

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.020721
REVIEW
Distribution, Etiology, Molecular Genetics and Management Perspectives of Northern Corn Leaf Blight of Maize (Zea mays L.)
1Mountain Research Centre for Field Crops, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, Jammu & Kashmir, 192124, India
2Dry Land Agriculture Research Station Rangreth, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, Jammu & Kashmir, 191132, India
3School of Agricultural Biotechnology, Punjab Agricultural University, Ludhiana, 141001, India
4Indian Institute of Maize Research, PAU Campus, Ludhiana, 141001, India
5College of Temperate Sericulture, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, Jammu & Kashmir, 191132, India
6Department of Agronomy, Faculty of Agriculture, Kafrelsheikh University, Kafr El-Sheikh, 33516, Egypt
7Department of Field Crops, Faculty of Agriculture, Siirt University, Siirt, 56100, Turkey
8Department of Field Crops, Faculty of Agriculture, University of Çukurova, Adana, 01330, Turkey
9Department of Field Crops, Faculty of Agriculture, Hatay Mustafa Kemal University, Hatay, 31060, Turkey
10Department of Genetics and Plant Breeding, Bangladesh Agricultural University, Mymensingh, 2202, Bangladesh
*Corresponding Authors: Shabir Hussain Wani. Email: shabirhwani@skuastkashmir.ac.in; Mohammad Anwar Hossain. Email: anwargpb@bau.edu.bd
Received: 08 December 2021; Accepted: 23 March 2022
Abstract: Maize is cultivated extensively throughout the world and has the highest production among cereals. However, Northern corn leaf blight (NCLB) disease caused by Exherohilum turcicum, is the most devastating limiting factor of maize production. The disease causes immense losses to corn yield if it develops prior or during the tasseling and silking stages of crop development. It has a worldwide distribution and its development is favoured by cool to moderate temperatures with high relative humidity. The prevalence of the disease has increased in recent years and new races of the pathogen have been reported worldwide. The fungus E. turcicum is highly variable in nature. Though different management strategies have proved effective to reduce economic losses from NCLB, the development of varieties with resistance to E. turcicum is the most efficient and inexpensive way for disease management. Qualitative resistance for NCLB governed by Ht genes is a race-specific resistance which leads to a higher level of resistance. However, some Ht genes can easily become ineffective under the high pressure of virulent strains of the pathogen. Hence, it is imperative to understand and examine the consistency of the genomic locations of quantitative trait loci for resistance to NCLB in diverse maize populations. The breeding approaches for pyramiding resistant genes against E. turcicum in maize can impart NCLB resistance under high disease pressure environments. Furthermore, the genome editing approaches like CRISPR-cas9 and RNAi can also prove vital for developing NCLB resistant maize cultivars. As such this review delivers emphasis on the importance and current status of the disease, racial spectrum of the pathogen, genetic nature and breeding approaches for resistance and management strategies of the disease in a sustainable manner.
Keywords: Northern corn leaf blight; etiology; Exherohilum turcicum; pathogenic variability; disease resistance; management strategies
Northern corn leaf blight (NCLB) caused by the fungus Exherohilum turcicum (synonym Setosphaeria turcica), is a destructive foliar disease of maize, sorghum, and related grass species [1]. The disease is widely distributed and economically the most important foliar disease of maize [2,3]. The disease has a worldwide distribution predominantly in areas with 75%–90% relative humidity and 22°C–25°C temperature during the cropping season [4,5]. NCLB causes enormous damage to the maize crops, and grain yield losses range from 24% to 91% [6,7], depending on the growth stage of the crop at which infection occurs, the severity of the outbreak, the resistance of the host plant and the virulence of the pathogen. The disease is more destructive if it appears prior to silk emergence. Disease development during the early growth stages results in the premature death of leaves. Hence the loss of photosynthetic area affects grain yield as well as fodder quality; this is of particular significance under temperate climatic conditions since fodder is fed to cattle during the lean season [8,9]. Different races of S. turcica have been identified throughout the world such as the races 0, 1, 2, 3, 12, 13 23, N, 1N, 2N, 3N, 13N, 23N and 123N based on their virulence against various resistant genes (Ht1, Ht2, Ht3, HtM, Htn1, ht4, HtP, HtNB) in maize [10,11]. Host plant tolerance relies on the efficacy of resistance against all virulent pathogen races in the region. The fungus S. turcica is considered to be extremely variable in cultural features, pathogenicity and genetic traits. Hence, the lack/loss of significant durable resistance in the maize genotypes is due to the presence of variability and continuous change in the racial spectrum of the pathogen [12]. Genetic diversity and pathogenicity of the pathogen are important factors in the resistance of the host plant. Hence, identifying the heterogeneity of pathogen isolates is an important step in the creation of a programme for disease management for a specific area and development of multi-racial disease-resistant cultivars. Deployment of resistant cultivars is by far the most successful and cost-effective way to manage the NCLB. Resistance to NCLB in maize can be obtained by breeding with qualitative and quantitative resistance, either independently or in combination. The Ht (Helminthosporium turcicum) genes are recognised for conferring qualitative resistance controlled by a single gene (and mostly dominant) which contributes to a higher level of resistance. Quantitative resistance is regulated by many genes and shows a significant reduction in NCLB disease severity, particularly in areas where race population of S. turcica is very high. Quantitative trait loci (QTL) or linkage mapping is an important approach to study polygenic and complex forms of disease tolerance. QTL for NCLB resistance has been established in many populations [13–16]. The present review lays out the rationale for NCLB disease development, variability and population structure of causal pathogen, genetics of resistance, the progress of gene identification against NCLB, and management strategies.
2 Distribution and Importance of NCLB
NCLB, also known as turcicum leaf blight (TLB) is among the most prevalent diseases of maize disseminated worldwide, particularly in regions with high humidity and moderate temperature [17–19]. The disease was first reported in 1876 from Italy by Passerine. The disease is distributed in the continents of Asia, Europe, North America, Africa, Oceania and South America (Fig. 1; CABI, 2019). Presently NCLB is a potential threat to maize cultivation in Europe, Australia, North-Eastern United States, Sub-Saharan Africa, and in areas of North Korea, India, and China [20,21–23]. Butler [24] first reported the NCLB in India on sorghum, and Mitra [25] reported it from Punjab on sorghum and maize. The disease is most prevalent in all the major maize growing regions of India during the rainy (Kharif) as well as winter (Rabi) season [26]. Almora, Bajaura, Mandya, Dharward, Imphal and Kashmir are the hot spots for NCLB in India. This disease occurs sporadically in most temperate, humid maize-grown areas and is of particular concern in the tropical highlands, where conditions favour disease development [27]. The disease was also found to be the major restraint of maize production under the temperate climatic conditions of Ahangar et al. [28].
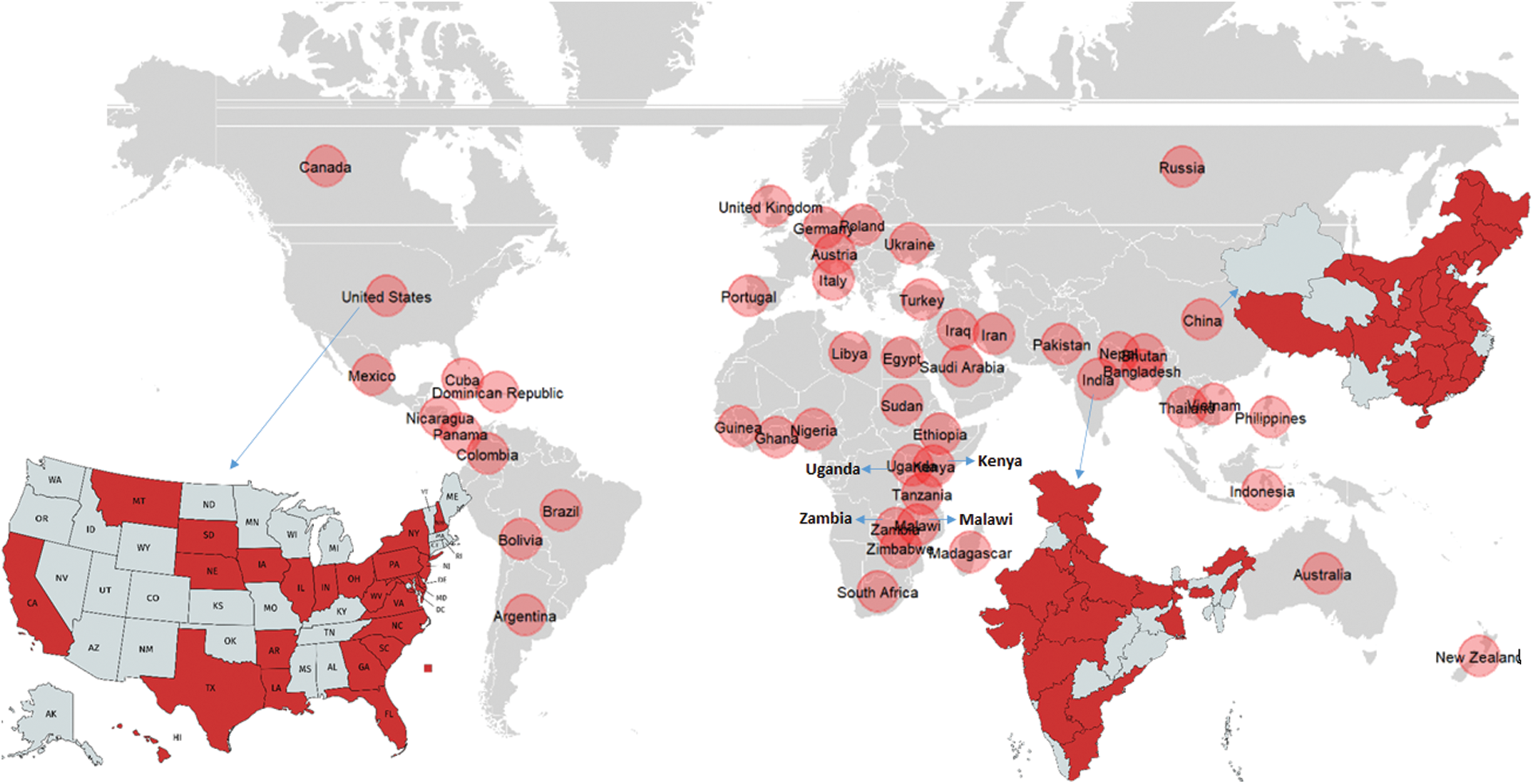
Figure 1: Worldwide distribution of NCLB along with a major focus of the distribution in USA, India and China (Data source: https://www.cabdirect.org/cabdirect/abstract/20046500257)
In maize, NCLB is a widespread foliar disease mainly found in temperate and tropical environments that cause yield reductions of up to 70% [29]. The disease also induces qualitative changes in the seed, such as reduced sugar content and germination potential, in addition to predisposition of infected plants to stalk rot [30,31]. The degree of yield losses due to NCLB depends on the growth stage of the crop at which infection occurs, the severity of the outbreak, the resistance of the host plant and the virulence of the pathogen. If the disease occurs before silking, a 40% yield decrease can occur [32], but if infection deferred until 6–8 weeks after silking, yield losses are minimal [33]. However, yield losses reach up to 50% when the disease occurs severely at 2–3 weeks after pollination [17]. The disease can substantially reduce the grain yield of maize over a wide range from 28% to 91% [7,32]. Average losses of 60% have been reported in Kenya, Uganda, Ethiopia, South Africa and Zambia [34]. Maize crops in the temperate belt of Kashmir are ravaged by this destructive disease with losses in the range of 27.6%–90.7% of total grain yield, particularly if the disease develops prior to silk emergence [35].
