-
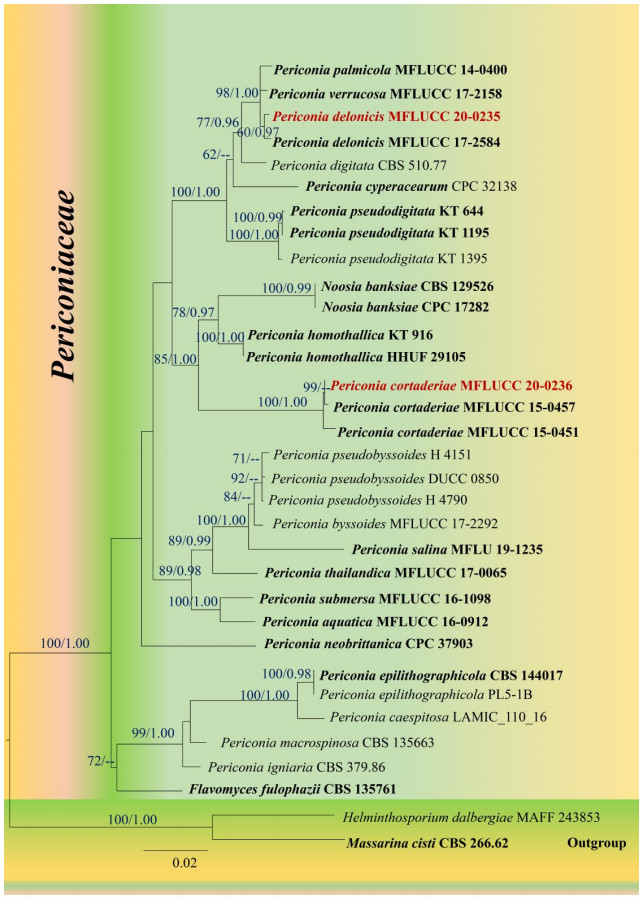
Figure 1.
Maximum likelihood tree revealed by RAxML from an analysis of a concatenated SSU, LSU, ITS and TEF sequence dataset of the species in Periconiaceae, showing the phylogenetic position of Periconia delonicis (MFLUCC 20–0235) and P. cortaderiae (MFLUCC 20–0236). ML bootstrap supports (≥ 60%) and Bayesian posterior probabilities (≥ 0.95 BYPP) are given above the branches as ML/BYPP. The tree is rooted with Helminthosporium dalbergiae (MAFF 243853) and Massarina cisti (CBS 266.62). Strains generated in this study are indicated in red bold. Ex–type strains are indicated in black bold. The scale bar 0.02 represents the expected number of nucleotide substitutions per site.
-

Figure 2.
Maximum likelihood tree (RAxML) revealed by an analysis of a concatenated SSU, LSU, ITS, TEF and RPB2 sequence dataset of the species in Torulaceae, showing the phylogenetic position of Torula chromolaenae (MFLUCC 20–0237), T. fici (MFLUCC 20–0238) and T. masonii (MFLUCC 20–0239). ML bootstrap supports (≥ 60%) and Bayesian posterior probabilities (≥ 0.95 BYPP) are given above the branches as ML/BYPP. The tree is rooted with Roussoella nitidula (MFLUCC 11–0182), R. scabrispora (MFLUCC 11–0624) and Roussoellopsis tosaensis (KT 1659). Strains generated in this study are indicated in red bold. Ex–type strains are indicated in black bold. The scale bar 0.04 represents the expected number of nucleotide substitutions per site.
-

Figure 3.
Periconia delonicis (MFLU 20-0696). a-c Conidiophores bearing conidia on host. d-f Conidia and conidiophores. g, h Conidiogenous cells bearing conidia. i Base of the conidiophore. j-s Mature and immature conidia. t Colonies on PDA after 14 days. Scale bars: a, b = 500 μm, c, d = 100 μm, e, f, i = 20 μm, g, h = 10 μm, j-s = 5 μm.
-

Figure 4.
Periconia cortaderiae (MFLU 20-0697). a-c Colonies on host substrate. d, f Conidiophores bearing conidia. e, g, h Conidial masses. i, j, m-o Conidiogenesis from terminal mother cells. l Monoblastic, annellidic conidiogenous cells bearing conidial chains. k, p, q, r Conidial chains. s Conidia. Scale bars: a = 3 mm, b = 3.5 mm, c = 50 μm, d, f = 50 μm, e, g = 20 μm, h-s = 5 μm.
-

Figure 5.
Torula chromolaenae (MFLU 20–0698). a-c Colonies on leaf vein of Musa sp. d, e Conidia arranged in multi planes. f Conidiophore and conidiogenous cell. g-p Conidia and the formation of immature conidia. q Colonies on PDA after 21 days. Scale bars a = 1000 μm, b = 500 μm, c = 50 μm, d-h, k, o, p = 10 μm, l-n, i, j = 5 μm.
-

