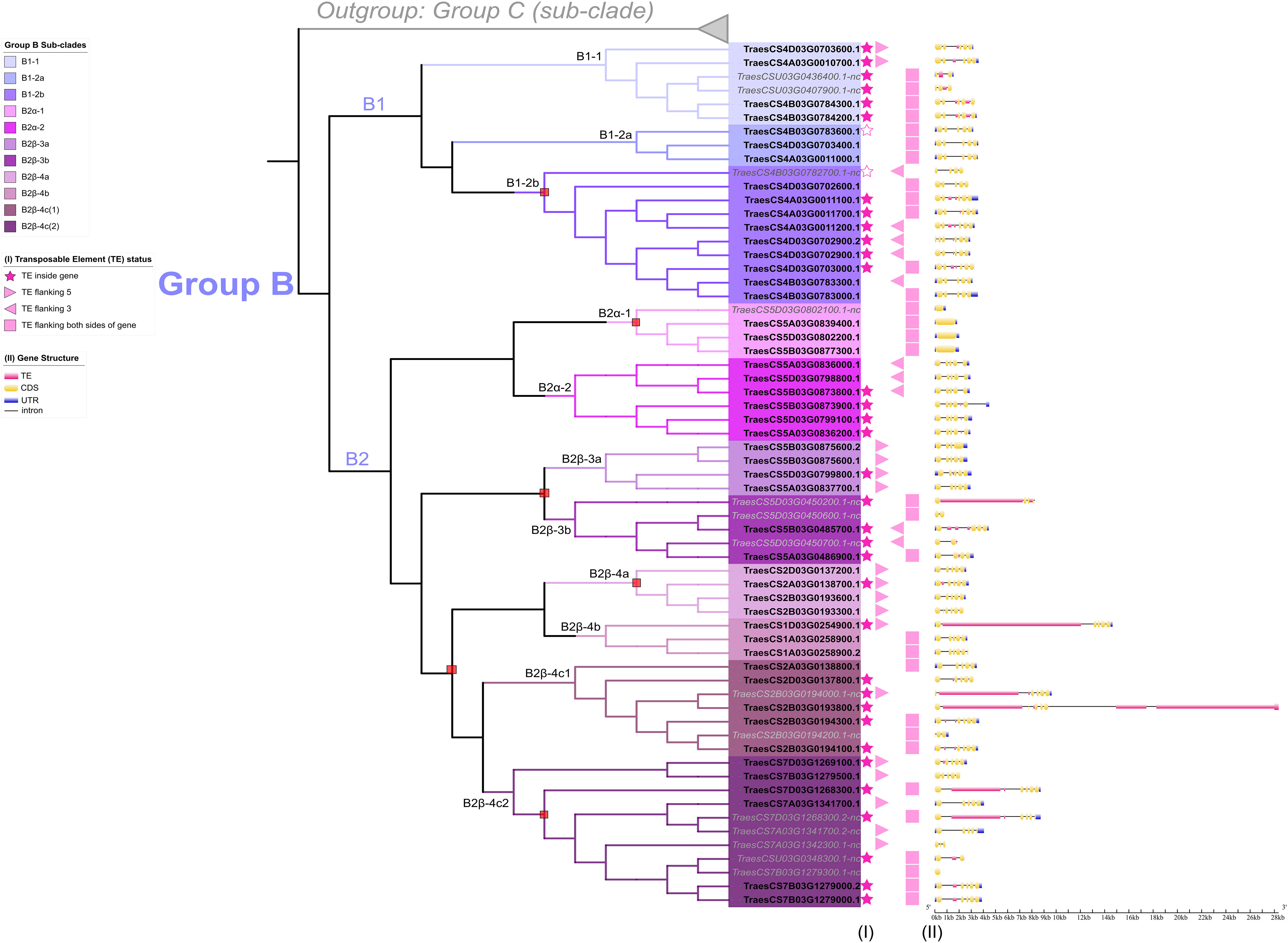
Figure 7 Graphical display of the gene structure and transposable element analysis for TaFMOs in Group B Polytomies are denoted by a red box at respective nodes on the ML tree. Status of TE distribution is described in (I) for filled star: TE insertion inside gene; unfilled star: non-coding region of TE spanning entire gene; filled triangle: TE flanking either 5’ or 3’ UTR of gene; and filled square: TE flanking both sides of the gene. A 2 kb region upstream of the start codon and downstream of the stop codon were scanned for flanking TEs. (II) The gene structure is displayed for each TaFMO, with graphical depiction of TE insertion, if present. The X-axis scale describes the size in kb of nucleotides for each gene region. In Group B, exon numbers vary from three to five (B1), one (B2α-1), and one to five (all rest of sub-clade B2).