NCLB/TLB is caused by Exherohilum turcicum [Pass.] Leonard and Suggs, synonym Setosphaeria turcica (Luttrell). The fungus is heterothallic ascomycete that belongs to the subclass Loculoascomycetidae, order Pleosporales. Phylogenetic studies based on different loci indicated that Exserohilum belongs to the family Pleosporaceae, order Pleosporales [36]. The pathogen is a polycyclic, facultative parasite of maize. Leonard and Suggs have proposed the nomenclature of the organism as Exherohilum turcicum (Pass.) K. J. Leonard and E. G. Suggs, as an imperfect stage and teleomorphic phase was described in 1957 as Trichometasphaeria turcica by Luttrell and later modified to Setosphaeriaturcica (Luttrell) Leonard and Suggs. Normally, the causal agent of NCLB is defined by its imperfect stage Exherohilum turcicum in which a conidial hilum is strongly protuberant (Fig. 2e). The use of the name Exserohilum over Setosphaeria was claimed according to the Article 57.2 of the International Code of Nomenclature for algae, fungi and plants [37]. 38 taxa in Exserohilum have been listed by MycoBank which are distinguished on the basis of morphological features [38]. Hernández-Restrepo et al. [39] described 11 Exserohilum phylogenetic species based on nine nuclear loci, viz., ITS, LSU, act, tub2, cam, gapdh, his, tef1 and rpb2, as well as phenotypic data. Setosphaeria differs from Trichometasphaeria by the production of non-clypeateascomata which can be erumpent or superficial and produce larger ascospores [40]. Eight Setosphaeria species have been described by mating of compatible isolates [40,41]. The sexual stage of the fungus, Setosphaeria turcica rarely occurs under natural conditions [42]. The fungus exists with three distinct mating forms present in nature [43]. Mating of S. turcica is attained by inoculating compatible strains onto culture media with sterilized trashes of natural substrates such as maize leaf or wheat straw. Bunkoed et al. [44,45] first conducted the study on sexual reproduction of S. turcica in Thailand. Pseudothecia were found on highly infected corn leaves from natural fields. Conidiophores are simple, cylindrical, olive brown, shaped individually or in groups of two to four from stomata in necrotic leaf lesions (Fig. 2c). Single conidium is formed terminally on the conidiophores (Fig. 2d) which then resumes growth to one side of the conidial attachment and eventually produce another conidium at the new tip [40,41,46]. The vegetative hyphae remain mostly immersed, septate, branched, olivaceous brown, smooth about 3–7.5 µm wide. The conidia of the fungi are olivaceous-grey, elongated and spindle-shaped often less curved on one side (Fig. 2f) compared with the conidia of Helminthosporium maydis, which are more curved. The average size of conidia is about 20–150 µ with one to nine septa. A conspicuous spore feature of the conidia that distinguishes it from other more common species attacking maize is the protruding hilum [17,45].
About 14 days after infection, the disease symptoms appear as small, oval, greyish green and water-soaked spots which grow into elongated, spindle-shaped necrotic lesions [38]. NCLB lesions are elongated elliptical greyish, measuring up to 12 mm wide and 2.5–15 cm long which run parallel to leaf margin [46,47] (Figs. 2a and 2b). On mature lesions, distinct dark grey areas develop associated with fungal spores [48]. Spore formation causes the appearance of lesions to olive, dark, grey or black in colour [49]. NCLB is essentially a leaf disease and symptoms usually appear at any stage of the crop on the lower leaves spreading upwards [50,51]. The disease spots first appear on lower leaves and the number of spots increases and spreads up with the development of plant, leading to a complete blighting of the foliage. Symptoms range from cigar-shaped lesions on the lower leaves to the total loss of the foliage (Fig. 2b), thereby reducing the amount of leaf area required for photosynthesis [47]. Lesions of NCLB vary in shape and size depending on the race of E. turcicum and tolerance level of the genotype. Lesionfeatures vary among maize genotypes based on their resistance and interaction with different races of the pathogen. Race 1 develops oval to circular, tan lesions on leaves about 1.2 to 2.5 cm in size while Race 2 develops about 0.5 to 2.5 cm long oblong, brown spots. Race 3 and Race 4 causes narrow, long, bordered grey lesions on leaves and Race 0 develops only small flecks or spots. E. turcicum develops appressoria and penetrates the leaf surface directly. In incompatible reactions, the pathogen penetrates into xylem vessels and causes leaf chlorosis while as in compatible interactions it strongly colonizes the mesophyll and results on leaf necrosis and development of typical NCLB symptoms.

Figure 2: Symptoms and etiology of S. turcica. Elongated elliptical greyish NCLB lesions (2a), blighting and death of leaves (2b), cylindrical conidiophores (2c), conidial formation on conidiophore (2d), conidia with protruding hilum (2e), elongated to spindle-shaped conidia (2f), bipolar germination of conidium (2g) and dormant conidiophores (2h)
This fungus has saprophytic survival which over-winters as dormant mycelia, conidia and chlamydospores (Fig. 2h) on maize residues left on the soil surface. Mycelia and conidia of this fungus from infected crop residues, in or on the soil, serve as the primary inoculum for the next crop [52,53]. The secondary inoculum is caused by disease lesions on leaves on which the fungus develops conidia that are spread over long distances by wind and rain [54,55]. Seed-borne nature of the fungus has been also reported which remains viable in the seed for 28 months [56]. The NCLB is favoured by mild temperatures (between 15°C to 25°C), high relative humidity (90%–100%), extended periods of leaf wetness (rain or dew at least 4 h) and frequent light showers [17,57–59]. Germination of E. turcicum conidia is bipolar (Fig. 2g) and develops infection 3–6 h after inoculation. Germ tubes grow at an angle and generate simple or forked terminal appressoria from which penetration pegs grow [60]. Penetration occurs directly through leaf cuticle and epidermis and occurs rarely through stomata [61]. The pathogen produces a range of secondary metabolites and toxins to allow penetration and colonization. Two pathogenicity related genes of the S. turcica genome encode xylanase enzymes that destroy the arabinoxylan in the plant cell wall responsible for its integrity thus leading to pathogen infiltration [62]. Infection pegs expand into or between, the epidermal cells of the dorsal or ventral sides of the leaf [2,63]. Penetration usually occurs 12–18 h after inoculation [50,64]. After penetration, the fungus develops a vesicle-like structure within the epidermal cells, giving rise to secondary hyphae that appear intracellular in the mesophyll tissue in different directions [63,65]. The hyphae begin to progress within the chlorenchyma tissue, culminating in cell death and lesion development. The cells later become devoid of all cytoplasm, separate and disorganized [60]. The hyphae grow from the xylem to the underlying healthy tissues, infiltrate the normal bundle sheath and grow quickly in neighbouring mesophyll cells resulting in the enlargement of the lesions. Inside the tissue, the hyphae secrete a HT toxin called Monocerin [66] which comprises low molecular weight, water-soluble compounds that inhibit chlorophyll synthesis [67]. Mycelial threads aggregate into pseudoparenchymatous masses in sub-stomatal chambers. Conidiophores produced by these thick masses arise through the stomata and grow conidia extensively [63,64]. The incidence and severity of NCLB vary from site to site and year over year depending upon the virulence of the pathogen, response of the plants and prevailing environmental conditions [5]. Disease lesions on maize leaves develops at a faster rate during the night than day period. Thus, day lengths shorter than 12 h increases disease development. NCLB is commonly considered to be sporadic in frequency, depending on the environmental factors and the disease tolerance of the plant [68,69]. In general, the increase in the prevalence of the disease might be attributed to mono-cropping practices, high humidity, morning fogs, extended dew periods, minimum tillage and the use of uniform and highly susceptible varieties [69–72].
4 Variability and Population Structure of E. turcicum
Genetic diversity and pathogenicity of the microorganism are important factors for the host resistance and production of effective disease control strategies. Novel races of the pathogen are being frequently developed and the pathogen is being shifted to new regions. The fungus E. turcicum is considered to be extremely diverse in terms of cultural traits, pathogenicity and genetic composition. Molecular diversity of E. turcicum isolates varies considerably from region to region [73]. Higher molecular variability has been observed among-populations of E. turcicum from different hosts compared to the populations from different locations [74]. Genotypic diversity and gametic phase equilibrium in S. turcica populations develop more in tropical regions than populations from temperate regions. Higher sexual recombination rates have also been observed in tropical climates, whereas populations in temperate areas tend to be more clonal. S. turcica populations are highly adaptable in both temperate and tropical climates, as an extensive migration was also found within agro-ecological zones [73]. Isolates of E. turcicum, from different locations, varies in parasitic fitness in terms of the effectiveness of invasion, sporulation and lesion size, as well as in colour, type of mycelium, growth rate and sporulation in culture [75–78]. Highly virulent isolates exhibit more infection on different differentials. In a study by Muiru et al. [79], E. turcicum isolates from Germany, Kenya and Austria showed a varied response on the differentials indicating a high virulence complexity and variability of the pathogen. The aggressiveness of the various E. turcicum isolates differs in terms of lesion density, area under the disease progress curve (AUDPC), lesion size, length of incubation period and rate of lesion expansion [73,80]. Assefa [81] indicated significant differences among E. turcicum isolates in their virulence and the mean virulence rating was significantly correlated with spore length and rate of germination. Wathaneeyawech et al. [22] found substantial variability among 478 isolates of E. turcicum collected from Thailand. Significant morphological variability was also detected among the E. turcicum isolates from Argentina and Brazil for all measured variables viz., length, width and number of septa [53]. Substantial variations in morphology [28,82,83,84], pathogenicity [77,79,85], and genetic diversity [86,87] have been observed among the E. turcicum isolates from different agro-ecological regions. Knox-Davies et al. [88] reported ample evidence of heterokaryons and their perpetuation by conidia, and proposed that high variability in the fungus population might be related to heterokaryosis. Bunkoed et al. [44] first examined the sexual stage of S. turcica and proposed that sexual reproduction had induced genetic variation in this pathogen. In addition, virulence may be improved and new physiological races might have been produced by sexual hybridization. Two mating types MAT-1 and MAT-2 of S. turcica have been reported. These mating types are regulated by a single locus having two highly dissimilar alleles. In tropical environments, sexual hybridization is responsible for greater adaptation ability due to the existence of equal fractions of MAT-1 and MAT-2 [73]. Li et al. [89] reported three mating types, ‘Aa’, ‘a’, and ‘A’, among which ‘a’ was the dominant type in China’s Heilongjiang Province.