Figure 6.
Torula fici (MFLU 20-0699). a-c Colonies on dead leaf of Musa sp. d-n Conidiogenesis. o, p Conidial masses. q Conidia (in chain). r-u Disposed conidia. v Colonies on PDA after 28 days. Scale bars: a = 500 μm, b = 100 μm, c = 20 μm, d-u = 5 μm.
-

Figure 7.
Torula masonii (MFLU 20–0700). a Colonies on dead leaf of Musa sp. b, c Masses of conidia on host surface. d–h, j–o Conidia. i Conidiophore, conidiogenous cell and budding of conidia. p Colonies on PDA after 28 days. Scale bars: a = 50 μm, b = 500 μm, c = 1000 μm, d, f–o = 10 μm, e = 5 μm.
-
Taxa Culture collection/Voucher no. GenBank accession numbers LSU SSU ITS TEF Flavomyces fulophazii CBS 135761T KP184040 KP184082 KP184001 NA F. fulophazii MF09 MN515261 NA MN537663 MN535259 Helminthosporium H 4628 AB807521 AB797231 LC014555 AB808497 dalbergiae Massarina cisti CBS 266.62T AB807539 AB797249 NA AB808514 Noosia banksiae CPC: 17282 JF951167 NA JF951147 NA N. banksiae CBS 129526 MH878062 NA NA NA Periconia aquatica MFLUCC 16–0912T KY794705 NA KY794701 KY814760 P. byssoides MFLUCC 17–2292 MK347968 MK347858 MK347751 MK360069 P. byssoides MFLUCC 18–1548 MK348013 MK347902 MK347794 MK360070 P. caespitosa LAMIC_110_16 MH051907 NA MH051906 NA P. cortaderiae MFLUCC 15–0457T NG_068238 NG_068373 NR_165853 KY310703 P. cortaderiae MFLUCC 15–0451 KX954403 KX986346 KX965734 KY429208 P. cortaderiae MFLUCC 20–0236 MW406971 MW406969 MW406973 MW422156 P. cyperacearum CPC: 32138T NG_064549 NA NR_160357 MH327882 P. delonicis MFLUCC 17–2584T MK347941 MK347832 NA MK360071 P. delonicis MFLUCC 20–0235 MW406970 MW406968 MW406972 MW422155 P. digitata CBS 510.77 AB807561 AB797271 NA AB808537 P. epilithographicola CBS 144017T NA NA NR_157477 NA P. epilithographicola PL5–1B NA NA MF422162 NA P. homothallica HHUF 29105 NG_059397 NG_064851 NR_153446 AB808541 P. homothallica KT 916 AB807565 AB797275 AB809645 NA P. igniaria CBS 379.86 AB807566 AB797276 LC014585 AB808542 P. igniaria CBS 845.96 AB807567 AB797277 LC014586 AB808543 P. macrospinosa CBS 135663 KP184038 KP184080 KP183999 NA P. neobrittanica CPC 37903T NG_068342 NA NR_166344 NA P. palmicola MFLUCC 14–0400T NG_068917 MN648319 NA MN821070 P. pseudobyssoides DLUCC 0850 MG333494 NA MG333491 MG438280 P. pseudobyssoides H 4151 AB807568 AB797278 LC014587 AB808544 P. pseudobyssoides H 4790 AB807560 AB797270 LC014588 AB808536 P. pseudodigitata KT 644 AB807562 AB797272 LC014589 AB808538 P. pseudodigitata KT 1195A AB807563 AB797273 LC014590 AB808539 P. pseudodigitata KT 1395 AB807564 AB797274 LC014591 AB808540 P. salina MFLU 19–1235T MN017846 MN017912 MN047086 NA P. submersa MFLUCC 16–1098T KY794706 NA KY794702 KY814761 P. thailandica MFLUCC 17–0065T KY753888 KY753889 KY753887 NA Table 1.
Taxa used in the phylogenetic analyses of Periconiaceae with the corresponding GenBank accession numbers. Type strains are superscripted with "T" and newly generated strains are indicated in black bold.
-
