Abstract
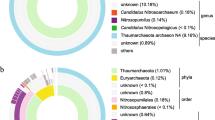
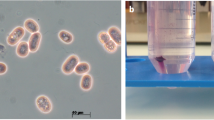
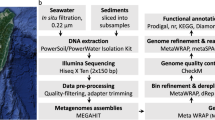
In this study, three metagenome-assembled genomes of a sediment sample were constructed. A Bin1 (JB001) genome was identified as a photo-litho-auto/heterotroph (purple sulfur bacteria) bacterium with the ability to fix nitrogen, tolerate salt, and to produce bacteriochlorophyll a. It has a genome length of 4.1 Mb and a G + C content of 64.9%. Phylogenetic studies based on concatenated 92 core genes and photosynthetic genes (pufLM and bchY) showed that Bin JB001 is related to Thiococcus pfennigii, “Thioflavicoccus mobilis” and to the Lamprocystis purpurea lineage. Bin JB001 and its closely related members were subjected to the genome-based study of phenotypic and phylogenomic analysis. Genomic similarity indices (dDDH and ANI) showed that Bin JB001 could be defined as a novel species. The average amino acid identity (AAI) and percentage of conserved proteins (POCP) values were below 60 and 50%, respectively. The pan-genome analysis indicated that the pan-genome was an open type wherein Bin JB001 had 855 core genes. This study shows that the binned genome, Bin JB001 could represent a novel species of a new genus under the family Chromatiaceae, for which the name “Candidatus Thioaporhodococcus sediminis” gen. nov. sp. nov. is proposed.


Similar content being viewed by others
Abbreviations
- APB:
-
Anoxygenic phototrophic bacteria
- MAG:
-
Metagenome assemble genome
- CTAB:
-
Cetyl trimethyl ammonium bromide
- MG-RAST:
-
Metagenomic Rapid Annotations using Subsystems Technology
- RAST:
-
Rapid Annotation using Subsystem Technology
- KEGG:
-
Kyoto Encyclopaedia of Genes and Genomes
- KAAS:
-
KEGG Automatic Annotation Server
- GTDB:
-
Genome Taxonomy Database
- AAI:
-
Average Amino Acid Identity
- cAAI:
-
Core Average Amino Acid Identity
- BPGA:
-
Bacterial Pan-genome Pipeline
- POCP:
-
Percentage of Conserved Proteins
- CAM:
-
Crassulacean Acid Metabolism
- TCA:
-
Tricarboxylic Acid Cycle
- WL:
-
Wood–Ljundahl
References
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Arkin AP, Cottingham RW, Henry CS, Harris NL, Stevens RL, Maslov S, Dehal P, Ware D, Perez F et al (2018) KBase: the United States department of energy systems biology knowledgebase. Nat Biotech 36(7):566–569. https://doi.org/10.1038/nbt.4163
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S et al (2008) The RAST server: rapid annotations using subsystems technology. BMC Genom 9:75
Bell E, Lamminmäki T, Alneberg J, Andersson AF, Qian C, Xiong W, Hettich RL, Balmer L et al (2018) Biogeochemical Cycling by a Low-Diversity Microbial Community in Deep Groundwater. Front Microbiol 9:2129. https://doi.org/10.3389/fmicb.2018.02129
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinform 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Britt RD, Rao G, Tao L (2020) Bioassembly of complex iron-sulfur enzymes: hydrogenases and nitrogenases. Nat Rev Chem 4(10):542–549
Bryant DA, Frigaard NU (2006) Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol 14(11):488–496. https://doi.org/10.1016/j.tim.2006.09.001
Bryant DA, Costas AM, Maresca JA, Chew AG, Klatt CG, Bateson MM, Tallon LJ, Hostetler J et al (2007) Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic Acidobacterium. Science 317(5837):523–526
Bryantseva I, Gorlenko VM, Kompantseva EI, Imhoff JF, Süling J, Mityushina L (1999) Thiorhodospira sibirica gen. nov., sp. Nov., a new alkaliphilic purple sulfur bacterium from a Siberian soda lake. Int J Syst Bacteriol 49(2):697–703. https://doi.org/10.1099/00207713-49-2-697
Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J (2021) eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol 38(12):5825–5829. https://doi.org/10.1093/molbev/msab293