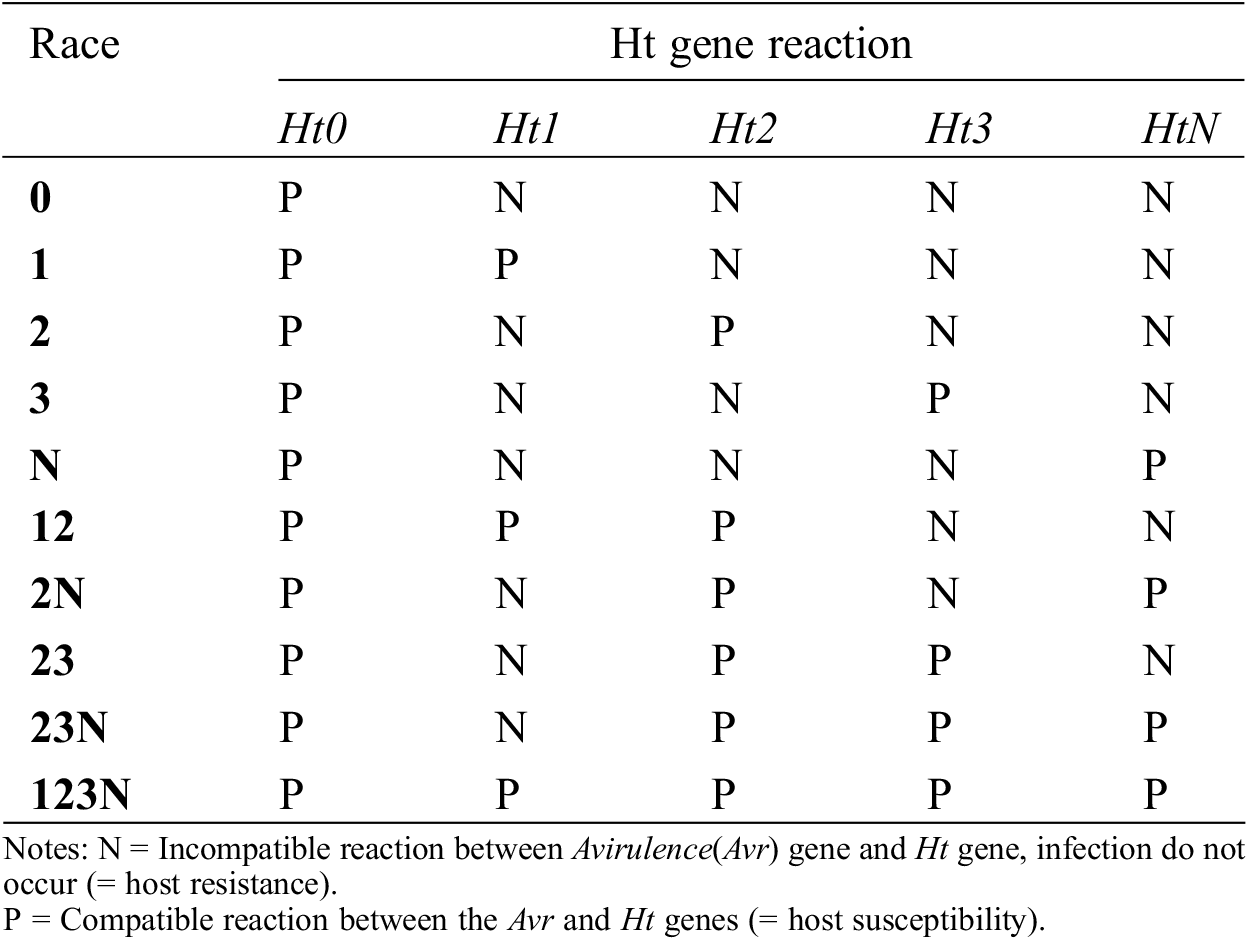
The prevalence of NCLB has increased in recent years and new races of the pathogen have been reported worldwide. NCLB intimidation to maize production is mainly due to the presence of S. turcicum races and the potential of the pathogen for the development of new races. Vidal-Villarejo et al. [90] reported that new pathogen lineages of S. turcica are not generated by race-specific virulence. High mutation rates of the pathogen may be the frequent origin of new races. Identification of races of the pathogen present in an area and the recognition of their spatial distribution are important steps in the generation of resistant cultivars in different pathogen systems where major genes regulate resistance. S. turcica races are described based on their phenotypic reaction whenever inoculated into a series of different maize lines [91]. The disease has spread around the world with a variety of distinct races such as Races 0, 1, 2, 3, 12, 13 23, N, 1N, 2N, 3N, 13N, 23N and 123N [11,79]. An increased number of S. turcica races identified from different regions of the world contributed to the quick loss in tolerance of many hybrids containing Ht genes [92]. Race designations are based on resistance genes and their corresponding virulence matches. For example, S. turcica Race 0 is avirulent for all the Ht genes, while Race 1 is only effective (virulent) for the Ht1 gene; Race 23N is virulent in response to Ht2, Ht3, and Htn1 genes; Races 3 and 4 are virulent in response to Ht2, Ht3, Htn genes, and Race 12 is virulent against Ht1 and Ht2 genes [Table 1]. Races 0 and 1 are more prevalent, whereas Races 2N, 23, and 23N are rarely found [93]. The Race 123N with the highest virulence complexity tends to infect all of the cultivars with matching Ht genes [94]. The race distribution of S. turcica is considerably variable in different regions of the world. Fourteen races of S. turcica have been reported from diverse geographic locations of China with a dominancy of Race 0 and 1 [11]. A total of 12 races have been found in samples from Germany, Kenya and Austria, with Race 2 appearing more frequently [79]. From Turkey eight diverse races of S. turcica were found, namely 0, 1, 2, 123, N, 3N, 12N, 1N, among which Race 0 and Race 1 were the most common [95]. Similarly, 17 races of S. turcica were identified in Canada with Race 0, 1M, 1N and 1MN being the most prevalent [96]. Race 0, 1 and 23N of S. turcica are found in Argentina and Brazil with Race 0 in abundance at a frequency of 83% and 65%, respectively. Therefore, Race 0 has the highest abundance throughout the world [97]. The physiological race of S. turcica present in the Indian subcontinent has been determined to be Race 2 [98]. Studies have also shown that S. turcica migration is likely over long distances, which could shift virulence to new regions [99]. Selection pressure, sexual recombination within the pathogen, and the capability to migrate long distances may create more virulent populations and contribute to spatial and temporal race population shifts. Distribution of S. turcica races on larger geographical regions of the world impresses upon the monitoring of pathogen diversity on large scales and over time to fully understand factors influencing the evolution of pathogen races.
5 Genetic Nature and Breeding Approaches for NCLB Resistance: Qualitative and Quantitative Genes
NCLB resistance is either qualitative which is usually race-specific and inherited from a single gene (monogenic) although quantitative resistance is race nonspecific and polygenic [100,101]. Monogenic or race-specific resistance for NCLB is controlled by Ht1, Ht2, Ht3 and Htn genes. Gene Ht1 (Ht for Helminthosporium turcicum) confers a chlorotic lesion type and was the first single gene resistance, identified by Hooker [102] from the inbred line, GE339 and popcorn cv ‘Ladyfinger’. In maize lines with Htn gene, lesion formation is delayed in such way that plants in the field remain free from lesion until shortly after pollination while the expression of Ht1, Ht2 and Ht3 resistant genes occur as chlorotic lesions with minimum sporulation [91]. Symptoms produced in resistance responses by the different Ht genes are considerably variable. Ht1 shows necrotic lesions with chlorosis, maize lines with Ht2 resistance gene exhibits chlorosis and small lesions, Ht3 shows chlorotic spots while as in Htn1 resistance reaction there is no lesion development. E. turcicum forms appressoria and penetrates the leaf surface directly in both compatible as well as in incompatible reaction types. However, the pathogen penetration in xylem vessels and colonization to mesophyll restrict in resistant interactions [103]. Six dominant genes (Ht2, Ht3, Ht1, Htn1, Htm1, and HtNN) and two recessive genes (ht4 and rt) have been identified to provide resistance to the various races of S. turcica [13,59,104–106]. HtP and Ht1 are found on chromosome 2L (Bin 2.08) and mapped 10 cM from one another [58]. Htn1 and Ht2 were found on chromosome 8L [107,108] and rt is positioned on chromosome 3L (bin 3.06) [59]. Htn1 encodes for a wall-associated receptor-like kinase which functions as core element of innate immune response by detecting pathogens or host-derived elicitors [109].
The specific region of chromosome 8 (bin 8.05–8.06) of the maize genome harbours a locus responsible for a substantial level of NCLB tolerance in maize germplasm. This is because in various biparental populations, several NCLB QTL and two major gene loci, Ht2 and Htn1 have been mapped to bin 8.05-8.06 [108,110,111]. The introgression of Ht3 gene from Tripsacumfloridanum was carried out in maize [112] and mapped on bin 7.04 [113]. The ht4 gene which provides race-specific resistance was found near the centromere on the chromosome 1 [114]. HtP has been mapped on chromosome 2 on bin 2.08. The gene HtNB which confers non-lesion resistance is located on chromosome 8.07 bin, flanked by MAC216826-4 and umc2218 at distances of 3.3 and 3.4 cM, respectively. Bins 1.05/7 and 9.05 were found to possess population-specific genes for resistance to S. turcica [13]. Among the up-regulated maize transcripts, one coiled coil (CC) forming, nucleotide-binding site (NB), leucine-rich repeats (LRR) [CC-NB-LRR] factors are found to be encoded by a gene GRMZM2G005347. The new plant resistance gene was designated as St referring to S. turcica. Comparative genomics showed that the CC-NB-LRR encoding Stgenes in maize were found on chromosome 2, and chromosome 5 in sorghum [115]. Recently, three NCLB resistance candidate genes viz., CDPK21, HEX9 and MKKK18 were identified with annotation functions of sugar signalling, calcium signalling, and MAPK signalling pathways [116].
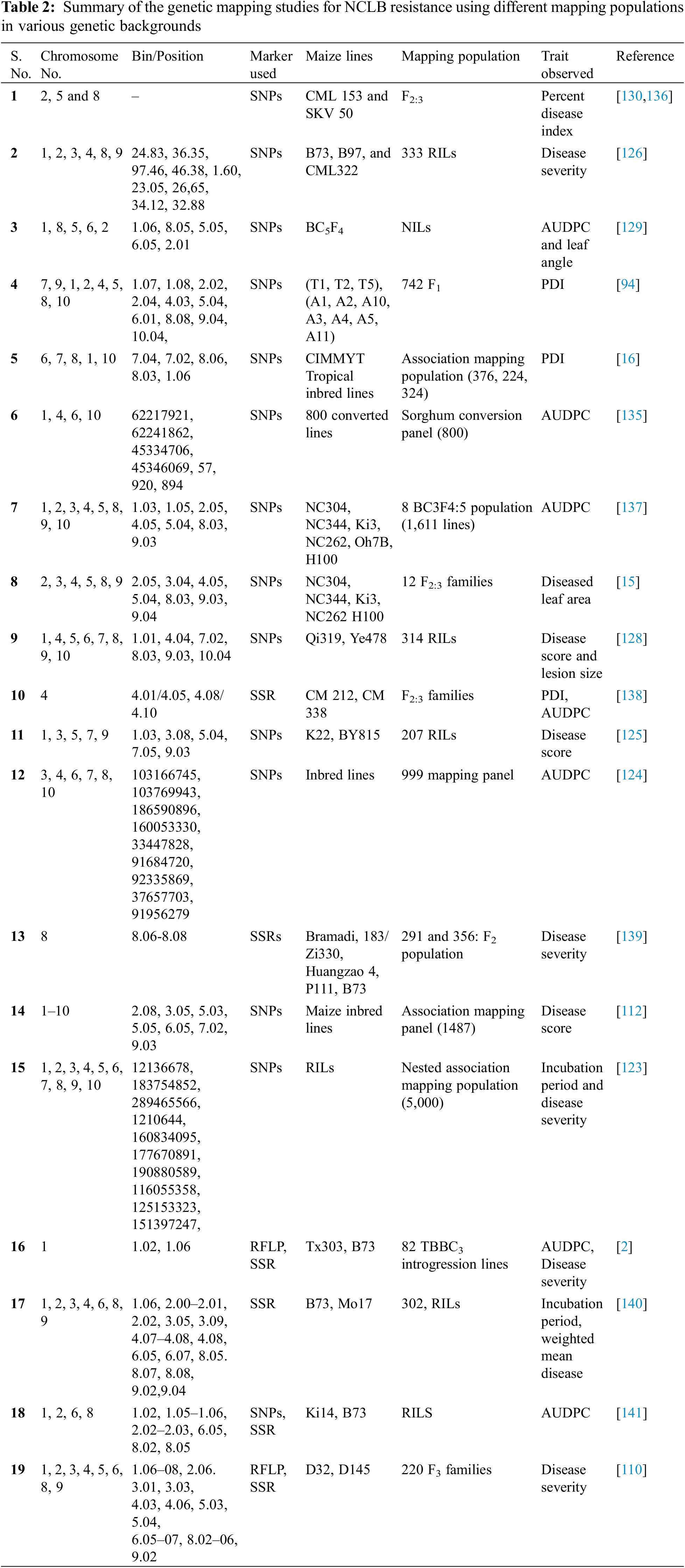
Quantitative resistance also called as horizontal or non-race specific resistance shows a substantial reduction in the incidence of NCLB disease and is controlled by several genes (polygenic). QTL is a chromosomal region that is associated with a quantitative trait. QTLs usually include genes that regulate the quantitative trait of a plant like yield and NCLB resistance. The mapping of the QTL and followed by its introgression into cultivars is regarded as QTLian breeding. The QTL or marker-trait associations (MTAs) can be identified through QTL mapping and genome-wide association studies (GWAS) approaches, respectively [117]. The former differs from the latter in that it uses constructed mapping populations (GWAS is based on diverse natural accessions called association panels). Numerous QTLs for NCLB resistance associated traits have been identified through QTL mapping and GWAS approaches (Table 2). Quantitative NCLB resistance is characterised by fewer, often smaller, lesions and a lengthy incubation period [114,118]. The extended incubation duration is usually expressed in young plants and is closely associated with the disease severity in adult plants of the region [119]. QTL which confers lesions length, width, and area have been mapped on chromosomes 1, 3, 5, and 8 [120], while the QTL conferring incubation period and area under the disease progression curve (AUDPC) have often been located on chromosome 8L [121,122]. Numerous QTLs for NCLB resistance were identified from different populations and distributed throughout the genome [14,112,122–126]. Four QTLs associated with NCLB resistance and a candidate gene, GRMZM2G024612 were also identified by association mapping [123]. One QTL at chromosome 1 (qNLB1.06) and another QTL at chromosome 8 (qNLB8.06) were found to be closely linked and functionally related to Ht2 [2,127]. The QTL qNCLB5.04 was found to be located at chromosome 5 and associated with NCLB disease scores and the width of the lesion [125]. Wang et al. [128] identified a new QTL, qNCLB7.02 for resistance to NCLB in maize. Recently, Rashid et al. [16] found 21 significant SNPs across three panels, some of them found to be co-located with major genes like Ht2, Ht3 and Htn1 and previously reported QTL for NCLB. Another study [129], by using nested near-isogenic line library revealed the role of liguleless1 for resistance to NCLB. The mutants for liguleless1 were found to be susceptible to NLB as a lack of ligule in maize resulted into highly erect leaves. However, this fact still needs confirmation as contrasting results were obtained for the correlation of leaf angle with NCLB resistance in different populations [129]. Galiano-Carneiro et al. [94] identified 17 QTL for NCLB resistance governing 3.57%–30.98% of the phenotypic variation. Moderate to high genomic prediction accuracies were observed between 0.58 and 0.83 based on population and continent. Recently, Ranganatha et al. [130] evaluated CML153 (susceptible) and SKV50 (resistant) based 344 F2:3 population for NLCB resistance and identified two major QTLs namely qNCLB-8-2 (phenotypic variation of 16.34%) and qNCLB-5 (phenotypic variation of 10.24%). Nevertheless, numerous QTLs have been identified in different populations by different research groups, but integrating the data of all the QTLs is an efficient approach to identify the consensus or stable regions harbouring multiple QTLs. Such studies were carried out using the meta-QTL analysis approach which declares regions (within certain confidence intervals) possessing two or more QTLs as meta-QTL. This helps in pinpointing the multi-allelic QTLs for NCLB and even multiple disease resistance [131]. Martins et al. [132] tested and advocated the use of this approach for identification of multiple disease resistance QTLs in maize. Genomic selection is a promising approach for simultaneous detection of favourable alleles in training sets and prediction of genotypic performance on basis of genome estimated breeding values [133]. Technow et al. [134] carried out the genomic prediction for NCLB resistance using the flint and dent training sets. The study revealed the prediction accuracies of 0.706 (dent) and 0.690 (flint). This indicates the potential use of genomic selection for NCLB resistance. Furthermore, the prediction accuracies can be further improved by the use of large diverse training sets. The detection of more than one QTL supports the theory that quantitative genes control resistance to S. turcica. Shared genetic regions were also identified conferring resistance to E. turcicum in both maize and sorghum. Several promising candidate genes have been identified with known roles in resistance to leaf blight including genes related to R-gene mediated resistance [135]. Novel identification/evaluation of NCLB-resistant quantitative trait loci and genes has been done which could improve the maize varieties (Table 2).