Taxa Culture collection/Voucher no. ITS LSU SSU RPB2 TEF Dendryphion aquaticum MFLUCC 15–0257T KU500566 KU500573 KU500580 NA NA D. comosum CBS 208.69T MH859293 MH871026 NA NA NA D. europaeum CPC 23231 KJ869145 KJ869202 NA NA NA D. hydei KUMCC 18–0009T MN061343 MH253927 MH253929 NA NA Neotorula aquatica MFLUCC 15–0342T KU500569 KU500576 KU500583 NA NA N. submersa HKAS 92660 NR_154247 KX789217 NA NA NA Rostriconidium aquaticum KUMCC 15–0297 MG208165 MG208144 NA MG207975 MG207995 R. aquaticum MFLUCC 161113T MG208164 MG208143 NA MG207974 MG207994 R. pandanicola KUMCC 17–0176T MH275084 MH260318 MH260358 MH412759 MH412781 Roussoella nitidula MFLUCC 11–0182T KJ474835 KJ474843 NA KJ474859 KJ474852 R. scabrispora MFLUCC 11–0624T KJ474836 KJ474844 NA KJ474860 KJ474853 Roussoellopsis tosaensis KT 1659 NA AB524625 AB524484 AB539104 AB539117 Rutola graminis CPC 33267 MN313814 MN317295 NA NA NA R. graminis CPC 33695 MN313815 MN317296 NA NA NA R. graminis CPC 33715T MN313816 MN317297 NA NA NA Sporidesmioides thailandica MFLUCC 13–0840 MN061347 NG_059703 NG_061242 KX437761 KX437766 S. thailandica KUMCC 16–0012T MN061348 KX437758 KX437760 KX437762 KX437767 Thyridaria broussonetiae TB1 KX650569 NA KX650515 KX650586 KX650539 Thyridariella mahakoshae NFCCI 4215 MG020435 MG020438 MG020441 MG020446 MG023140 Th. mangrovei PUFD 17–98T MG020434 MG020437 MG020440 MG020445 MG020443 Torula acaciae CPC 29737T NR_155944 NG_059764 NA KY173594 NA T. aquatica DLUCC 0550 MG208166 MG208145 NA MG207976 MG207996 T. aquatica MFLUCC 16–1115T MG208167 MG208146 NA MG207977 NA T. breviconidiophora KUMCC 18–0130T MK071670 MK071672 MK071697 NA MK077673 T. camporesii KUMCC 19–0112T MN507400 MN507402 MN507401 MN507404 MN507403 T. chiangmaiensis KUMCC 16–0039T MN061342 KY197856 KY197863 NA KY197876 T. chromolaenae MFLUCC 20–0237 MW412524 MW412518 MW412515 MW422161 MW422158 T. fici CBS 595.96T KF443408 KF443385 KF443387 KF443395 KF443402 T. fici KUMCC 15–0428 MG208172 MG208151 NA MG207981 MG207999 T. fici KUMCC 16–0038 MN061341 KY197859 KY197866 KY197872 KY197879 T. fici MFLUCC 20–0238 MW412525 MW412519 MW412516 NA MW422159 T. gaodangensis MFLUCC 17–0234T MF034135 NG_059827 NG_063641 NA NA T. goaensis MTCC 12620T NR_159045 NG_060016 NA NA NA T. herbarum CPC 24414 KR873260 KR873288 NA NA NA T. hollandica CBS 220.69 NR_132893 NG_064274 KF443389 KF443393 KF443401 T. hydei KUMCC 16–0037T MN061346 MH253926 MH253928 NA MH253930 T. mackenziei MFLUCC 13–0839T MN061344 KY197861 KY197868 KY197874 KY197881 T. masonii CBS 245.57T NR_145193 NG_058185 NA NA NA T. masonii DLUCC 0588 MG208173 MG208152 NA MG207982 MG208000 T. masonii MFLUCC 20–0239 MW412523 MW412517 MW412514 MW422160 MW422157 T. pluriseptata MFLUCC 14–0437T MN061338 KY197855 KY197862 KY197869 KY197875 T. polyseptata KUMCC 18–0131T MK071671 MK071673 MK071698 NA MK077674 Abbreviations of culture collections: CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. CPC: Working collection of Pedro Crous housed at CBS. DLUCC: Dali University Culture Collection, China. H: University of Helsinki, Helsinki, Finland. HKAS: Herbarium of Cryptogams, Kunming Institute of Botany, Academia Sinica, China. KT: K. Tanaka. LAMIC: Laboratorio Asociaciones suelo, planta microorganismos, Pontificia Universidad Javeriana, Bogotá, D.C., Colombia. MTCC: Microbial Type Culture Collection, CSIR–Institute of Microbial Technology, Sector 39–A, Chandigarh – 160036, India. KUMCC: Kunming Institute of Botany Culture Collection, China. MFLU: Mae Fah Luang University, Chiang Rai, Thailand. MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand. NFCCI: National Fungal Culture Collection of India. NA: DNA sequence data are not available in GenBank. Table 2.
Taxa used in the phylogenetic analyses of Torulaceae with the corresponding GenBank accession numbers. Type strains are superscripted with "T" and newly generated strains are indicated in black bold.
Figures
(7)
Tables
(2)