Focusing on the Group B TaFMOs, analysis of the CAREs revealed involvement in responsiveness to major phytohormones and conditions (Figure 6; Appendix S5). Jasmonic acid (JA)-responsive CAREs were present in all TaFMO promoters at one or multiple sites; JA is a phytohormone known for involvement in many plant processes ranging broadly from photosynthesis to heat stress-responsiveness (Per et al., 2018), but most widely documented for defense against necrotrophic pathogens (Qi et al., 2016) and induced systemic resistance (Heil, 2002). In wheat, JA-signaling has also been implicated in seedling salt-stress tolerance (Qiu et al., 2014). Other notable phytohormone-responsive CAREs in Group B are for salicylic acid (SA), gibberellic acid (GA) and ethylene (ETH) (Supplementary Tables S3, 4, Figure 6). SA is a major phytohormone involved in SAR and pathogen defense signaling pathways (Dey et al., 2014; Ding et al., 2018). SA-responsive CAREs were found in higher abundance and closer to the start codons of the triads from B2α-2 (Figure 6, Supplementary Table S4), whose genes were more transcriptionally active in Zt- and Pst-infected wheat (Figure 5 – III); this may indicate that number and location of these CAREs influence which TaFMOs are activated under pathogen stress. GA is a phytohormone that was documented in wheat for its involvement in enhancing disease resistance against F. graminearum infection, by modulating both primary and specialized metabolism during early plant defense signaling (Buhrow et al., 2021); GA-responsive CAREs are found predominantly in subclade B2, further supporting that TaFMOs in this subclade are likely involved in microbial disease resistance. Auxin (IAA)-responsive CAREs are found to be abundant among many Group B TaFMOs (Supplementary Table S4), challenging the idea of Group C TaFMOs—the ‘YUCCA’ FMO clade—being largely responsible for auxin biosynthesis and growth regulation in plants for underreported crop species (Kendrew, 2001).
3.8 Transposable elements, gene structure diversity, and differential TaFMO expression
We explored transposable element (TE) distribution in or near the identified TaFMO genes and affecting intron-exon structure to find support for relationships among TaFMO family members (Figure 7, Supplementary Figures S10, 11; Appendix S6). Focusing again on the Group B TaFMOs, we observed much variation in the number and spatial distribution of introns and exons; high similarity of gene structures was found among certain syntenic triads highlighting distinct TaFMO subfamilies that possibly remain conserved for functional importance. In contrast, extensive intron-exon shuffling following gene duplication events (Kolkman and Stemmer, 2001; França et al., 2012) may indicate lower selective pressure and neofunctionalization, such as in the case for subclades B2β-4c1 and B2β-4c2 (Figure 7).
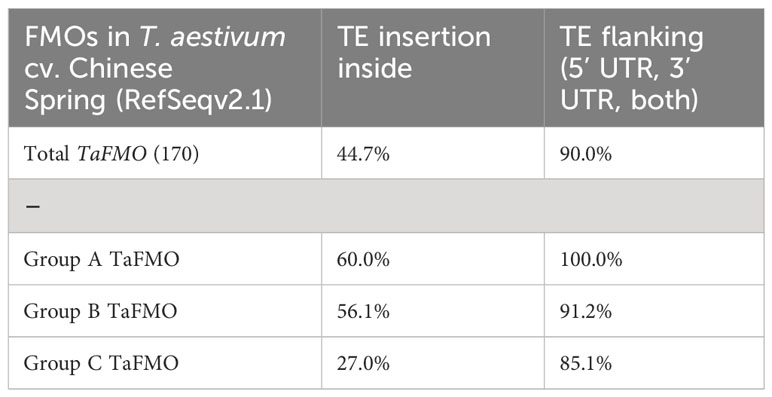
Nearly all TaFMOs (90%) are flanked by TEs within a 2 kb region upstream and/or downstream of the start or stop codons (Figure 7, Table 4) in accordance with a report of TEs representing over 80% of the wheat genome (Choulet et al., 2014; Bariah et al., 2020). 44.7% of all TaFMO genes show disruption by TE insertion, mostly occurring inside introns, inside 5’- or 3’-UTRs, or, very rarely, inside exons (Figure 7). TE insertions are present in 56.1% of Group B TaFMO, with the largest TE interruption in TraesCS2B03G0193800 increasing its total gene length to upwards of 29 kb (Figure 7). Most TE disruptions in subclade B2 occur in the B- and D-copy homeologs and appear to be correlated with differential expression patterns among homoeologous triads. Varying locations of TE insertions in the highly tandemly duplicated B-copy TaFMOs in B2β-4c(1) correlate with no transcriptional expression compared to the A- and D-copies, and only an A-copy TaFMO in B2β-4c(2) lacking TE insertions was transcriptionally active (Figure 5 – III, IV; Figure 7). In contrast, one of the triads in B2α-2 (TraesCS5A03G0836200, TraesCS5B03G0873900, TraesCS5D03G0799100) shares the same TE insertion in its three members, which are all more transcriptionally active than gene members in the other B2α-2 triad lacking this TE insertion, particularly upregulated towards Pst infection and cold stress responses (Figure 5 – III, IV; Figure 7).
Yang et al. (2021) reported on the gene structure of a candidate ‘YUCCA’ FMO in wheat, TaYUCCA5-D (TraesCS1D03G0254900.1, Group B), containing a first intron size of approximately 13 kb. We now know this is due to a TE insertion (Figure 7), underscoring a need for expanding on factors driving gene structure diversity in large gene-family analyses. We see that TE insertions in the intronic and 3’-UTR regions may play a role in influencing gene expression of triads on a sub-genome level, possibly leading to eventual neofunctionalization of FMO family members. Several studies indicate that TEs are important drivers in polyploid plant evolution (Vicient and Casacuberta, 2017; Gill et al., 2021), such as in wheat where distinct TE families are correlated with up- or down-regulation of genes in their vicinity (Bariah et al., 2023). Thus, exploring classes of TE elements that lie in or around TaFMOs could aid in predicting gene expression regulation in the different wheat sub-genomes.