6 Genomics and Proteomics of S. turcica
Developments in high-throughput and cheaper sequencing platforms resulted in the sequencing of a number of fungal genomes (https://www.fungalgenomes.org). Fungi typically have small genome size, ranging from 20 to 50 Mbp and low volume of long repeats unlike eukaryotes. The complete genome of S. turcica was assembled in 2011 using Roche (454), Sanger Fosmids, and shredded consensus from Illumina assembled data (https://www.jgi.doe.gov). The size of its genome is 43 Mbp and comprised 11,702 predicted gene models. Later, the race 23N of S. turcica strain Et28A was sequenced again using IlluminaHiSeq and PacBio Sequel technologies, and assembled to approximately 43,480,261 bp on 30 scaffolds [142]. In total, 13,183 protein-coding genes were predicted, 13,142 of them were well annotated. This S. turcica genome resource is important for understanding the genetics behind pathogen evolution and infection mechanisms. The culturing of S. turcica on artificial media is feasible and the fungus is amenable to genetic alteration using Agrobacterium tumefaciens-mediated transformation [62]. Considering the availability of genome size and ease of genetic alteration, genomics should prove vital for thorough understanding of the S. turcicum.
Proteomic analysis is an effective approach for investigating the gene products to better understand the gene expression of resistant genes or mode of action of plant against pathogens. Leaf proteins were isolated from control and S. turcica infected leaves (inoculated for 72 h) and tested for differentially expressing proteins using two-dimensional electrophoresis and mass spectrometry-based recognition. A total of 137 proteins displayed more than 2-fold variations in abundance, including 50 up-regulated proteins and 87 down-regulated proteins [113]. About 48 protein spots were successfully identified by MS analysis, including 10 unique up-regulated, 20 down-regulated protein spots. These proteins were further grouped into nine functional classes considered to be involved in several functions, including energy metabolism (46%), protein destination and storage (12%) and disease protection (18%). The expression of photosynthesis-related proteins and metabolism-related proteins were found to be decreased by inoculation with S. turcica. The findings showed that the dynamic regulatory network functioned through the relationship between the A619 Ht2 and S. turcica resistant lines. The resistance mechanisms of A619 Ht2 consisted primarily via the direct release of defensive proteins and the regulation of primary metabolism, mainly photosynthesis and carbohydrate metabolism.
7 Genome Editing for NCLB Resistance
The first reference genome sequence for maize was published (B73 RefGen-v1) during year 2009 based on the sequencing of fosmids and bacterial artificial chromosomes [143]. Since release of B73 reference genome sequence, it had been widely used in functional genomics of maize. Plant diseases are amongst the major factors for yield losses in maize that necessitates the adoption of economically feasible novel genome editing technologies for production of genetically engineered disease resistant maize. A loss/gain of function mutation in these genes via genome editing methods such as CRISPR-Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats-Cas9), could shed light on the function of these genes and their role in disease resistance [144]. Techniques for the cloning of quantitative disease-resistance genes and subsequent analysis have been explained by Yang et al. [145]. Recently two genes namely ZmCCT10 and ZmAuxRp1 were cloned for disease-resistance in maize. RNAi based silencing of ZmAuxRP1 results in stalk rot resistant plants, while plants which shows overexpression of ZmAuxRP1 gene becomes more vulnerable to Gibberella stalk rot. This gene controls resistance by influencing the synthesis of indole-3-acetic acid and benzoxazinoid acid [146]. Major QTL, qRfg1 and qRfg2 that provides resistance to Gibberella stalk rot [147] has been found to be the part of ZmCCT10. Altered histone modification in the cis regulatory region of ZmCCT10 is the main cause for imparting the disease resistance in maize [148].
The use of RNAi to hinder specific genes in the pathogen relies on the availability of the information about the pathogenesis pathways to be targeted. In such instances, a double-stranded RNA molecule, complements a particular pathogen gene, is expressed in the host and transmitted to the pathogen during infection and causes silencing of specific genes. Micro RNAs (miRNAs) are regulators for gene expression involved with certain biotic stress reactions. Wu et al. [149] showed that miR811 and miR829 confer high tolerance to NCLB. Genome editing facilitates the functional study of genes and the integration of novel traits into major crop plants. Site-specific endonuclease-based systems allow site-specific modifications of the genome by producing double-stranded DNA breaks in genes of interest with a low chance of off-target results. Subsequently, cellular DNA repair machine homologous recombination and non-homologous end-joining pathways repair the cut end. By understanding the molecular pathways involved in disease resistance, breeders will be able to develop crop varieties with durable resistance to plant pathogens through adoption of RNAi and CRISPR-Cas9.
8 Management of NCLB: Need for an Integrated Approach
A plant’s inherent resistance to infection by pathogen could most likely be a safe, inexpensive and eco-friendly disease control strategy. However, to develop disease-resistant cultivars, it is necessary to consider the structure of the population and the evolutionary potential in pathogens. In environments where there is a continuous change in the racial spectrum of the pathogen and the population is highly diverse, exploiting quantitative resistances are recommended. This can be achieved by using cultivar mixture and production of complex hybrids, like three-way and double-cross hybrids, with inbred lines varying in tolerance level [150–153]. The qualitative resistance is typically recommended in locations where the pathogen diversity is low. Marker assisted backcrossing can result in the pyramiding of Ht genes in maize. Planting hybrids with good NCLB resistance is an economical, effective and sustainable method of avoiding yield losses in maize. Great efforts have been made worldwide to develop, identify and utilize germplasm with TLB resistance. Extensive evaluation of maize germplasm revealed new and durable resistance sources against NCLB [36,61,98,154–157]. Although the primary approach to manage NCLB is to use plant resistant genotypes, in some situations, farmers may consider the application of fungicide a useful approach, where environmental factors are favourable for NCLB. Selecting the proper timing for application is essential in determining the efficacy of fungicides and economic benefit. The efficiency of different fungicide applications to manage NCLB has been studied extensively [158,159,160–162]. The disease management by use of fungicides seems to be cost-effective when used on NCLB-susceptible maize varieties, and when applied during tasseling or flowering [163]. The best growth stages for trifloxystrobin + epoxiconazole fungicide applications to reduce NCLB were between V10 and V14, showing coincidence with the disease onset [164]. Demethylation inhibitor fungicides (DMI), had shown greater efficiency for controlling NCLB [165]. Among the DMI fungicides, propiconazole is the most effective in reducing the severity of the disease. The quinine oxidation inhibitor (QoI) fungicides known as strobilurin induce favourable physiological activities of plants like improved stalk strength and sustained green leaf tissue by delayed leaf senescence [164]. The fungicides prothioconazole + trifloxystrobin exhibited the highest chemical control efficiency for NCLB [166]. Though the chemical application has been proved very effective in the management of NCLB, excessive chemical usage has hazardous effects on human health and the environment [167]. Hence, minimum dosage of fungicides in combination with other cultural practices and moderate levels of host plant resistance is the best approach for the control of NCLB.
9 Conclusion and Future Perspectives
NCLB is the most important re-emerging foliar disease of maize, limiting maize production. The disease has a worldwide distribution and its development is favoured by cool to moderate temperature with high relative humidity. Disease development usually originate from mycelia and conidia of the causal fungus from infected crop residues left in farm fields. Early detection of the disease development is of great importance in the management of NCLB. The prevalence of the disease has increased in recent years and new races of the pathogen have been reported worldwide. The fungus E. turcicum is highly variable in nature. Identification of races of the pathogen present in the area and ability to understand their geographical distribution are important steps for the development of disease resistant cultivars. Distribution of S. turcica races on larger geographical regions of the world impresses upon the monitoring of pathogen diversity on large scales and over time to fully understand factors influencing the evolution of pathogen races. Though chemical measures are available for the control of the TLB. They are difficult to sustain and have not been adopted particularly in marginal farming systems under high altitude rainfed conditions. Most efficient and cost-effective ways to manage the disease is to develop varieties with resistance against E. turcicum. Ht genes confer race specific qualitative resistance against NCLB inherited by single gene; however, the resistance tends to break down under the pressure of high virulent races of S. turcica. Hence, pyramiding of multiple Ht genes will play a crucial role for the development of durable resistance against a number of pathogen races. Quantitative resistance to NCLB is favoured in high disease pressure environments. The discovery, validation and introgression of NCLB resistant genomic regions would surely prove vital to achieve improved genetic gains for grain yield. The utilization of genome editing technologies like CRIPSR-Cas9 will help in the development of resistant cultivars that would be relatively easy to release unlike transgenics. Furthermore, omics approaches such as proteomics and metabolomics would pave the way for a better understanding of the molecular mechanism of NCLB and defence response of the host plant.