Chaudhari NM, Gupta VK, Dutta C (2016) BPGA- an ultra-fast pan-genome analysis pipeline. Sci Rep 6:24373. https://doi.org/10.1038/srep24373
Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH (2020) GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36(6):1925–1927. https://doi.org/10.1093/bioinformatics/btz848
Chen YH, Shyu YT, Lin SS (2018) Characterization of candidate genes involved in halotolerance using high-throughput omics in the halotolerant bacterium Virgibacillus chiguensis. PLoS ONE 13(8):e0201346. https://doi.org/10.1371/journal.pone.0201346
Cheng JE, Su P, Zhang ZH, Zheng LM, Wang ZY, Hamid MR, Dai JP, Du XH et al (2022) Metagenomic analysis of the dynamical conversion of photosynthetic bacterial communities in different crop fields over different growth periods. PLoS One 17(7):e0262517. https://doi.org/10.1371/journal.pone.0201346
Chew AG, Bryant DA (2007) Chlorophyll biosynthesis in bacteria: the origins of structural and functional diversity. Annu Rev Microbiol 61:113–129. https://doi.org/10.1146/annurev.micro.61.080706.093242
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68(1):461–466. https://doi.org/10.1099/ijsem.0.002516
Cui YX, Biswal BK, Guo G, Deng YF, Huang H, Chen GH, Wu D (2019) Biological nitrogen removal from wastewater using sulphur-driven autotrophic denitrification. Appl Microbiol Biotechnol 103(15):6023–6039. https://doi.org/10.1007/s00253-019-09935-4
Daum M, Herrmann S, Wilkinson B, Bechthold A (2009) Genes and enzymes involved in bacterial isoprenoid biosynthesis. Curr Opin Chem Biol 13(2):180–188. https://doi.org/10.1016/j.cbpa.2009.02.029
Deb S, Das SK (2022) Phylogenomic analysis of metagenome-assembled genomes deciphered novel acetogenic nitrogen-fixing Bathyarchaeota from hot spring sediments. Microbiol Spectr 10(3):e0035222. https://doi.org/10.1128/spectrum.00352-22
Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, Haft DH, Hoffmann M, Pettengill JB et al (2021) AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep 11(1):12728. https://doi.org/10.1038/s41598-021-91456-0
Frigaard NU, Dahl C (2009) Sulfur metabolism in phototrophic sulfur bacteria. Adv Microb Physiol 54:103–200. https://doi.org/10.1016/S0065-2911(08)00002-7
George DM, Vincent AS, Mackey HR (2020) An overview of anoxygenic phototrophic bacteria and their applications in environmental biotechnology for sustainable Resource recovery. Biotechnol Rep (amst) 28:e00563. https://doi.org/10.1016/j.btre.2020.e00563
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57(81):91. https://doi.org/10.1099/ijs.0.64483-0
Gorlenko VM, Bryantseva IA, Rabold S, Tourova TP, Rubtsova D, Smirnova E, Thiel V, Imhoff JF (2009) Ectothiorhodospira variabilis sp. nov., an alkaliphilic and halophilic purple sulfur bacterium from soda lakes. Int J Syst Evol Microbiol 59:658–664. https://doi.org/10.1099/ijs.0.004648-0
Hanada S (2016) Anoxygenic photosynthesis-A photochemical reaction that does not contribute to oxygen reproduction. Microbes Environ 31(1):1–3. https://doi.org/10.1264/jsme2.ME3101rh
Hedlund BP, Chuvochina M, Hugenholtz P, Konstantinidis KT, Murray AE, Palmer M, Parks D, Probst AJ, Reysenbach AL et al (2022) SeqCode: a nomenclatural code for prokaryotes described from sequence data. Nat Microbiol. https://doi.org/10.1038/s41564-022-01214-9
Higuchi-Takeuchi M, Numata K (2019) Marine purple photosynthetic bacteria as sustainable microbial production hosts. Front Bioeng Biotechnol 7:258. https://doi.org/10.3389/fbioe.2019.00258
Imhoff J (2004) Taxonomy and physiology of phototrophic purple bacteria and green sulfur bacteria. Anoxygenic photosynthetic bacteria. Springer, Berlin, pp 1–15
Imhoff JF, Rahn T, Kunzel S, Neulinger SC (2019) Phylogeny of anoxygenic photosynthesis based on the sequences of photosynthetic reaction center proteins and a key enzyme in bacteriochlorophyll biosynthesis, the chlorophyllide reductase. Microorganisms 7(11):576