4 Concluding remarks
We present here a comprehensive phylogenetic analysis of the FMO superfamily in wheat and integrate various publicly available transcriptomic and proteomic data to shed light on the breadth of expansion for this superfamily in wheat and predict potential roles for the members of this family for hypothesis generation and further functional studies. We show that TaFMOs segregate into three distinct groups, and that unique domain architectures and protein motifs may indicate ligand-binding specificities among sub-groups of TaFMOs. We corroborate the expression data from existing literature of certain candidate TaFMOs with their possible biological functions as discussed in various studies. This study provides a solid foundation for the further exploration and functional characterization of the FMO gene family in common wheat.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
SS: Data curation, Formal analysis, Investigation, Software, Visualization, Writing – original draft. GB: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Funding was provided by the Natural Sciences and Engineering Research Council of Canada, CREATE grant (509257-2018) and Natural Sciences and Engineering Research Council of Canada, Canada Graduate Scholarship – Doctoral award (CGSD-579396-2023).
Acknowledgments
We thank our funding support by the Natural Sciences and Engineering Research Council of Canada (NSERC) CREATE program, as well as the NSERC CGS-D program. The authors are indebted to Dr. Sean Graham for help reviewing the phylogenetic analyses, manuscript editing and valuable discussions. Additionally, we thank Dr. Matthias Kretschmer for his constructive feedback on the original manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1369299/full#supplementary-material
References
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bariah, I., Gribun, L., Kashkush, K. (2023). Transposable elements are associated with genome-specific gene expression in bread wheat. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1072232
Bariah, I., Keidar-Friedman, D., Kashkush, K. (2020). Where the wild things are: transposable elements as drivers of structural and functional variations in the wheat genome. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.585515
Birchler, J. A., Yang, H. (2022). The multiple fates of gene duplications: Deletion, hypofunctionalization, subfunctionalization, neofunctionalization, dosage balance constraints, and neutral variation. Plant Cell 34, 2466–2474. doi: 10.1093/plcell/koac076
Boden, S. A., McIntosh, R. A., Uauy, C., Krattinger, S. G., Dubcovsky, J., Rogers, W. J., et al. (2023). Updated guidelines for gene nomenclature in wheat. Theor. Appl. Genet. 136, 72. doi: 10.1007/s00122-023-04253-w
Buhrow, L. M., Liu, Z., Cram, D., Sharma, T., Foroud, N. A., Pan, Y., et al. (2021). Wheat transcriptome profiling reveals abscisic and gibberellic acid treatments regulate early-stage phytohormone defense signaling, cell wall fortification, and metabolic switches following Fusarium graminearum-challenge. BMC Genomics 22, 798. doi: 10.1186/s12864-021-08069-0
Cang, W., Sheng, Y.-X., Evivie, E. R., Kong, W.-W., Li, J. (2018). Lineage-specific evolution of flavin-containing monooxygenases involved in aliphatic glucosinolate side-chain modification: Evolution of flavin-containing monooxygenases. J. Systematics Evol. 56, 92–104. doi: 10.1111/jse.12289
Cantu, D., Segovia, V., MacLean, D., Bayles, R., Chen, X., Kamoun, S., et al. (2013). Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. Sp. Tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genomics 14, 270. doi: 10.1186/1471-2164-14-270
Cao, X., Yang, H., Shang, C., Ma, S., Liu, L., Cheng, J. (2019). The roles of Auxin biosynthesis YUCCA gene family in plants. Int. J. Mol. Sci. 20, 6343. doi: 10.3390/ijms20246343
Castano-Duque, L., Ghosal, S., Quilloy, F. A., Mitchell-Olds, T., Dixit, S. (2021). An epigenetic pathway in rice connects genetic variation to anaerobic germination and seedling establishment. Plant Physiol. 186, 1042–1059. doi: 10.1093/plphys/kiab100
Catalá, R., López-Cobollo, R., Berbís, M.Á., Jiménez-Barbero, J., Salinas, J. (2021). Trimethylamine N -oxide is a new plant molecule that promotes abiotic stress tolerance. Sci. Adv. 7, eabd9296. doi: 10.1126/sciadv.abd9296
Chandler, J. W. (2015). “Auxin biosynthesis,” in Amino acids in higher plants, 1st ed. Ed. D’Mello, J. P. F. (CAB International), 340–361. doi: 10.1079/9781780642635.0340
Chen, K., Durand, D., Farach-Colton, M. (2000). NOTUNG: A program for dating gene duplications and optimizing gene family trees. J. Comput. Biol. 7, 429–447. doi: 10.1089/106652700750050871
Chen, Y.-C., Holmes, E. C., Rajniak, J., Kim, J.-G., Tang, S., Fischer, C. R., et al. (2018). N -hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. 115 (21), E4920–E4929. doi: 10.1073/pnas.1805291115