Author’s Contribution: Study conception and design: M.A.A; S.H.W; M.C; Literature collection: Z.A.D; J.R; F.M; M.B; Draft manuscript preparation: M.C; S.K.A; S.A.W; K.A.D; A.E.S; Review, editing: C.B; O.K; M.A.H; S.H.W; M.A.A; All authors have read and approved the final version of the manuscript.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Agrios, G. N. (2005). Plant diseases caused by fungi. In: Plant pathology (5th editionpp. 385–614. San Diego, CA: Academic Press. [Google Scholar]
2. Chung, C. L., Longfellow, J. M., Walsh, E. K., Kerdieh, Z., van Esbroeck, G. et al. (2010). Resistance loci affecting distinct stages of fungal pathogenesis: Use of introgression lines for QTL mapping and characterization in the maize Setosphaeria turcica pathosystem. BMC Plant Biology, 10, 103. DOI 10.1186/1471-2229-10-103. [Google Scholar] [CrossRef]
3. Sibiya, J., Tongoona, P., Derera, J., Makanda, I. (2013). Smallholder farmers’ perceptions of maize diseases, pests, and other production constraints, their implications for maize breeding and evaluation of local maize cultivars in KwaZulu-Natal, South Africa. African Journal of Agricultural Research, 8(17), 1790–1798. DOI 10.5897/AJAR12.1906. [Google Scholar] [CrossRef]
4. Khatri, N. K. (1993). Influence of temperature and relative humidity on the development of Helminthosporium turcicum on maize in Westeren Georgia. Indian Journal of Mycology and Plant Pathology, 23, 35–37. [Google Scholar]
5. Navarro, B. L., Campos, R. A., Gasparoto, M. C., Tiedemann, A. V. (2021). In vitro and in planta studies on temperature adaptation of Exherohilum turcicum isolates from maize in Europe and South America. Pathogens, 10, 154. DOI 10.3390/pathogens10020154. [Google Scholar] [CrossRef]
6. Pant, S. K., Kumar, P., Chauhan, V. S. (2000). Effect of turcicum leaf blight on photosynthesis in maize. Indian Phytopathology, 54, 251–252. [Google Scholar]
7. Nwanosike, M. R., Mabagala, R. (2017). Influence of metrological parameters on the development of Exserohilum turcicum (Pass.) Leonard and Suggs on maize in Tanzania. International Journal of Agricultural and Food Research, 6(3), 1–9. [Google Scholar]
8. Payak, M. M., Renfro, B. L. (1968). Combating maize disease. Indian Farmer Disease, 1, 53–58. [Google Scholar]
9. Ahangar, M. A., Bhat, Z. A., Sheikh, F. A., Dar, Z. A., Ajaz, A. et al. (2016). Pathogenic variability in Exherohilum turcicum and identification of resistant sources to turcicum leaf blight of maize (Zea mays L.). Journal of Applied and Natural Science, 8(3), 1523–1529. DOI 10.31018/jans.v8i3.994. [Google Scholar] [CrossRef]
10. Galiano-Carneiro, A. L., Miedaner, T. (2017). Genetics of resistance and pathogenicity in the maize Setosphaeria turcica pathosystem and implications for breeding. Frontier of Plant Science, 8, 1490. DOI 10.3389/fpls.2017.01490. [Google Scholar] [CrossRef]
11. Ma, Z., Liu, B., He, S., Gao, Z. (2020). Analysis of physiological races and genetic diversity of Setosphaeria turcica (Luttr.) K.J. Leonard & Suggs from different regions of China. Canadian Journal of Plant Pathology, 42(3), 1–12. DOI 10.1080/07060661.2019.1679261. [Google Scholar] [CrossRef]
12. Pandurangegowda, K. T., Shetty, H. S., Gowda, B. J., Prakash, H. S., Sangam, L. (1993). Comparison of two methods for assessment of yield losses due to turcicum leaf blight of maize. Indian Phytopathology, 45, 316–320. [Google Scholar]
13. Welz, H. G., Geiger, H. H. (2000). Genes for resistance to northern corn leaf blight in diverse maize populations. Plant Breeding, 119(1), 1–14. DOI 10.1046/j.1439-0523.2000.00462.x. [Google Scholar] [CrossRef]
14. Wisser, R. J., Balint-Kurti, P. J., Nelson, R. J. (2006). The genetic architecture of disease resistance in maize: A synthesis of published studies. Phytopathology, 96, 120–129. DOI 10.1094/PHYTO-96-0120. [Google Scholar] [CrossRef]
15. Kessel, B., Presterl, T., Miedaner, T. (2021). Multi-parental QTL mapping of resistance to white spot of maize (Zea mays) in Southern Brazil and relationship to QTLs of other foliar diseases. Plant Breeding, 140(5), 801–811. DOI 10.1111/pbr.12964. [Google Scholar] [CrossRef]
16. Rashid, Z., Sof, M., Harlapur, S. I., Kachapur, R. M., Dar, Z. A. et al. (2020). Genome‐wide association studies in tropical maize germplasm reveal novel and known genomic regions for resistance to Northern corn leaf blight. Scientific Reports, 10(1), 21949. DOI 10.1038/s41598-020-78928-5. [Google Scholar] [CrossRef]
17. Shurtleff, M. C. (1980). Compendium of corn diseases second edition. St. Paul Minnesota: The American Phytopathological Society. [Google Scholar]
18. Ceballos, H., Deutsch, J. A., Gutierrez, H. (1991). Recurrent selection for resistance to Exherohilum turcicum in eight sub-tropical maize populations. Crop Science, 31, 964–971. DOI 10.2135/CROPSCI1991.0011183X003100040025X. [Google Scholar] [CrossRef]
19. Juliana, B. O., Marco, A. G., Isaias, O. G., Luis, E. A. C. (2005). New resistance gene in Zea mays Exherohilum turcicum patho-system. Genetics and Molecular Biology, 28, 435–439. DOI 10.1590/S1415-47572005000300017. [Google Scholar] [CrossRef]
20. Adipala, E., Lipps, P. E., Madden, L. V. (1993). Reaction of maize cultivars from Uganda to Exherohilum turcicum. Phytopathology, 83, 217–223. [Google Scholar]
21. Kim, S. K., Kim, H. W., Lee, J. S. (2012). Tolerance expression of maize genotypes to Exherohilum turcicum in North and South Korea. Korean Journal of Crop Science and Plant Biotechnology, 57(2), 113–126. DOI 10.7740/kjcs.2012.57.2.113. [Google Scholar] [CrossRef]
22. Wathaneeyawech, S., Smitamana, P., Smitamana, P. (2015). Study of the host range of northern corn leaf blight disease and effect of Exherohilum turcicum toxin on sweet corn. Journal of Agricultural Technology, 11(4), 953–963. [Google Scholar]
23. Sartori, M. A., Nescia, A., Formentoc, A., Etcheverry, M. (2015). Selection of potential biological control of Exherohilum turcicum with epiphytic microorganisms from maize. Revista Argentina de Microbiologia, 47(1), 62–71. DOI 10.1016/j.ram.2015.01.002. [Google Scholar] [CrossRef]
24. Butler, E. J. (1918). Fungi and disease in plants, pp. 547. Calcutta, India: Thacker, Spink & Co. [Google Scholar]
25. Mitra, M. (1923). Helminthosporium species on cereals and sugarcane in India. Part-1 (Disease of Zea mays and Sorghum vulgare) caused by species of Helminthosporium. Memoirs of the Department of Agriculture in India, Botanical Series, 11(10), 219–242. [Google Scholar]
26. Lal (1991). Genetics of Helminthosporium leaf blight resistance in maize. Maize Genetics Prospectus Symposium TNAU, pp. 231–237. Coimbatore. [Google Scholar]
27. Lim, S. M., Kinsey, J. G., Hooker, A. L. (1974). Inheritance of virulence in Helminthosporium turcicum to monogenic resistance corn. Phytopathology, 64, 1150–1151. [Google Scholar]
28. Ahangar, M. A., Bhat, Z. A., Najeeb, S., Dar, Z. A., Reyaz, M. et al. (2016). Variability of Exherohilum turcicum (Pass.) Leonard and Suggs, causing Turcicum leaf blight of maize. SKUAST Journal of Research, 18(2), 96–101. [Google Scholar]
29. Yeshitila, D. (2003). Cloning and characterization of xylanase genes from phytopathogenic fungi with a special reference to Helminthosporium turcicum the cause of northern leaf blight of maize (academic dissertation). Department of Applied Biology, University of Helsinki-Finland. [Google Scholar]
30. Gowda, K. T. P., Shetty, H. S., Gowda, B. J., Prakash, H. S., Sangam, L. (1992). Comparison of two methods for assessment of yield loss due to turcicum leaf blight of maize. Indian Phytopathology, 45, 319–320. [Google Scholar]
31. Cardwell, K. F., Schulthess, F., Ndemah, R., Ngoko, Z. A. (1997). Systems approach to assess crop health and maize yield losses due to pests and diseases in Cameroon. Agriculture Ecosystem and Environment, 65(1), 33– 47. DOI 10.1016/S0167-8809(97)00056-X. [Google Scholar] [CrossRef]
32. Raymond, A. D., Hooker, A. L. (1981). Measuring the relationship between northern leaf blight and yield losses. Plant Disease, 65, 325–327. DOI 10.1094/PD-65-325. [Google Scholar] [CrossRef]
33. Raymond, A. D. (1978). Epidemiology of northern corn leaf blight as affected by host resistance and yield losses following simulated epidemics (Ph.D. Thesis), pp. 111. University of Illinois, Urbana Champaign. Research Institute, New Delhi. [Google Scholar]
34. Nwanosike, M. R. O., Mabagala, R. B., Kusolwa, P. M. (2013). Effect of northern leaf blight (Exherohilum turcicum) severity on yield of maize (Zea mays L.) in Morogoro, Tanzania. International Journal of Science and Research, 4, 466–474. [Google Scholar]
35. Chenula, V. V., Hora, T. S. (1962). Studies on losses due to Heliminthosporium blight of maize. Indian Phytopathology, 15, 235–237. [Google Scholar]
36. Zhang, Y., Crous, P. W., Schoch, C. L., Hyde, K. D. (2012). Pleosporales. Fungal Diversity, 53(1), 1–221. DOI 10.1007/s13225-011-0117-x. [Google Scholar] [CrossRef]
37. McNeill, J., Barrie, F. R., Buck, W. R., Demoulin, V., Greuter, W. et al. (2012). International code of nomenclature for algae, fungi, and plants (Melbourne Code), pp. 154. Königstein: Koeltz Scientific Books. [Google Scholar]
38. Sivanesan, A., Abdullah, S. K., Abbas, B. A. (1993). Exserohilum curvisporum sp. nov., a new hyphomycete from Iraq. Mycological Research, 97, 1486–1488. [Google Scholar]
39. Hernández-Restrepo, M., Madrid, N., Tan, Y. P., Cunha, K. C., Gené, J. et al. (2018). Multi-locus phylogeny and taxonomy of Exserohilum. Persoonia, 41(1), 71–108. DOI 10.3767/persoonia.2018.41.05. [Google Scholar] [CrossRef]
40. Leonard, K. J., Suggs, E. G. (1974). Setosphaeriaprolata is the ascigenous state of Exserohilumprolata. Mycologia, 66, 181–297. [Google Scholar]
41. Alcorn, J. L. (1988). The taxonomy of Helminthosporium species. Annual Review of Phytopathology, 26, 37–56. DOI 10.1146/annurev.py.26.090188.000345. [Google Scholar] [CrossRef]
42. Lutrell, E. S. (1958). The perfect stage of Helminthosporium turcicum. Phytopathology, 48, 281–287. [Google Scholar]