Imhoff JF, Kyndt JA, Meyer TE (2022) Genomic comparison, phylogeny and taxonomic reevaluation of the Ectothiorhodospiraceae and description of Halorhodospiraceae fam. Nov. and Halochlorospira gen. nov. Microorganisms 10(2):295. https://doi.org/10.3390/microorganisms10020295
Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S (2018) High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. https://doi.org/10.1038/s41467-018-07641-9
Karthikeyan S, Rodriguez-R LM, Heritier-Robbins P, Kim M, Overholt WA, Gaby JC, Hatt JK, Spain JC et al (2019) “Candidatus Macondimonas diazotrophica”, a novel gammaproteobacterial genus dominating crude-oil-contaminated coastal sediments. ISME J 13(8):2129–2134. https://doi.org/10.1038/s41396-019-0400-5
Keegan KP, Glass EM, Meyer F (2016) MG-RAST, a metagenomics service for analysis of microbial community structure and function. Methods Mol Biol 1399:207–233. https://doi.org/10.1007/978-1-4939-3369-3_13
Konstantinidis KT, Rosselló-Móra R, Amann R (2017) Uncultivated microbes in need of their own taxonomy. ISME J 11(11):2399–2406. https://doi.org/10.1038/ismej.2017.113
Loh HQ, Hervé V, Brune A (2021) Metabolic potential for reductive acetogenesis and a novel energy-converting [NiFe] hydrogenase in Bathyarchaeia from termite guts - a genome-centric analysis. Front Microbiol 11:635786. https://doi.org/10.3389/fmicb.2020.635786
Lübbert C, Baars C, Dayakar A, Lippmann N, Rodloff AC, Kinzig M, Sörgel F (2017) Environmental pollution with antimicrobial agents from bulk drug manufacturing industries in Hyderabad, South India, is associated with dissemination of extended-spectrum beta-lactamase and carbapenemase-producing pathogens. Infection 45(4):479–491. https://doi.org/10.1007/s15010-017-1007-2
Masclaux-Daubresse C, Reisdorf-Cren M, Pageau K, Lelandais M, Grandjean O, Kronenberger J, Valadier MH, Feraud M et al (2006) Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol 140(2):444–456. https://doi.org/10.1104/pp.105.071910
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60
Meier-Kolthoff JP, Sardà Carbasse J, Peinado-Olarte RL, Göker M (2022) TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acid Res 50:D801–D807
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185. https://doi.org/10.1093/nar/gkm321
Mujakić I, Andrei AŞ, Shabarova T, Fecskeová LK, Salcher MM, Piwosz K, Ghai R, Koblížek M (2021) Common presence of phototrophic Gemmatimonadota in temperate freshwater lakes. mSystems. 6(2): e01241–20
Murray AE, Freudenstein J, Gribaldo S, Hatzenpichler R, Hugenholtz P, Kämpfer P, Konstantinidis KT, Lane CE et al (2020) Roadmap for naming uncultivated archaea and bacteria. Nat Microbiol 5(8):987–994. https://doi.org/10.1038/s41564-020-0733-x
Na SI, Kim YO, Yoon SH, Ha SM, Baek I, Chun J (2018) UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56(2):280–285. https://doi.org/10.1007/s12275-018-8014-6
Nurk S, Meleshko D, Korobeynikov A, Pevzner PA (2017) metaSPAdes: a new versatile metagenomic assembler. Genome Res 27(5):824–834
Oren A, Garrity GM (2021) Valid publication of the names of forty-two phyla of prokaryotes. Int J Sys Evol Microbiol. https://doi.org/10.1099/ijsem.005056
Palmer M, Sutcliffe I, Venter SN, Hedlund BP (2022) It is time for a new type of type to facilitate naming the microbial world. New Microbes New Infect 47:100991. https://doi.org/10.1016/j.nmni.2022.100991
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25(7):1043–1055. https://doi.org/10.1101/gr.186072.114
Parks DH, Rinke C, Chuvochina M, Chaumeil PA, Woodcroft BJ, Evans PN, Hugenholtz P, Tyson GW (2017) Recovery of nearly 8000 metagenome-assembled genomes substantially revises the tree of life. Nat Biotechnol 2(11):1533–1542