Chen, C., Wu, Y., Xia, R. (2022). A painless way to customize Circos plot: From data preparation to visualization using TBtools. iMeta 1, e35. doi: 10.1002/imt2.35
Choulet, F., Alberti, A., Theil, S., Glover, N., Barbe, V., Daron, J., et al. (2014). Structural and functional partitioning of bread wheat chromosome 3B. Science 345, 1249721. doi: 10.1126/science.1249721
Conrath, U. (2006). Systemic acquired resistance. Plant Signaling Behav. 1 (4), 179–84. doi: 10.4161/psb.1.4.3221
Curtis, T., Halford, N. G. (2014). Food security: The challenge of increasing wheat yield and the importance of not compromising food safety. Ann. Appl. Biol. 164, 354–372. doi: 10.1111/aab.12108
Czarnocka, W., Fichman, Y., Bernacki, M., Różańska, E., Sańko-Sawczenko, I., Mittler, R., et al. (2020). FMO1 is involved in excess light stress-induced signal transduction and cell death signaling. Cells 9, 2163. doi: 10.3390/cells9102163
Dey, S., Wenig, M., Langen, G., Sharma, S., Kugler, K. G., Knappe, C., et al. (2014). Bacteria-triggered systemic immunity in barley is associated with WRKY and ETHYLENE RESPONSIVE FACTORs but not with salicylic acid. Plant Physiol. 166, 2133–2151. doi: 10.1104/pp.114.249276
Ding, Y., Sun, T., Ao, K., Peng, Y., Zhang, Y., Li, X., et al. (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173 (6), P1454–1467.E15. doi: 10.1016/j.cell.2018.03.044
Dobon, A., Bunting, D. C. E., Cabrera-Quio, L. E., Uauy, C., Saunders, D. G. O. (2016). The host-pathogen interaction between wheat and yellow rust induces temporally coordinated waves of gene expression. BMC Genomics 17, 380. doi: 10.1186/s12864-016-2684-4
Duncan, O., Trösch, J., Fenske, R., Taylor, N. L., Millar, A. H. (2017). Resource: Mapping the Triticum aestivum proteome. Plant J. 89, 601–616. doi: 10.1111/tpj.13402
Eswaramoorthy, S., Bonanno, J. B., Burley, S. K., Swaminathan, S. (2006). Mechanism of action of a flavin-containing monooxygenase. Proc. Natl. Acad. Sci. 103, 9832–9837. doi: 10.1073/pnas.0602398103
Fennema, D., Phillips, I. R., Shephard, E. A. (2016). Trimethylamine and Trimethylamine N-oxide, a Flavin-containing Monooxygenase 3 (FMO3)-mediated host-microbiome metabolic axis implicated in health and disease. Drug Metab. Disposition 44, 1839–1850. doi: 10.1124/dmd.116.070615
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39 (4), 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Flamini, G. (2012). “Natural herbicides as a safer and more environmentally friendly approach to weed control: A review of the literature since 2000,” in Studies in Natural Products Chemistry, vol. 38. (Elsevier), 353–396. doi: 10.1016/B978-0-444-59530-0.00013-7
França, G. S., Cancherini, D. V., De Souza, S. J. (2012). Evolutionary history of exon shuffling. Genetica 140, 249–257. doi: 10.1007/s10709-012-9676-3
Gaba, Y., Bhowal, B., Pareek, A., Singla-Pareek, S. L. (2023). Genomic survey of Flavin monooxygenases in wild and cultivated rice provides insight into evolution and functional diversities. Int. J. Mol. Sci. 24, 4190. doi: 10.3390/ijms24044190
Gill, R. A., Scossa, F., King, G. J., Golicz, A. A., Tong, C., Snowdon, R. J., et al. (2021). On the role of transposable elements in the regulation of gene expression and subgenomic interactions in crop genomes. Crit. Rev. Plant Sci. 40, 157–189. doi: 10.1080/07352689.2021.1920731
Grzesiak, M. T., Hordyńska, N., Maksymowicz, A., Grzesiak, S., Szechyńska-Hebda, M. (2019). Variation among spring wheat (Triticum aestivum L.) genotypes in response to the drought stress. II—Root system structure. Plants 8, 584. doi: 10.3390/plants8120584
Hallgren, J., Tsirigos, K. D., Pedersen, M. D., Almagro Armenteros, J. J., Marcatili, P., Nielsen, H., et al. (2022). DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. Bioinformatics. doi: 10.1101/2022.04.08.487609. [Pre-print]
Hansen, B. G., Kliebenstein, D. J., Halkier, B. A. (2007). Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis: Identification of a flavin monooxygenase. Plant J. 50, 902–910. doi: 10.1111/j.1365-313X.2007.03101.x
Hartmann, M., Zeier, T., Bernsdorff, F., Reichel-Deland, V., Kim, D., Hohmann, M., et al. (2018). Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity. Cell 173, 456–469.e16. doi: 10.1016/j.cell.2018.02.049
Heil, M. (2002). Induced systemic resistance (ISR) against pathogens in the context of induced plant defences. Ann. Bot. 89, 503–512. doi: 10.1093/aob/mcf076
Holmes, E. C., Chen, Y.-C., Sattely, E. S., Mudgett, M. B. (2019). An engineered pathway for N -hydroxy-pipecolic acid synthesis enhances systemic acquired resistance in tomato. Sci. Signaling 12, eaay3066. doi: 10.1126/scisignal.aay3066
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huijbers, M. M. E., Montersino, S., Westphal, A. H., Tischler, D., van Berkel, W. J. H. (2014). Flavin dependent monooxygenases. Arch. Biochem. Biophysics 544, 2–17. doi: 10.1016/j.abb.2013.12.005
International Wheat Genome Sequencing Consortium, Wicker, T., Gundlach, H., Spannagl, M., Uauy, C., Borrill, P., et al. (2018a). Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biol. 19, 103. doi: 10.1186/s13059-018-1479-0
Islam, M. T., Croll, D., Gladieux, P., Soanes, D. M., Persoons, A., Bhattacharjee, P., et al. (2016). Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biol. 14, 84. doi: 10.1186/s12915-016-0309-7
Jermiin, L. S. (2017). Homo version 1.3 (CSIRO). Data Access Portal. [Computer software]. Canberra, Australia. doi: 10.4225/08/5a56c7a9e4c2a
Jermiin, L. S., Catullo, R. A., Holland, B. R. (2020). A new phylogenetic protocol: Dealing with model misspecification and confirmation bias in molecular phylogenetics. NAR Genomics Bioinf. 2, lqaa041. doi: 10.1093/nargab/lqaa041
John Lilly, J., Subramanian, B. (2019). Gene network mediated by WRKY13 to regulate resistance against sheath infecting fungi in rice (Oryza sativa L.). Plant Sci. 280, 269–282. doi: 10.1016/j.plantsci.2018.12.017
Kalamaki, M. S., Merkouropoulos, G., Kanellis, A. K. (2009). Can ornithine accumulation modulate abiotic stress tolerance in Arabidopsis? Plant Signaling Behav. 4, 1099–1101. doi: 10.4161/psb.4.11.9873
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., Jermiin, L. S. (2017). ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kamruzzaman, M., Beyene, M. A., Siddiqui, A., Ballvora, A., Léon, J., Naz, A. A. (2022). Pinpointing genomic loci for drought-induced proline and hydrogen peroxide accumulation in bread wheat under field conditions. BMC Plant Biol. 22 (1), 584. doi: 10.1186/s12870-022-03943-9
Katoh, K., Rozewicki, J., Yamada, K. D. (2019). MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinf. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kendrew, S. G. (2001). YUCCA: A flavin monooxygenase in auxin biosynthesis. Trends Biochem. Sci. 26, 218. doi: 10.1016/S0968-0004(01)01814-X
Kolkman, J. A., Stemmer, W. P. C. (2001). Directed evolution of proteins by exon shuffling. Nat. Biotechnol. 19, 423–428. doi: 10.1038/88084
Kong, W., Li, J., Yu, Q., Cang, W., Xu, R., Wang, Y., et al. (2016). Two novel flavin-containing monooxygenases involved in biosynthesis of aliphatic glucosinolates. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01292
Krithika, R., Marathe, U., Saxena, P., Ansari, Mohd., Z., Mohanty, D., et al. (2006). A genetic locus required for iron acquisition in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. 103, 2069–2074. doi: 10.1073/pnas.0507924103
Lenk, M., Wenig, M., Bauer, K., Hug, F., Knappe, C., Lange, B., et al. (2019). Pipecolic acid is induced in barley upon infection and triggers immune responses associated with elevated nitric oxide accumulation. Mol. Plant-Microbe Interactions® 32, 1303–1313. doi: 10.1094/MPMI-01-19-0013-R
Lescot, M. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Khedkar, S., Bork, P. (2021). SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Letunic, I., Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucl. Acids Res. 49 (W1), W293–W296. doi: 10.1093/nar/gkab301
Li, H.- Z., Gao, X., Li, X.- Y., Chen, Q.- J., Dong, J., Zhao, W.- C. (2013). Evaluation of assembly strategies using RNA-Seq data associated with grain development of wheat (Triticum aestivum L.). PLoS ONE 8 (12), e83530. doi: 10.1371/journal.pone.0083530
Li, J., Hansen, B. G., Ober, J. A., Kliebenstein, D. J., Halkier, B. A. (2008). Subclade of flavin-monooxygenases involved in aliphatic glucosinolate biosynthesis. Plant Physiol. 148, 1721–1733. doi: 10.1104/pp.108.125757
Li, J., Kristiansen, K. A., Hansen, B. G., Halkier, B. A. (2011). Cellular and subcellular localization of flavin-monooxygenases involved in glucosinolate biosynthesis. J. Exp. Bot. 62, 1337–1346. doi: 10.1093/jxb/erq369
Li, N., Yin, N., Niu, Z., Hui, W., Song, J., Huang, C., et al. (2014). Isolation and characterization of three TaYUC10 genes from wheat. Gene 546, 187–194. doi: 10.1016/j.gene.2014.06.020
Li, Q., Zheng, Q., Shen, W., Cram, D., Fowler, D. B., Wei, Y., et al. (2015). Understanding the biochemical basis of temperature-induced lipid pathway adjustments in plants. Plant Cell 27 (1), 86–103. doi: 10.1105/tpc.114.134338
Liu, Z., Xin, M., Qin, J., Peng, H., Ni, Z., Yao, Y., et al. (2015). Temporal transcriptome profiling reveals expression partitioning of homologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 15, 152. doi: 10.1186/s12870-015-0511-8
Luo, W., Xiao, N., Wu, F., Mo, B., Kong, W., Yu, Y. (2022). Genome-wide identification and characterization of YUCCA gene family in Mikania micrantha. Int. J. Mol. Sci. 23, 13037. doi: 10.3390/ijms232113037
Ma, H., Lin, J., Mei, F., Mao, H., Li, Q. Q. (2023). Differential alternative polyadenylation of homoeologous genes of allohexaploid wheat ABD subgenomes during drought stress response. Plant J. 114, 499–518. doi: 10.1111/tpj.16150
Maddison, W. P., Maddison, D. R. (2023). Mesquite: a modular system for evolutionary analysis. Version 3.7. Available at: http://www.mesquiteproject.org/.
Malito, E., Alfieri, A., Fraaije, M. W., Mattevi, A. (2004). Crystal structure of a Baeyer–Villiger monooxygenase. Proc. Natl. Acad. Sci. 101, 13157–13162. doi: 10.1073/pnas.0404538101
Mangini, G., Blanco, A., Nigro, D., Signorile, M. A., Simeone, R. (2021). Candidate Genes and quantitative trait loci for grain yield and seed size in durum wheat. Plants 10, 312. doi: 10.3390/plants10020312
Mascotti, M. L., Lapadula, W. J., Juri Ayub, M. (2015). The origin and evolution of Baeyer—Villiger monooxygenases (BVMOs): an ancestral family of flavin monooxygenases. PloS One 10, e0132689. doi: 10.1371/journal.pone.0132689
Matros, A., Liu, G., Hartmann, A., Jiang, Y., Zhao, Y., Wang, H., et al. (2017). Genome–metabolite associations revealed low heritability, high genetic complexity, and causal relations for leaf metabolites in winter wheat (Triticum aestivum). J. Exp. Bot. 68, erw441. doi: 10.1093/jxb/erw441
Mignone, F., Gissi, C., Liuni, S., Pesole, G. (2002). Untranslated regions of mRNAs. Genome Biol. 3, reviews0004.1. doi: 10.1186/gb-2002-3-3-reviews0004
Mishina, T. E., Zeier, J. (2006). The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 141 (4), 1666–1675. doi: 10.1104/pp.106.081257
Mitchell, A. J., Weng, J.-K. (2019). Unleashing the synthetic power of plant oxygenases: from mechanism to application. Plant Physiol. 179, 813–829. doi: 10.1104/pp.18.01223
Navathe, S., Pandey, A. K., Sharma, S., Chand, R., Mishra, V. K., Kumar, D., et al. (2022). New Genomic Regions Identified for Resistance to Spot Blotch and Terminal Heat Stress in an Interspecific Population of Triticum aestivum and T. spelta. Plants 11, 2987. doi: 10.3390/plants11212987
Nussbaumer, T., Warth, B., Sharma, S., Ametz, C., Bueschl, C., Parich, A., et al. (2015). Joint transcriptomic and metabolomic analyses reveal changes in the primary metabolism and imbalances in the subgenome orchestration in the bread wheat molecular response to Fusarium graminearum. G3 Genes|Genomes|Genetics 5, 2579–2592. doi: 10.1534/g3.115.021550
Per, T. S., Khan, M. I. R., Anjum, N. A., Masood, A., Hussain, S. J., Khan, N. A. (2018). Jasmonates in plants under abiotic stresses: Crosstalk with other phytohormones matters. Environ. Exp. Bot. 145, 104–120. doi: 10.1016/j.envexpbot.2017.11.004
Qi, P.-F., Balcerzak, M., Rocheleau, H., Leung, W., Wei, Y.-M., Zheng, Y.-L., et al. (2016). Jasmonic acid and abscisic acid play important roles in host–pathogen interaction between Fusarium graminearum and wheat during the early stages of fusarium head blight. Physiol. Mol. Plant Pathol. 93, 39–48. doi: 10.1016/j.pmpp.2015.12.004
Qin, M., Wang, J., Zhang, T., Hu, X., Liu, R., Gao, T., et al. (2020). Genome-wide identification and analysis on YUCCA gene family in isatis indigotica fort. And iiYUCCA6-1 functional exploration. Int. J. Mol. Sci. 21, 2188. doi: 10.3390/ijms21062188
Qiu, Z., Guo, J., Zhu, A., Zhang, L., Zhang, M. (2014). Exogenous jasmonic acid can enhance tolerance of wheat seedlings to salt stress. Ecotoxicology Environ. Saf. 104, 202–208. doi: 10.1016/j.ecoenv.2014.03.014
Ramírez-González, R. H., Borrill, P., Lang, D., Harrington, S. A., Brinton, J., Venturini, L., et al. (2018). The transcriptional landscape of polyploid wheat. Science 361, eaar6089. doi: 10.1126/science.aar6089
R Core Team. (2021). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Roosens, N. H. C. J., Thu, T. T., Iskandar, H. M., Jacobs, M. (1998). Isolation of the Ornithine-δ-Aminotransferase cDNA and Effect of Salt Stress on Its Expression in Arabidopsis thaliana 1. Plant Physiol. 117, 263–271. doi: 10.1104/pp.117.1.263
Rossner, R., Kaeberlein, M., Leiser, S. F. (2017). Flavin-containing monooxygenases in aging and disease: Emerging roles for ancient enzymes. J. Biol. Chem. 292, 11138–11146. doi: 10.1074/jbc.R117.779678
Rudd, J. J., Kanyuka, K., Hassani-Pak, K., Derbyshire, M., Andongabo, A., Devonshire, J., et al. (2015). Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle definition. Plant Physiol. 167, 1158–1185. doi: 10.1104/pp.114.255927
Sahu, R., Prabhakaran, N., Kundu, P., Kumar, A. (2021). Differential response of phytohormone signaling network determines nonhost resistance in rice during wheat stem rust (Puccinia graminis f. Sp. Tritici) colonization. Plant Pathol. 70, 1409–1420. doi: 10.1111/ppa.13376
Schall, P., Marutschke, L., Grimm, B. (2020). The flavoproteome of the model plant Arabidopsis thaliana. Int. J. Mol. Sci. 21, 5371. doi: 10.3390/ijms21155371
Schlaich, N. L. (2007). Flavin-containing monooxygenases in plants: Looking beyond detox. Trends Plant Sci. 12, 412–418. doi: 10.1016/j.tplants.2007.08.009
Schnake, A., Hartmann, M., Schreiber, S., Malik, J., Brahmann, L., Yildiz, I., et al. (2020). Inducible biosynthesis and immune function of the systemic acquired resistance inducer N-hydroxypipecolic acid in monocotyledonous and dicotyledonous plants. J. Exp. Bot. 71, 6444–6459. doi: 10.1093/jxb/eraa317
Schrödinger, L., DeLano, W. (2020). PyMOL. Available at: http://www.pymol.org/pymol.
Schweiger, W., Steiner, B., Vautrin, S., Nussbaumer, T., Siegwart, G., Zamini, M., et al. (2016). Suppressed recombination and unique candidate genes in the divergent haplotype encoding Fhb1, a major Fusarium head blight resistance locus in wheat. Theor. Appl. Genet. 129, 1607–1623. doi: 10.1007/s00122-016-2727-x
Shumayla and Upadhyay, S. K. (2023). “An overview of receptor-like kinases in plants,” in Plant Receptor-Like Kinases (Elsevier), 1–23. doi: 10.1016/B978-0-323-90594-7.00005-3
Song, C., Zhang, D., Zheng, L., Shen, Y., Zuo, X., Mao, J., et al. (2020). Genome-wide identification and expression profiling of the YUCCA gene family in Malus domestica. Sci. Rep. 10, 10866. doi: 10.1038/s41598-020-66483-y
Srivastava, A. K., Lu, Y., Zinta, G., Lang, Z., Zhu, J.-K. (2018). UTR-dependent control of gene expression in plants. Trends Plant Sci. 23, 248–259. doi: 10.1016/j.tplants.2017.11.003
Stehr, M., Diekmann, H., Smau, L., Seth, O., Ghisla, S., Singh, M., et al. (1998). A hydrophobic sequence motif common to N-hydroxylating enzymes. Trends Biochem. Sci. 23, 56–57. doi: 10.1016/S0968-0004(97)01166-3
Teufel, F., Almagro Armenteros, J. J., Johansen, A. R., Gíslason, M. H., Pihl, S. I., Tsirigos, K. D., et al. (2022). SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 40, 1023–1025. doi: 10.1038/s41587-021-01156-3
The International Wheat Genome Sequencing Consortium (IWGSC), Appels, R., Eversole, K., Stein, N., Feuillet, C., Keller, B., et al. (2018a). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191
Thodberg, S., Jakobsen Neilson, E. H. (2020). The “Green” FMOs: diversity, functionality and application of plant flavoproteins. Catalysts 10, 329. doi: 10.3390/catal10030329
Thodberg, S., Sørensen, M., Bellucci, M., Crocoll, C., Bendtsen, A. K., Nelson, D. R., et al. (2020). A flavin-dependent monooxygenase catalyzes the initial step in cyanogenic glycoside synthesis in ferns. Commun. Biol. 3, 507. doi: 10.1038/s42003-020-01224-5
Tichtinsky, G., Vanoosthuyse, V., Cock, J. M., Gaude, T. (2003). Making inroads into plant receptor kinase signaling pathways. Trends Plant Sci. 8, 231–237. doi: 10.1016/S1360-1385(03)00062-1
Vicient, C. M., Casacuberta, J. M. (2017). Impact of transposable elements on polyploid plant genomes. Ann. Bot. 120, 195–207. doi: 10.1093/aob/mcx078
Vikas, V. K., Pradhan, A. K., Budhlakoti, N., Mishra, D. C., Chandra, T., Bhardwaj, S. C., et al. (2022). Multi-locus genome-wide association studies (ML-GWAS) reveal novel genomic regions associated with seedling and adult plant stage leaf rust resistance in bread wheat (Triticum aestivum L.). Heredity 128, 434–449. doi: 10.1038/s41437-022-00525-1
Visca, P., Ciervo, A., Orsi, N. (1994). Cloning and nucleotide sequence of the pvdA gene encoding the pyoverdin biosynthetic enzyme L-ornithine N5-oxygenase in Pseudomonas aeruginosa. J. Bacteriology 176, 1128–1140. doi: 10.1128/jb.176.4.1128-1140.1994
Vlot, A. C., Sales, J. H., Lenk, M., Bauer, K., Brambilla, A., Sommer, A., et al. (2021). Systemic propagation of immunity in plants. New Phytol. 229 (3), 1234–1250. doi: 10.1111/nph.16953
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49–e49. doi: 10.1093/nar/gkr1293
Wong, T. K., Kalyaanamoorthy, S., Meusemann, K., Yeates, D., Misof, B., Jermiin, L. (2014). AliStat version 1.3 (CSIRO). Canberra, Australia. doi: 10.4225/08/59309DA8368E1
Xue, X., Liu, A., Hua, X. (2009). Proline accumulation and transcriptional regulation of proline biosynthesis and degradation in Brassica napus. BMB Rep. 42, 28–34. doi: 10.5483/BMBRep.2009.42.1.028
Yamamoto, Y., Kamiya, N., Morinaka, Y., Matsuoka, M., Sazuka, T. (2007). Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 143, 1362–1371. doi: 10.1104/pp.106.091561
Yang, F., Li, W., Jørgensen, H. J. L. (2013). Transcriptional Reprogramming of Wheat and the Hemibiotrophic Pathogen Septoria tritici during Two Phases of the Compatible Interaction. PloS One 8, e81606. doi: 10.1371/journal.pone.0081606
Yang, Y., Xu, T., Wang, H., Feng, D. (2021). Genome-wide identification and expression analysis of the TaYUCCA gene family in wheat. Mol. Biol. Rep. 48, 1269–1279. doi: 10.1007/s11033-021-06197-0
Yang, Z., Peng, Z., Wei, S., Liao, M., Yu, Y., Jang, Z. (2015). Pistillody mutant reveals key insights into stamen and pistil development in wheat (Triticum aestivum L.). BMC Genomics 16 (1), 211. doi: 10.1186/s12864-015-1453-0
Yoshimoto, N., Onuma, M., Mizuno, S., Sugino, Y., Nakabayashi, R., Imai, S., et al. (2015). Identification of a flavin-containing S -oxygenating monooxygenase involved in alliin biosynthesis in garlic. Plant J. 83, 941–951. doi: 10.1111/tpj.12954
Yoshimoto, N., Saito, K. (2015). “Identification of genes potentially encoding S-oxygenation enzymes for the biosynthesis of S-alk(en)yl-l-cysteine sulfoxides in onion,” in Molecular Physiology and Ecophysiology of Sulfur. Eds. De Kok, L. J., Hawkesford, M. J., Rennenberg, H., Saito, K., Schnug, E. (Cham: Springer International Publishing), 201–204. doi: 10.1007/978-3-319-20137-5_21
Yu, Y., Ouyang, Y., Yao, W. (2018). shinyCircos: An R/Shiny application for interactive creation of Circos plot. Bioinformatics 34, 1229–1231. doi: 10.1093/bioinformatics/btx763
Yu, J., Xuan, W., Tian, Y., Fan, L., Sun, J., Tang, W., et al. (2021). Enhanced OsNLP4-OsNiR cascade confers nitrogen use efficiency by promoting tiller number in rice. Plant Biotechnol. J. 19, 167–176. doi: 10.1111/pbi.13450
Zhang, C., Huang, L., Zhang, H., Hao, Q., Lyu, B., Wang, M., et al. (2019). An ancestral NB-LRR with duplicated 3′UTRs confers stripe rust resistance in wheat and barley. Nat. Commun. 10, 4023. doi: 10.1038/s41467-019-11872-9
Zhang, K., Zhang, J., Cui, C., Chai, L., Zheng, B., Jiang, J., et al. (2022). Genome-wide identification and expression profiling of the YUCCA gene family in Brassica napus. Oil Crop Sci. 7, 103–111. doi: 10.1016/j.ocsci.2022.07.001
Zhang, E. T., Zhang, H., Tang, W. (2021). Transcriptomic analysis of wheat seedling responses to the systemic acquired resistance inducer N-hydroxypipecolic acid. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.621336
Zhao, Y., Christensen, S. K., Fankhauser, C., Cashman, J. R., Cohen, J. D., Weigel, D., et al. (2001). A role for flavin monooxygenase-like enzymes in Auxin biosynthesis. Science 291, 306–309. doi: 10.1126/science.291.5502.306
Zhao, P., Ma, X., Zhang, R., Cheng, M., Niu, Y., Shi, X., et al. (2023). Integration of genome-wide association study, linkage analysis, and population transcriptome analysis to reveal the TaFMO1-5B modulating seminal root growth in bread wheat. Plant J. 116, 1385–1400. doi: 10.1111/tpj.16432
Zhou, C., Dong, Z., Zhang, T., Wu, J., Yu, S., Zeng, Q., et al. (2020). Genome-Scale Analysis of Homologous Genes among Subgenomes of Bread Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 21, 3015. doi: 10.3390/ijms21083015
Zhu, Y., Li, H., Su, Q., Wen, J., Wang, Y., Song, W., et al. (2019). A phenotype-directed chemical screen identifies ponalrestat as an inhibitor of the plant flavin monooxygenase YUCCA in auxin biosynthesis. J. Biol. Chem. 294, 19923–19933. doi: 10.1074/jbc.RA119.010480
Zhu, T., Wang, L., Rimbert, H., Rodriguez, J. C., Deal, K. R., De Oliveira, R., et al. (2021). Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 107, 303–314. doi: 10.1111/tpj.15289
Glossary
Keywords: disease resistance, FMO, gene family, phylogenetics, wheat
Citation: Sun S and Bakkeren G (2024) A bird’s-eye view: exploration of the flavin-containing monooxygenase superfamily in common wheat. Front. Plant Sci. 15:1369299. doi: 10.3389/fpls.2024.1369299
Received: 12 January 2024; Accepted: 19 February 2024;
Published: 12 April 2024.
Edited by:
Mingyue Gou, Henan Agricultural University, ChinaReviewed by:
Deepak Singla, Punjab Agricultural University, IndiaWei-Hua Tang, Chinese Academy of Sciences (CAS), China
Copyright © 2024 Sherry Sun and His Majesty the King in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada for the contribution of Guus Bakkeren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guus Bakkeren, guus.bakkeren@agr.gc.ca
†ORCID: Sherry Sun, orcid.org/0000-0002-9093-9171
Guus Bakkeren, orcid.org/0000-0002-3065-6989
 Sherry Sun
Sherry Sun Guus Bakkeren
Guus Bakkeren