43. Fan, Y. S., Ma, J. F., Gui, X. M., An, X. L., Sun, S. Q. et al. (2007). Distribution of mating types and genetic diversity induced by sexual recombination in Setosphaeria turcica in Northern China. Frontiers in Agriculture China, 1(6), 368–376. DOI 10.12691/wjar-5-6-2. [Google Scholar] [CrossRef]
44. Bunkoed, W., Kasam, S., Chaijuckam, P., Yhamsoongnern, J., Prathuang, W. (2014). Sexual reproduction of Setosphaeria turcica in natural corn fields in Thailand. Kasetsart Journal, 48, 175–182. [Google Scholar]
45. McGee, D. (1990). Maize diseases: A reference source for seed technologists. Minnesota. USA: APS Press. [Google Scholar]
46. de Rossi, R. L., Reis, E. M. (2014). Semi-selective culture medium for Exherohilum turcicum isolation from corn seeds. Summa Phytopathology, 40(2), 163–167. DOI 10.1590/0100-5405/1925. [Google Scholar] [CrossRef]
47. Li, B., Wilson, W. A. (2013). Composition and methods for enhancing resistance to northern leaf blight in maize. US Patent, 20,130,061,355. [Google Scholar]
48. Laxminarayan, C., Shankerlingam, S. (1983). Turcicum leaf blight of maize. Current Science, 52, 440–444. DOI 10.1016/j.cropro.2020.105386. [Google Scholar] [CrossRef]
49. King, S. B., Mukuru, S. Z. (1994). An overview of sorghum finger millet and pearl millet in Eastern Africa with special attention to diseases. In: Danial, D. L. (Ed.Breeding for disease resistance with emphasis on durability, pp. 24–34. Wageningen, Netherlands: Wageningen Agricultural University. [Google Scholar]
50. Sajeed, A., Chowdhury, A. K. (2014). Histological studies on turcicum leaf blight disease of maize in hill agro-ecological zones of west. Bengal Journal of Mycological Research, 113, 116. [Google Scholar]
51. Vieira, R. A., Moquino, R. M., Silva, C. N., Hata, F. T., Tessmann, D. J. et al. (2014). A new diagrammatic scale for the assessment of northern corn leaf blight. Journal of Crop Protection, 56, 55–57. DOI 10.1016/j.cropro.2011.04.018. [Google Scholar] [CrossRef]
52. Taken, J. P., Adipala, E., Ogenga-Latigo, M. W. (1994). Northern leaf blight progress and spread from Exherohilum turcicum infested maize residue. African Crop Science Journal, 2(2), 197–205. [Google Scholar]
53. De-Rossi, R. L., Reis, E. M., Brustolin, R. (2015). Conidial morphology and pathogenicity of Exherohilum turcicum isolates of corn from Argentina and Brazil. Summa Phytopathology, 41(1), 58–63. DOI 10.1590/0100-5405/1948. [Google Scholar] [CrossRef]
54. Shenoi, M. M., Ramalingam, A. (1983). Leaf blight of sorghum: Influence of meteorological factors and crop growth stages on the spread of inoculum and disease. Indian Phytopathology, 36(4), 700–706. [Google Scholar]
55. Ferguson, L. M., Carson, M. L. (2004). Spatial diversity of Setosphaeria turcica sampled from the Eastern United States. Phytopathology, 94(8), 892–900. DOI 10.1094/PHYTO.2004.94.8.892. [Google Scholar] [CrossRef]
56. Patil, S. J., Wali, M. C., Harlapur, S. I., Prasanth (2000). Maize research in North Karnataka, pp. 54. University of Agricultural Science, Dharwad. [Google Scholar]
57. Ullstrup, A. J. A. (1970). Comparison of monogenic and polygenic resistance to H. turcicum in corn. Phytopathology, 60, 1597–1599. DOI 10.1094/Phyto-60-1597. [Google Scholar] [CrossRef]
58. Bentolila, S., Guitton, C., Bouvet, N., Sailland, A., Nykaza, S. et al. (1991). Identification of an RFLP marker tightly linked to the Ht1 gene in maize. Theoretical and Applied Genetics, 82(4), 393–398. DOI 10.1007/BF00588588. [Google Scholar] [CrossRef]
59. Ogliari, J. B., Guimaraes, M. A., Camargo, L. E. A. (2007). Chromosomallocations of the maize (Zea mays L.) HtP and rt genes that confer resistance to Exherohilum turcicum. Genetics and Molecular Biology, 30(3), 630–634. DOI 10.1590/S1415-47572007000400021. [Google Scholar] [CrossRef]
60. Muiru, W. M. (2008). Histological studies and characterization of races of Exherohilum turcicum the causal agent of northern leaf blight of maize in Kenya (Ph.D Thesis), University of Nairobi, Kenya. [Google Scholar]
61. Setyawan, B., Irfan, S., Aswaldi, A., Etti, S. (2016). Resistance of eleven new hybrid maize genotypes to turcicum leaf blight (Exherohilum turcicum). Biodiversitas, 17(2), 604–608. DOI 10.13057/biodiv/d170230. [Google Scholar] [CrossRef]
62. Degefu, Y., Hanif, M. (2003). Agrobacterium tumefaciens mediated transformation of Helminthosporium turcicum, the maize leaf-blight fungus. Archives of Microbiology, 80(4), 279–284. DOI 10.1007/s00203-003-0589-5. [Google Scholar] [CrossRef]
63. Hilu, H. M., Hooker, A. L. (1964). Host pathogen relationship of Helminthosporium turcicum in resistant and susceptible corn seedlings. Phytopathology, 54, 570–575. [Google Scholar]
64. Lilian, Z. L., Watson, A. K., Paulitz, T. C. (2002). Reaction of rice (Oryza sativa) cultivars to penetration and infection by Curvularia tuberculata and C. oryzae. Plant Disease, 86(5), 470–476. DOI 10.1094/PDIS.2002.86.5.470. [Google Scholar] [CrossRef]
65. Stangarlin, J. R., Tartaro, E. L., Pascholati, S. F. (2022). Characterization of Exherohilum turcicum infection sites in maize genotypes. Revista Caatinga, 35, 1–13. DOI 10.1590/1983-21252022v35n101rc. [Google Scholar] [CrossRef]
66. Bashan, B., Levy, R. S., Cojocaru, M., Levy, Y. (1995). Purification and structural determination of a phytotoxic substance from Exherohilum turcicum. Physiology and Molecular Plant Pathology, 47, 225–235. DOI 10.1006/pmpp.1995.1054. [Google Scholar] [CrossRef]
67. Li, P., Gong, X., Jia, H., Fan, Y., Zhang, Y. et al. (2016). MAP kinase gene STK1 is required for hyphal, conidial, and appressorial development toxin biosynthesis, pathogenicity, and hypertonic stress response in the plant pathogenic fungus Setosphaeriaturcica. Journal of Integrated Agriculture, 15(12), 2786–2794. DOI 10.1016/S2095-3119(16)61472-7. [Google Scholar] [CrossRef]
68. Perkins, J. M., Pedersen, W. L. (1987). Disease development and yield losses associated with northern leaf blight on corn. Plant Disease, 71, 940–943. DOI 10.1094/PD-71-0940. [Google Scholar] [CrossRef]
69. Babu, R., Mani, V. P., Pandey, A. K., Pant, S. K., Rajeshsingh, K. S. (2004). Maize research at vivekan and Parvatiya Krishi Anusandhan Sansthan–An overview. In: Technical bulletin, vol. 21, pp. 31. Vivekanand Parvatiya Krishi Anusandhan Sansthan, Almora, 21, 31. [Google Scholar]
70. Harlapur, S. I. (2005). Epidemiology and management of turcicum leaf blight of maize caused by Exserohilum turcicium (pass.) Leonard and Suggs (Ph.D. Thesis), University of Agricultural Sciences, Dharwad. [Google Scholar]
71. Khedekar, S. A., Haralpur, S. I., Kulakarni, S., Benagi, V. I., Desphande, V. K. (2010). Survey of turcicum leaf blight of maize in Northern Karnataka. Journal of Plant Disease Science, 5(1), 249–250. [Google Scholar]
72. Reddy, T. R., Reddy, P. N., Reddy, R. R. (2013). Pathogenic variability of isolates of Exherohilum turcicum, incitant of leaf blight of maize. Indian Journal of Plant Protection, 41(1), 72–75. [Google Scholar]
73. Borchardt, D. S., Welz, H. G., Geiger, H. H. (1998). Genetic structure of Setosphaeriaturcica populations in temperate and tropical climates. Phytopathology, 88(4), 322–329. DOI 10.1094/PHYTO.1998.88.4.322. [Google Scholar] [CrossRef]
74. Nieuwoudt, A., Humana, M. P., Cravenb, M., Cramptona, B. G. (2018). Genetic differentiation in populations of Exherohilum turcicum from maize and sorghum in South Africa. Plant Pathology, 67, 1483–1491. DOI 10.1111/ppa.12858. [Google Scholar] [CrossRef]
75. Mwangi, S. M. (1998). Status of Northern leaf blight, Phaeosphaeria maydis leaf spot, Southern leaf blight, rust, maize streak virus and physiological specialization of E. turcicum in Kenya. (Ph.D. Thesis). Virginia Polytechnic Institute and State University. [Google Scholar]
76. Levy, Y. (1991). Variation in fitness among field isolates of Exherohilum turcicum in Israel. Plant Disease, 75(2), 1243–1245. DOI 10.1094/PD-75-0163. [Google Scholar] [CrossRef]
77. Abebe, D., Singburaudom, N. (2006). Morphological, cultural and pathogenicity variation of Exherohilum turcicum (Pass.) Leonard and Suggs isolates in maize (Zea mays L.). Kasetsart Journal of Natural Science, 40, 341–352. [Google Scholar]
78. Shree, U., Reddy, R. N., Mohan, S. M., Madhusudhana, R., Mather, K. et al. (2012). Genetic diversity and pathogenic variation in the isolates of Exherohilum turcicum causing common leaf blight of Sorghum. Indian Phytopathology, 65(4), 349–355. [Google Scholar]
79. Muiru, W. M., Koopmann, B., Tiedemann, A. V., Mutitu, E. W., Kimenju, J. W. (2010). Race typing and evaluation of Aggressiveness of Exherohilum turcicum isolates of Kenyan, German and Austrian origin. World Journal of Agricultural Sciences, 6(3), 277–284. [Google Scholar]
80. Yadav, O. P., Karjagi, C. G., Jat, S. L., Dhillon, B. S. (2014). Overview of maize improvement in India. Indian Farm, 64, 5–11. [Google Scholar]
81. Assefa, T. (1995). Recent outbreaks of turcicum leaf blight on maize in Ethiopia. Proceeding of the Third Annual Conference of the Crop Protection Society of Ethiopia, pp. 153–156. Addis Ababa and Ethiopia. [Google Scholar]
82. Harlapur, S. I., Kulkarni, M. S., Hegde, Y., Kulkarni, S. (2007). Variability in Exherohilum turcicum (Pass.). Leonard and Suggs. Causal agent of turcicum leaf blight of maize. Karnataka Journal, 21(1), 55–60. [Google Scholar]
83. Bunker, R. N., Rathore, R. S., Kumawat, D. K. (2011). Pathogenic and morphological variability of Bipolaris maydis incitant of maydis leaf blight in maize. Journal of Mycology and Plant Pathology, 41, 418–421. [Google Scholar]
84. Kutawa, A. B., Kamaruzaman, S., Khairulmazmi, A., Zulkifli, A. S., Firdaus, M. S. et al. (2017). Characterisation and pathological variability of Exherohilum turcicum responsible for causing northern corn leaf blight (NCLB) disease in Malaysia. Malaysian Journal of Microbiology, 13(1), 41–49. DOI 10.21161/mjm.83016. [Google Scholar] [CrossRef]
85. Bunker, R. N., Mathur, K. (2010). Pathogenic and morphological variability of Exherohilum turcicum isolates causing leaf blight in Sorghum (Sorghum bicolor). Indian Journal of Agricultural Science, 80(10), 888–892. [Google Scholar]
86. Eschholz, T. W., Stamp, P., Peter, R., Leipner, J., Hund, A. (2010). Genetic structure and history of Swiss maize (Zea mays L. ssp. mays) landraces. Genetic Research and Crop Evolution, 57, 71–84. DOI 10.1007/s10722-009-9452-0. [Google Scholar] [CrossRef]