Pérez-Cataluña A, Salas-Massó N, Diéguez AL, Balboa S, Lema A, Romalde JL, Figueras MJ (2018) Revisiting the Taxonomy of the Genus Arcobacter: Getting Order From the Chaos. Front Microbiol 9:2077. https://doi.org/10.3389/fmicb.2018.02077
Qin Q, Xie B, Zhang X, Chen X, Zhou B, Zhou J, Oren A, Zhang YZ (2014) A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196(1):2210–2215. https://doi.org/10.1128/jb.01688-14
Rodriguez-R LM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints. https://doi.org/10.7287/peerj.preprints.1900v1
Saini MK, Sebastian A, Shirotori Y, Soulier NT, Garcia Costas AM, Drautz-Moses DI, Schuster SC, Albert I et al (2021) Genomic and phenotypic characterization of Chloracidobacterium isolates provides evidence for multiple species. Front Microbiol 12:704168. https://doi.org/10.3389/fmicb.2021.704168
Setubal JC (2021) Metagenome-assembled genomes: concepts, analogies, and challenges. Biophys Rev 13(6):905–909
Tank M, Bryant DA (2015) Chloracidobacterium thermophilum gen. nov, sp. Nov.: an anoxygenic microaerophilic chlorophotoheterotrophic acidobacterium. Int J Syst Evol Microbiol 65(5):1426–1430. https://doi.org/10.1099/ijs.0.000113
Tsukatani Y, Yamamoto H, Harada J, Yoshitomi T, Nomata J, Kasahara M, Mizoguchi T, Fujita Y et al (2013) An unexpectedly branched biosynthetic pathway for bacteriochlorophyll b capable of absorbing near-infrared light. Sci Rep 3:1217
Ward LM, Cardona T, Holland-Moritz H (2019) Evolutionary implications of anoxygenic phototrophy in the bacterial phylum Candidatus Eremiobacterota (WPS-2). Front Microbiol 23(10):1658
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsk MI et al (1984) International committee on systematic bacteriology. report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Wirth JS, Whitman WB (2018) Phylogenomic analyses of a clade within the roseobacter group suggest taxonomic reassignments of species of the genera Aestuariivita, Citreicella, Loktanella, Nautella, Pelagibaca, Ruegeria, Thalassobius, Thiobacimonas and Tropicibacter and the proposal of six novel genera. Int J Syst Evol Microbiol 68(7):2393–2411
Zeng Y, Feng F, Medová H, Dean J, Koblížek M (2014) Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum Gemmatimonadetes. Proc Natl Acad Sci USA 111(21):7795–7800. https://doi.org/10.1073/pnas.1400295111
Acknowledgements
We are grateful to Bernhard Schink, University of Konstanz, Germany, and Prof. Aharon Oren (The Hebrew University of Jerusalem) for correcting the protologues.
Funding
Amulya thanks IOE-UoH project for the JRF fellowship, and Anusha thanks DST, Government of India for the award of INSPIRE SRF. Ramana thanks the Department of Biotechnology, Government of India, for the award of Tata Innovation Fellowship and IoE-UoH for the project grant. Financial support received from CSIR is acknowledged. Sasikala thanks UGC for the award of the mid-career grant. Infrastructural facilities funded by DST-FIST (UoH and JNTUH), UGC-SAP (DRS; UoH), TEQIP, and AICTE (from JNTUH) are acknowledged.
Author information
Authors and Affiliations
Contributions
AS designed and planned the studies under the supervision of CS and CVR. AR and AS performed the genomic and phylogenetic analysis and tabulated the data. AR and AR wrote the manuscript. CVR and CS supervised the study and contributed to text preparation and revised the manuscript. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Amulyasai, B., Anusha, R., Sasikala, C. et al. Phylogenomic analysis of a metagenome-assembled genome indicates a new taxon of an anoxygenic phototroph bacterium in the family Chromatiaceae and the proposal of “Candidatus Thioaporhodococcus” gen. nov. Arch Microbiol 204, 688 (2022). https://doi.org/10.1007/s00203-022-03298-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-03298-7