87. Aci, M. M., Revilla, P., Morsli, A., Djemel, A., Belalia, N. et al. (2013). Genetic diversity in Algerian maize (Zea mays L.) landraces using SSR markers. Maydica, 58, 304–310. [Google Scholar]
88. Knox-Davies, P., Dickson, J. (1960). Cytology of Helminthosporium turcicum and its ascogenous stage, Trichometasphaeria turcica. American Journal of Botany, 47(2), 328–339. DOI 10.2307/2440108. [Google Scholar] [CrossRef]
89. Li, Y. G., Jiang, W. Y., Zhang, Q. F., Ali, E., Ji, P. (2019). Population structure and genetic diversity of Setosphaeria turcica from corn in Heilongjiang Province, China. Journal of Applied Microbiology, 127(6), 1814–1823. DOI 10.1111/jam.14449. [Google Scholar] [CrossRef]
90. Vidal-Villarejo, M., Freund, F., Hanekamp, H., Tiedemann, A., Schmid, K. (2020). Population history of the Northern corn leaf blight fungal pathogen Setosphaeriaturcica in Europe. bioRxiv. DOI 10.1101/2020.09.18.303354. [Google Scholar] [CrossRef]
91. Leonard, K. J., Levy, Y., Smith, D. R. (1989). Proposed nomenclature for pathogenic races of Exherohilum turcicum on corn. Plant Diseases, 73, 776–777. [Google Scholar]
92. Welz, H. G., Wagner, R., Geiger, H. H. (1993). Virulence in Setosphearia turcica populations collected from maize in China, Mexico, Uganda and Zambia. Phytopathology, 83, 1356. [Google Scholar]
93. Fallah Moghaddam, P., Pataky, J. K. (1994). Reactions for isolates from mating of races 1 and 23N of Exherohilum turcicum. Plant Diseases, 78, 767–771. [Google Scholar]
94. Galiano-Carneiro, A. L., Kessel, B., Presterl, T., Miedaner, T. (2020). Intercontinental trials reveal stable QTL for northern corn leaf blight resistance in Europe and in Brazil. Theoretical and Applied Genetics, 134, 1–17. DOI 10.1007/s00122-020-03682-1. [Google Scholar] [CrossRef]
95. Turgey, E. B., Buyuk, O., Tunali, B., Helvacioglu, O., Kurt, S. (2019). Detection of the race of Exherohilum turcicum [(Pass.)K.J. Leonard & Suggs] causing northern leaf blight diseases of corn in Turkey. Journal of Plant Pathology, 102, 387–393. DOI 10.1007/s42161-019-00440-1. [Google Scholar] [CrossRef]
96. Jindal, K. K., Tenuta, A. U., Woldemariam, T., Zhu, X., Hooker, D. C. et al. (2019). Occurrance and distribution of physiological races of Exherohilum turcicum in Ontario Canada. Plant Disease, 103, 1450–1457. DOI 10.1094/PDIS-06-18-0951-SR. [Google Scholar] [CrossRef]
97. Welz, H. G. (1998). Genetics and epidemiology of the pathosystem Zea mays/Setosphaeria turcica (Doctoral Thesis). University of Hohenheim. [Google Scholar]
98. Payak, M. M., Sharma, R. C. (1985). Maize diseases and their approach to their management. Tropical Pest Management, 31, 302–310. DOI 10.1080/09670878509371006. [Google Scholar] [CrossRef]
99. Navarro, B. L., Ramos Romero, L., Kistner, M. B., Iglesias, J., Tiedemann, A. (2021). Assessment of physiological races of Exherohilum turcicum isolates from maize in Argentina and Brazil. Tropical Plant Pathology, 46, 371–380. DOI 10.1007/s40858-020-00417-x. [Google Scholar] [CrossRef]
100. Geiger, H. H., Heun, M. (1989). Genetics of quantitative resistance to fungal diseases. Annual Review of Phytopathology, 27, 317–341. DOI 10.1146/annurev.py.27.090189.001533. [Google Scholar] [CrossRef]
101. Pataky, J. K., Raid, R. N., Du Toit, L. J., Schueneman, T. J. (1998). Disease severity and yield of sweet corn hybrids with resistance to Northern leaf blight. Plant Diseases, 82, 57–63. DOI 10.1094/PDIS.1998.82.1.57. [Google Scholar] [CrossRef]
102. Hooker, A. L. (1963). Inheritance of chlorotic lesion resistance to Helminthosporium turcicum in seedling corn. Phytopathology, 53, 660–662. [Google Scholar]
103. Navarro, B. L., Hanekamp, H., Koopmann, B., von Tiedemann, A. (2020). Diversity of expression types of Htgenes conferring resistance in maize to Exherohilum turcicum. Frontiers in Plant Science, 11, 607850. DOI 10.3389/fpls.2020.607850. [Google Scholar] [CrossRef]
104. Gevers, H. (1975). A new major gene for resistance to Helminthosporium turcicum leaf blight of maize. Plant Disease Replication, 59, 296–299. [Google Scholar]
105. Hooker, A. L. (1977). 2nd major gene locus in corn for chlorotic-lesion resistance to Helminthosporium turcicum. Crop Science, 17, 132–135. DOI 10.2135/cropsci1977.0011183X001700010035x. [Google Scholar] [CrossRef]
106. Hooker, A., Tsung, Y. (1980). Relationship of dominant genes in corn for chlorotic lesion resistance to Helminthosporium turcicum. Plant Disease, 64, 387–388. [Google Scholar]
107. Zaitlin, D., Demars, S. J., Gupta, M. (1992). Linkage of a second gene for NCLB resistance to molecular markers in maize. Maize Genetics Cooperation (News Letter), 66, 69. [Google Scholar]
108. Simcox, K. D., Bennetzen, J. L. (1993). The use of molecular markers to study Setosphaeria turcica resistance in maize. Phytopathology, 83, 1326–1330. [Google Scholar]
109. Hurni, S., Scheuermann, D., Krattinger, S. G., Kessel, B., Wicker, T. (2015). The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proceedings of the National Academy of Sciences, 112, 8781–8785. DOI 10.1073/pnas.1502522112. [Google Scholar] [CrossRef]
110. Welz, H. G., Schechert, A. W., Geiger, H. H. (1999). Dynamic gene action at QTLs for resistance to Setosphaeria turcica in maize. Theoretical and Applied Genetics, 98, 1036–1045. DOI 10.1007/s001220051165. [Google Scholar] [CrossRef]
111. Yin, X., Wang, Q., Yang, J., Jin, D., Wang, F. et al. (2003). Fine mapping of the Ht2 (Helminthosporium turcicum) resistance 2) gene in maize. Chinese Science Bullitin, 48, 165–169. DOI 10.1360/03tb9034. [Google Scholar] [CrossRef]
112. van Inghelandt, D., Melchinger, A. E., Martinant, J. P., Stich, B. (2012). Genome-wide association mapping of flowering time and northern corn leaf blight (Setosphaeria turcica) resistance in a vast commercial maize germplasm set. BMC Plant Biology, 12, 56. DOI 10.1186/1471-2229-12-56. [Google Scholar] [CrossRef]
113. Zhang, X. L., Si, B. W., Fan, C. M., Li, H. J., Wang, X. M. (2014). Proteomics identification of differentially expressed leaf proteins in response to Setosphaeria turcica infection in resistant maize. Journal of Integrative Agriculture, 13, 789–803. DOI 10.1016/S2095-3119(13)60513-4. [Google Scholar] [CrossRef]
114. Carson, M. L. (1995). A new gene in maize conferring the “Chlorotic Halo” reaction to infection by Exherohilum turcicum. Plant Disease, 79, 717–720. DOI 10.1094/PD-79-0717. [Google Scholar] [CrossRef]
115. Martin, T., Biruma, M., Fridborg, I., Okori, P., Dixelius, C. (2011). A highly conserved NB-LRR encoding gene cluster effective against Setosphaeria turcica in sorghum. BMC Plant Biology, 11(1), 151. DOI 10.1186/1471-2229-11-151. [Google Scholar] [CrossRef]
116. Li, C., Ling, F., Su, G., Sun, W., Liu, H. (2020). Location and mapping of the NCLB resistance genes in maize by bulked segregant analysis (BSA) using whole genome re-sequencing. Molecular Breeding, 40, 92. DOI 10.1007/s11032-020-01171-3. [Google Scholar] [CrossRef]
117. Pritchard, J. K., Stephens, M., Rosenberg, N. A., Donnelly, P. (2000). Association mapping in structured populations. American Journal of Human Genetics, 67(1), 170–181. DOI 10.1086/302959. [Google Scholar] [CrossRef]
118. Smith, D. R., Kinsey, J. G. (1993). Latent period-a possible selection tool for Exherohilum turcicum resistance in corn (Zea mays). Maydica, 38, 205–208. [Google Scholar]
119. Schechert, A., Geiger, H. H., Welz, H. G. (1997). Generation means and combining ability analysis of resistance to Setosphaeria turcica in African maize. In: Maize productivity gains through research and technology dissemination, pp. 212–218. Proceedings of the Fifth Eastern and Southern Africa Regional Maize Conference. [Google Scholar]
120. Huang, T., Duman, J. G. (2002). Cloning and characterization of a thermal hysteresis (antifreeze) protein with DNA-binding activity from winter bittersweet nightshade, Solanum dulcamara. Plant Molecular Biology, 48, 339–350. DOI 10.1023/a:1014062714786. [Google Scholar] [CrossRef]
121. Dingerdissen, A. L., Geiger, H. H., Lee, M., Schechert, A., Welz, H. G. (1996). Interval mapping of genes for quantitative resistance of maize to Setosphaeria turcica, cause of northern leaf blight, in a tropical environment. Molecular Breeding, 2, 143–156. DOI 10.1007/BF00441429. [Google Scholar] [CrossRef]
122. Schechert, A. W., Welz, H. G., Geiger, H. H. (1999). QTL for resistance to Setosphaeria turcica in tropical African maize. Crop Science, 39, 514–523. DOI 10.2135/cropsci1999.0011183X003900020036x. [Google Scholar] [CrossRef]
123. Poland, J. A., Bradbury, P. J., Buckler, E. S., Nelson, R. J. (2011). Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proceedings of the National Academy of Sciences, 108, 6893–6898. DOI 10.1073/pnas.1010894108. [Google Scholar] [CrossRef]
124. Ding, J., Ali, F., Chen, G., Li, H., Mahuku, G. et al. (2015). Genome-wide association mapping reveals novel sources of resistance to northern corn leaf blight in maize. BMC Plant Biology, 206(1), 276. DOI 10.1186/s12870-015-0589-z. [Google Scholar] [CrossRef]
125. Chen, G., Wang, X., Long, X., Jaqueth, S., Li, J. et al. (2016). Mapping of QTL conferring resistance to northern corn leaf blight using high-density SNPs in maize. Molecular Breeding, 36, 1–9. DOI 10.1007/s11032-015-0421-3. [Google Scholar] [CrossRef]
126. Xia, H., Gao, W., Qu, J., Dai, L., Gao, Y. et al. (2020). Genetic mapping of northern corn leaf blight-resistant quantitative trait loci in maize. Medicine, 99, 31. DOI 10.1097/MD.0000000000021326. [Google Scholar] [CrossRef]
127. Jamann, T. M., Luo, X., Morales, L., Kolkman, J. M., Chung, C. L. et al. (2016). A remorin gene is implicated in quantitative disease resistance in maize. Theoretical and Applied Genetics, 129, 591–602. DOI 10.1007/s00122-015-2650-6. [Google Scholar] [CrossRef]
128. Wang, J., Xu, Z., Yang, J., Lu, X., Zhou, Z. et al. (2018). qNCLB7.02, a novel QTL for resistance to northern corn leaf blight in maize. Molecular Breeding, 38, 54. DOI 10.1007/s11032-017-0770-1. [Google Scholar] [CrossRef]
129. Kolkman, J. M., Strable, J., Harline, K., Kroon, D. E., Wiesner-Hanks, T. et al. (2020). Maize introgression library provides evidence for the involvement of liguleless1 in resistance to northern leaf blight. G3: Genes, Genome, Genetics, 10(10), 3611–3622. DOI 10.1534/g3.120.401500. [Google Scholar] [CrossRef]
130. Ranganatha, H. M., Lohithaswa, H. C., Pandravada, A. (2021). Mapping and validation of major quantitative trait loci for resistance to northern corn leaf blight along with the determination of the relationship between resistances to multiple foliar pathogens of maize (Zea mays L.). Frontiers in Genetics, 11, 548407. DOI 10.3389/fgene.2020.548407. [Google Scholar] [CrossRef]
131. Ali, F., Pan, Q., Chen, G., Zahid, K. R., Yan, J. (2013). Evidence of multiple disease resistance (MDR) and implication of meta-analysis in marker assisted selection. PLoS One, 8(7), 68150. DOI 10.1371/journal.pone.0068150. [Google Scholar] [CrossRef]
132. Martins, L. B., Rucker, E., Thomason, W., Wisser, R. J., Holland, J. B. et al. (2019). Validation and characterization of maize multiple disease resistance QTL. G3: Genes Genome Genetics, 9(9), 2905–2912. DOI 10.1534/g3.119.400195. [Google Scholar] [CrossRef]
133. Goddard, M. E., Hayes, B. J. (2007). Genomic selection. Journal of Animal Breeding and Genetics, 124(6), 323–330. DOI 10.1111/j.1439-0388.2007.00702.x. [Google Scholar] [CrossRef]
134. Technow, F., Bürger, A., Hechinger, A. E. (2013). Genomic prediction of northern corn leaf blight resistance in maize with combined or separated training sets for heterotic groups. G3: Genes Genome Genetics, 3(2), 197–203. DOI 10.1534/g3.112.004630. [Google Scholar] [CrossRef]
135. Zhang, X., Fernandes, S. B., Kaiser, C., Adhikari, P., Brown, P. J. et al. (2020). Conserved defence responses between maize and sorghum to Exherohilum turcicum. BMC Plant Biology, 20, 67. DOI 10.1186/s12870-020-2275-z. [Google Scholar] [CrossRef]
136. Balasundra, D. C., Lohithaswa, H. C., Rahul, M., Ravikumar, R. L., Anand, P. et al. (2021). Genetic mapping and genomic prediction for Northern Corn Leaf Blight (Exserohilum Turcicum (Pass.) Leonard and Suggs) resistance. Research Square. DOI 10.21203/rs.3.rs-618501/v1. [Google Scholar] [CrossRef]
137. Lopez-Zuniga, L. O., Wolters, P., Davis, S., Weldekidan, T., Kolkman, J. M. et al. (2019). Using maize chromosome segment substitution line populations for the identification of loci associated with multiple disease resistance. G3: Genes Genomes Genetics, 9(1), 189–201. DOI 10.1534/g3.118.200866. [Google Scholar] [CrossRef]
138. Singh, S. R. (2017). New quantitative trait loci (QTL) for turcicum leaf blight in maize. Crop Science, 6, 44–52. [Google Scholar]
139. Wang, H., Xiao, Z. X., Wang, F. G., Xiao, Y. N., Zhao, J. R. et al. (2012). Mapping of HtNB, a gene conferring non-lesion resistance before heading to Exherohilum turcicum (Pass.in a maize inbred line derived from the Indonesian variety Bramadi. Genetic Molecular Research, 11, 2523–2533. DOI 10.4238/2012.July.10.7. [Google Scholar] [CrossRef]
140. Zwonitzer, J. C., Coles, N. D., Krakowsky, M. D., Arellano, C., Holland, J. B. (2010). Mapping resistance quantitative trait loci for three foliar diseases in a maize recombinant inbred line population-evidence for multiple disease resistance? Phytopathology, 100(1), 72–79. DOI 10.1094/PHYTO-100-1-0072. [Google Scholar] [CrossRef]
141. P., J., Yang, J., van Esbroeck, G., Jung, J., Smith, M. E. (2010). Use of a maize advanced intercross line for mapping of QTL for northern leaf blight resistance and multiple disease resistance. Crop Science, 50, 458–466. DOI 10.2135/cropsci2009.02.0066. [Google Scholar] [CrossRef]
142. Cao, Z., Zhang, K., Guo, X., Turgeon, B. G., Dong, J. A. (2020). Genome resource of Setosphaeria turcica, causal agent of northern leaf blight of maize. Phytopathology, 110(12), 2014–2016. DOI 10.1094/PHYTO-06-20-0225-A. [Google Scholar] [CrossRef]
143. Schnable, S. P., Ware, D., Fulton, R. S., Stein, J. C., Wei, F. et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science, 326, 1112–1115. DOI 10.1126/science.1178534. [Google Scholar] [CrossRef]
144. Knott, G. J., Doudna, J. A. (2018). CRISPR-Cas guides the future of genetic engineering. Science, 361(6405), 866–869. DOI 10.1126/science.aat5011. [Google Scholar] [CrossRef]
145. Yang, N., Xu, X. W., Wang, R. R., Peng, W. L., Cai, C. et al. (2017). Contributions of Zea mays subspecies mexicana haplotypes to modern maize. Natural Communication, 8, 1874. DOI 10.1038/s41467-017-02063-5. [Google Scholar] [CrossRef]
146. Ye, J., Zhong, T., Zhang, D., Ma, C., Wang, L. (2019). The auxin-regulated protein ZmAuxRP1 coordinates the balance between root growth and stalk rot disease resistance in maize. Molecular Plant, 12, 360–373. DOI 10.1016/j.molp.2018.10.005. [Google Scholar] [CrossRef]
147. Yang, Q., Yin, G., Guo, Y., Zhang, D., Chen, S. et al. (2010). Major QTL for resistance to Gibberella stalk rot in maize. Theoretical and Applied Genetics, 121, 673–687. DOI 10.1007/s00122-010-1339-0. [Google Scholar] [CrossRef]
148. Wang, C., Yang, Q., Wang, W., Li, Y., Guo, Y. et al. (2017). Transposon-directed epigenetic change in ZmCCT underlies quantitative resistance to Gibberella stalk rot in maize. New Phytologist, 215, 1503–1515. DOI 10.1111/nph.14688. [Google Scholar] [CrossRef]
149. Wu, F., Shu, J., Jin, W. (2014). Identification and validation of miRNAs associated with the resistance of maize (Zea mays L.) to Exherohilum turcicum. PLoS One, 9(1), e87251. DOI 10.1371/journal.pone.0087251. [Google Scholar] [CrossRef]
150. McDonald, B. A., Linde, C. (2002). Pathogen population genetics, evolutionary potential, and durable resistance. Annual Review of Phytopathology, 40, 349–379. DOI 10.1146/annurev.phyto. [Google Scholar] [CrossRef]
151. Najar, Z. A., Farooq, A. S., Najeeb, S., Shikari, A. B., Ahangar, M. A. et al. (2018). Genotypic and morphological diversity analysis in high altitude maize (Zea mays L.) inbreds under Himalayan temperate ecologies. Maydica, 63, 1–7. [Google Scholar]
152. Shikari, A. B., Zafar, G. (2009). Evaluation and identification of maize for turcicum leaf blight resistance under cold temperate conditions. Maize Genetics Cooperation Newsletter, 83, 1–8. [Google Scholar]
153. Kumar, S., Gowd, P. K. T., Pant, S. K., Shekhar, M., Kumar, B. et al. (2011). Sources of resistance to Exherohilum turcicum (Pass.) and Pucciniapolysora (Underw.) incitant of Turcicum leaf blight and polysora rust of maize. Plant Protection, 44, 528–536. DOI 10.1080/03235400903145558. [Google Scholar] [CrossRef]
154. Chandrashekara, C., Jha, S. K., Arukumar, R., Agrawal, P. K. (2014). Identification of new sources of resistance to turcicum leaf blight and maydis leaf blight in maize (Zea mays L.). SABRAO Journal of Breeding and Genetics, 46(1), 44–55. [Google Scholar]
155. Singh, R., Srivastava, R. P., Mani, V. P., Khandelwal, R. S., Ram, L. (2014). Screening of maize genotypes against northern corn leaf blight. Supplement on Genetics and Plant Breeding, 9(4), 1689–1693. DOI 10.30848/PJB2019-5(10). [Google Scholar] [CrossRef]
156. Muiru, W. M., Charles, A. K., Kimenju, J. W., Njoroge, K., Miano, D. W. (2015). Evaluation of resistance reaction of maize germplasm to common foliar diseases in Kenya. Journal of Natural Sciences Research, 5(1), 140–145. [Google Scholar]
157. Garomaa, B., Bitew, T., Midekssa, D., Temesgen, D., Girma, D. et al. (2016). Evaluation of quality protein maize inbred lines for resistance to turcicum leaf blight and grey leaf spot disease under field condition at mid altitude sub-humid agro-ecology of Ethiopia. Scientific Journal of Crop Science, 5(11), 137–145. DOI 10.14196/sjcs.v5i11.2252. [Google Scholar] [CrossRef]
158. Munkvold, G. P., Gorman, D. (2006). Foliar fungicide use in corn. In: Crop insights. Johnston, IA: Pioneer Hi-Bred. [Google Scholar]
159. Shah, D. A., Dillard, H. R. (2010). Managing foliar diseases of processing sweet corn in New York with Strobilurin fungicides. Plant Disease, 94(2), 213–220. DOI 10.1094/PDIS-94-2-0213. [Google Scholar] [CrossRef]
160. Kumar, S., Archana, R., Jha, M. M. (2009). Efficacy of fungicide against Helminthosporium maydis of maize. Annual Plant Protection Science, 17, 255–256. [Google Scholar]
161. Arcibal, S. M. (2013). Integrated management of southern corn rust and northern corn leaf blight using hybrids and fungicides(MSc. Thesis), pp. 105. University of Georgia, Athens. [Google Scholar]
162. Couretot, L., Parisi, L., Ferraris, G., Magnone, G. (2014). Efecto de Fungicidas Foliares y Momento de Aplicación sobre la Severidad de Tizón Foliar y Enfermedades de Raíz y Tallo en Maíz. Rosario, Argentina: Abstracts of XIV Jornadas Fitosanitarias. [Google Scholar]
163. Carpane, P. D., Peper, A. M., Kohn, F. (2019). Management of northern corn leaf blight using nativo (trifloxistrobin + epoxiconazole) fungicide applications. Crop Protection, 127, 104982. DOI 10.1016/j.cropro.2019.104982. [Google Scholar] [CrossRef]
164. Veerabhadraswamy, A. L., Pandurangegowda, K. T., Prasanna, K. M. K. (2014). Efficacy of strobilurin group fungicides against turcicum leaf blight and polysora rust in maize hybrids. International Journal of Agriculture and Crop Science, 7, 100–106. [Google Scholar]
165. Blandino, M., Galeazzi, M., Savoia, W., Reyneri, A. (2012). Timing of azoxystrobin + propiconazole application on maize to control northern corn leaf blight and maximize grain yield. Field Crop Research, 139, 20–29. DOI 10.1016/j.fcr.2012.09.014. [Google Scholar] [CrossRef]
166. Camera, J. N., Forcelini, C. A., Koefender, J., Golle, D. P., Schoffe, A. (2019). Reaction of maize hybrids to Northern corn leaf blight and common rust, and chemical control of Northern corn leaf blight. Plant Pathology, 86, 1–10. DOI 10.1590/1808-1657000082018. [Google Scholar] [CrossRef]
167. Avenot, H. F., Michailides, T. J. (2010). Progress in understanding molecular mechanisms and evolution of resistance to succinate dehydrogenase inhibiting (SDHI) fungicides in phytopathogenic fungi. Crop Protection, 29(7), 643–651. DOI 10.1016/j.cropro.2010.02.019. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |