Abstract
Main conclusion
Tripsacum dactyloides is closely related to Zea mays since Zea perennis and the MTP tri- species hybrid have four possible reproductive modes.
Eastern gamagrass (Tripsacum dactyloides L.) and tetraploid perennial teosinte (Zea perennis) are well known to possess genes conferring resistance against biotic and abiotic stresses as well as adaptation to flood and aluminum toxic soils. However, plant breeders have been hampered to utilize these and other beneficial traits for maize improvement due to sterility in their hybrids. By crossing a tetraploid maize-inbred line × T. dactyloides, a female fertile hybrid was produced that was crossed with Z. perennis to yield a tri-genomic female fertile hybrid, which was backcrossed with diploid maize to produce BC1 and BC2. The tri-genomic hybrid provided a new way to transfer genetic material from both species into maize by utilizing conventional plant breeding methods. On the basis of cytogenetic observations using multi-color genomic in situ hybridization, the progenies were classified into four groups, in which chromosomes could be scaled both up and down with ease to produce material for varying breeding and genetic purposes via apomixis or sexual reproduction. In the present study, pathways were found to recover maize and to obtain specific translocations as well as a speedy recovery of the T. dactyloides–maize addition line in a second backcross generation. However, phenotypes of the recovered maize were in most cases far from maize as a result of genetic load from T. dactyloides and Z. perennis, and could not be directly used as a maize-inbred line but could serve as an intermediate material for maize improvement. A series of hybrids was produced (having varying chromosome number, constitution, and translocations) with agronomic traits from all three parental species. The present study provides an application of overcoming the initial interspecific barriers among these species. Moreover, T. dactyloides is closely related to Z. mays L. ssp. mays since Z. perennis and the MTP tri- species hybrid have four possible reproductive modes.
Similar content being viewed by others
Introduction
The narrow gene pool in maize results in retardation of its genetic improvement mainly due to invariability. The broadening of the maize gene pool is essential to boost biotic and abiotic stresses tolerance as well as to ensure food supply to a growing population. Plant breeders and geneticists are restrained mostly in exploitation of the genetic variability in the primary gene pool. Yet several interesting traits lie in secondary and tertiary gene pools but their transfer into cultivated crops is not so straight forward. Although gene transfer from diverse species is possible through transgenic technologies nowadays, it is effective only for dominant and oligogenic traits. The recessive and quantitative traits are still the domain of conventional plant breeding, and most of the economically important traits providing abiotic stress tolerance, yield, nutrition, and quality are quantitative in nature. Therefore, we are bound to utilize available genetic resource and crop-wild relatives to improve these traits. No doubt, wild relatives are a rich source of new beneficial genes but linkage of unwanted traits and sterility reduces their effectiveness in boosting germplasm’s genetic diversity (Duvick 1984). The genetic bridges (usage of a third parent in between two parents) can break crossing barriers in between divergent species (Hussain and Williams 1997; Reagon and Snow 2006) and choice of small DNA fragments (translocations) can be advantageous to get rid of unwanted traits. Polyploid provides an effective bridge for a controlled induction of alien genes into cultivated crops and the choice of an allopolyploid bridge plant is preferable due to its genome plasticity (Stebbins 1971; Cheng et al. 2016), gene redundancy, asexual reproduction (Comai 2005) and allopolyploid’s capability to seldom disrupt self-incompatibility barriers (Miller and Venable 2000). Polyploidy and distinct hybridization have already enriched the genetic background of important agricultural crops (Hajjar and Hodgkin 2007), for example wheat (Jiang et al. 2017), rice (Shakiba and Eizenga 2014; Ramos et al. 2016), maize (Maazou et al. 2017), Brassica (Li et al. 2014) and tobacco (Hancock and Lewis 2017).
Maize (Zea mays L. ssp. mays, 2n = 20) is the only cultivated species of the genus Zea. All other Zea species are mainly wild grasses, collectively known as teosinte, widely distributed in Mexico and Central America and serve as an important source of beneficial alien genetic material for maize improvement. All species in genus Zea are diploid (2n = 2x = 20) and cross compatible with maize except the tetraploid perennial teosinte Z. perennis (2n = 2x = 40) which belongs to the section Luxuriantes, which is distinctly related to maize as later belong to section Zea (Doebley and Iltis 1980). Emerson and Beadle (1930) were the first who crossed maize with Z. perennis. Natural gene flow between diploid maize and Z. perennis is tremendously low, if of any kind (Doebley 1990). However, artificial hybridization is possible (Tang et al. 2005) but the triploid nature of F1 hybrids (2n = 3x = 30) makes them sterile (Shaver 1964; Cheng et al. 2016). The cross between colchicine-treated tetraploid maize (2n = 4x = 40)and Z. perennis (2n = 4x = 40) could give rise to allotetraploid F1 (2n = 4x = 40) that showed normal meiosis and higher pollen viability (Shaver 1964). Previous studies have confirmed that five chromosomes from “A” genome of maize shared higher homology with 10 chromosomes from Z. perennis (Schnable et al. 2011; Cheng et al. 2016).
Eastern gamagrass (Tripsacum dactyloides), also called “ice cream grass”, predominantly found at diploid (2n = 2x = 36) and tetraploid (2n = 4x = 72) levels (Farquharson 1955), belongs to the genus Tripsacum, which resides in the tertiary gene pool of maize and owns many interesting genes for maize improvement (Hardin 1994; Maazou et al. 2017). It is a warm-season C4 facultative apomictic perennial forage grass having a remarkable ability to withstand various biotic and abiotic stresses (Eubanks 2006). Artificial crosses are possible between corn and T. dactyloides with more or less success but generally involve embryo rescue (Leblanc et al. 2009; Belova et al. 2010). Although F1 hybrids between maize and T. dactyloides showed an abnormal meiosis, pairing between chromosomes from both parental species indicated the possibility of genetic recombination between them (Molina et al. 2006). Previous studies reported that maize chromosomes 2, 5, 8, and 9 have a potential for genetic exchange with T. dactyloides chromosomes (Galinat 1973). The Mz9S region (small arm of maize chromosome no. 9; Mz stands for Z. mays chromosome; S and L for short and long arms, respectively) showed pairing and recombination with chromosome no. 5 of T. dactyloides (Kindiger and Beckett 1990) and sub-terminal regions of Mz2S, Mz6L, and Mz8L shared homology with T. dactyloides chromosomes (Poggio et al. 1999). Previously, various pathways for the Tripsacum introgression in maize have been reported but till today only two recombination/translocation instances have been verified (Maguire 1962; Kindiger et al. 1996a). By crossing a maize (2n = 20) x T. dactyloides (2n = 36), a hybrid was produced that was treated with colchicine to induce chromosome doubling and fertilized with pollen from Z. diploperennis (2n = 20), Walton Galinat produced an individual containing genomes of all three species (W. Galinat, personal communication), which was later studied by Lamb and Birchler (2006) and confirmed that tri-species hybrid was 2n = 37 with missing of chromosome number 2 from maize genome. The novelty of our research is to create a hybrid by crossing autotetraploid maize (2n = 40) × tetraploid T. dactyloides (2n = 72) that was later fertilized with pollen from Z. perennis (2n = 40) to produce a tri-genomic hybrid (2n = 74) with a different genetic constitution than previously reported from hybrids. Furthermore, we broadened the understanding of meiotic behavior, genetic relationships, and co-segregations of three distinct genome chromosomes in case they co-habitat one nucleus.
In this study, an allopolyploid tri-genomic bridge plant between T. dactyloides, tetraploid maize and Z. perennis has been produced, which was backcrossed with maize to produce BC1 and BC2 with the following objectives; (1) to create maize germplasm with variable agronomic and genetic characteristics, (2) to obtain hybrids with varying genomic composition and translocation, (3) to observe distinct species chromosomal segregation behavior in one nucleus. Various cytological groups among different tested progenies have been established. Together, our results support the notion that this route can give rise to cytologically variable progeny that could be utilized to breed new inbred lines and to obtain various types of addition and substitution lines. Such cytologically diverse groups could serve as raw material for the production of new species.
Materials and methods
Plant material and creation of tri-genomic hybrid bridge plant
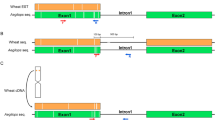
Interspecific hybrid named as MT (2n = 4x = 56) was introduced by United States Department of Agriculture (USDA), which was produced by crossing autotetraploid maize-inbred line V182 (2n = 4x = 40) and T. dactyloides (2n = 4x = 72). Seeds of Z. perennis (2n = 4x = 40, accession no. 9475) were obtained from the International Maize and Wheat Improvement Center (CIMMYT) and a diploid maize-inbred line Mo17 (2n = 2x = 20) was provided by USDA. The plant material was raised at the experimental farm of Sichuan Agricultural University, Wenjiang, China. The vegetatively propagated hybrid MT was hybridized using pollens of Z. perennis to produce a tri-genomic interspecific hybrid MTP (M stands for maize, T for T. dactyloides, and P for Z. perennis). In next growing season, MTP was backcrossed using pollens of Mo17 to produce BC1, BC2 and so on. The schematic representation of material creation is illustrated in Fig. 1.
Phenotypes of interspecific hybrids
Agronomic traits (tillers number, plant height, stem diameter, lateral branch number, spike length, number of spikes), ear traits (ear length, ear diameter, number of rows/ear, number of seed/ear, seed setting percentage), pollen viability, total days of tasseling, silking and anthesis were recorded for BC1 and BC2 plants. The pollen fertility was assessed using the I2–KI staining method. Three anthers were taken, squashed, and stained on a glass slide, observed under the light microscope and three images of each were taken for analysis. The 75% of stained pollen were counted as fertile ones. The seed setting percentage was calculated by dividing the total number of kernels on an ear by the total number of seeds.
Meiotic observation in pollen mother cells (PMCs)
Pre-mature anthers of BC1 plants were collected and treated with Carnoy’s solution for a minimum of 12 h, then saved in 70% ethanol solution at 4 °C. The saved anthers were rinsed with distilled water, dissected on a clean glass slide, stained with carbol fuchsin, covered with cover glass and observed under phase-contrast light microscope (Olympus BX-41). The images were captured with Media Cybernetics CCD 700 (charge coupled device).
Mitotic chromosome and DNA preparation
The roots collected from interspecific hybrid MTP, BC1 and BC2 plants were fixed immediately in a saturated solution of α-bromonaphthalene for 3 h and then transferred in Carnoy’s solution I (3:1 ethanol: glacial acetic acid, v/v) for 24 h, finally submerged in 70% ethanol solution and saved at 4 °C. The saved root tips were transferred to distilled water to remove effects of ethanol and then treated with an enzymatic solution comprising 6% cellulase (R-10, Yakult Biochemicals, Nishinomiya, Japan) and 1% pectinase (Y-23, Yakult) for 2.5–5.0 h at 37 °C. Root tips were thoroughly cleaned with distilled water. Dissection, staining, and image capture were the same procedures as for meiotic chromosomes. For GISH, enzyme-treated root tips were washed with distilled water, squashed onto glass slides in a drop of Carnoy’s solution I and dried with ethanol flame. The preparations showing well-spread and clean mitotic chromosomes were selected by phase-contrast light microscopy (Olympus BX-41) and stored at − 20 °C for in situ hybridization. Total genomic DNA from young leaves of maize and Z. perennis was extracted following a modified 2 × CTAB method (Fu et al. 2015).
Genomic in situ hybridization
The genomic DNA of maize and Z. perennis was labeled with DIG-Nick Translation and Biotin-Nick Translation Mix (Roche, Basel, Switzerland), respectively, according to the manufacturer’s protocol. The selected slides were preheated in an air-blowing oven at 60 °C for 1 h and then transferred into 0.1 µg/ml RNase in 2 × SSC (Solarbio, Beijing, China) solutions in a thermostat water bath at 37 °C for 1 h. Afterwards, slides were washed twice with 2 × SSC for 5 min each at room temperature, followed by chromosome denaturation in 70% deionized formide (FAD) solution at 70 °C for 2.5 min. Then the slides were immediately dehydrated in chilled 70, 95, and 100% ethyl alcohol series, respectively, at − 20 °C and air-dried at room temperature. The hybridization mixture involved 150 µl 50% FAD, 60 µl 10% dextran sulfate (DS), 30 µl 2 × SSC, 15 µl 0.5% sodium dodecyl sulfate (SDS), 30 μg salmon sperm DNA (SSDNA) and 18 µl labeled probes for six slides. Hybridization mixture was denatured in a thermostat at 85 °C for 10 min, followed by quick cooling in ice for 10 min. An aliquot of 46 µl hybridization mixture was loaded on each slide and hybridization was accomplished in an incubator at 37 °C for 20–24 h. After hybridization, slides were immersed in 20% FAD, 2 × SSC, 0.1 × SSC for 15 min each, respectively, at 42 °C. Subsequently, slides were washed in 0.1% TritonX-100 once and in 1 × PBS thrice for 5 min each and then air-dried at room temperature. All further procedure was performed in darkness, 50 µl antibody diluent, which contained anti-digoxigenin–fluorescein (0.6 µg/µl in 1 × PBS, Roche) and CY-3 fluorescein (0.6% in 1 × PBS, Sigma, St. Louis, MO, USA) were applied onto air-dried slides and hybridization was done at 37 °C for 1 h in an incubator. The slides were then washed in 1 x PBS thrice for 5 min each at room temperature and air-dried finally. The chromosome counterstaining was performed by 4, 6-diamidino-2-phenylindole (DAPI), and slides were observed with a fluorescence microscope (Olympus BX-61). Images were captured with Media Cybernetics CCD 700 (Charge Coupled Device) and Image Pro Plus 6.0 (Media Cybernetics, Inc.). Captured images were processed with Adobe Photoshop 5.1.
Results
Creation of interspecific hybrid and backcross progenies
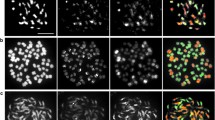
The artificially produced tri-genomic interspecific hybrid MTP was a male sterile perennial plant showing intermediate phenotype of parental species (Fig. 2a). Multi-color Genomic in Situ-Hybridization (mcGISH) with Cent-C probe was carried out to verify the genomic constitution of MTP. The results confirmed that the MTP was an allohexaploid having 20 chromosomes from maize, 20 chromosomes from Z. perennis and 34 chromosomes from T. dactyloides (Fig. 2b–e). The MTP was backcrossed to diploid maize inbred Mo17 and seeds from F1 stalks were harvested. At sowing, 34 seeds could germinate (germination rate was 88.23%), of which 30 plants could complete life cycle and only one plant, named as BC1-31, was fertile. The plants of BC1 were backcrossed with maize using pollen of Mo17 to produce BC2 with an average seed-setting rate of 3.80%. On the basis of chromosome distribution in BC1 plants, 16 lineages were selected and placed into 4 groups. The germination rate of BC2 seeds was 71.47%, and 218 seedlings for BC2 were obtained. From BC2, a total of 6 groups were backcrossed and 5 groups were self-crossed. The seed-setting rate for backcrossed plants varied from 0 to 31.48%, with an average of 3.67%, while that of self-crossed plants varied from 0 to 35.11%, with an average of 4.55%.
Phenotypic observations
First backcross generation (BC1)
Phenotypes of BC1 plants are shown in Fig. 3a–d. The phenotypes of interspecific hybrids could be divided into two forms: wild types with profound tillers and branches, and types with ear and grain morphologies closer to the wild type except of the plant stature that was comparatively closer to maize. The BC1 plants had lower tiller numbers and plant heights but a greater stem diameter and higher number of lateral branches than the parent (MTP). Each lateral branch was bearing 1–3 ears at the tip. 90% of BC1 plants were sterile and the average pollen viability was 1.86% with a maximum range of 37.5%. Coefficient of variation for analyzed traits was higher, depicting large variations among plant populations (Suppl. Table S1). The spikelet on cob was arranged opposite to each other and more than two-third of the seeds were covered with hard cupules (Fig. 3h). All the measured ear traits were nearer to maize than to the parent MTP (Suppl. Table S2).
Plant phenotypes of backcross progenies. a–d Plant phenotypes of BC1-11 (2n = 35), BC1-40 (2n = 45), BC1-16 (2n = 76) and BC1-6 (2n = 86), respectively. e–g Phenotypes of BC2-25-10 (2n = 22), BC2-40 family and BC2-40-1 (2n = 21), respectively. h, i Ears of BC1 and BC2 plants, respectively. Boxes in h and i show pollen viability
Second backcross generation (BC2)
Plant phenotypes of BC2 (Fig. 3e–g) were comparatively more similar to maize than to BC1 (Fig. 3e–g). Most of the plants had no tillers and lateral branches. Maximum number of tiller was reduced from 48 in BC1 to 7 in BC2 (Suppl. Table S1). The tassels of 51.78% BC2 plants blossom normally and 4.57% plants had only one tassel. The average pollen viability was 29.07% with a maximum range of 97% (CV > 1, Suppl. Table S1). Most of the seeds were without cupules and were larger than that of BC1. Average ear length, ear diameter, seed rows, and number of seeds/ear were higher in BC2 than in BC1 (Suppl. Table S2; Fig. 3i).
Analysis of variance among 15 families of BC2 showed highly significant differences in all agronomic traits (Suppl. Table S3). Among them, a family (BC2-10) had a significantly higher average plant height, stem diameter, male spike length and tassel branches, while the number of tillers and lateral branches was significantly lower. The comprehensive analysis of BC2 families showed that the BC2-10 has excellent aspects for breeding and found nearer to cultivated corn. Thus, it had excessive potential for further corn breeding.
Growth period of BC1 and BC2
Plant growth period variation among different individuals in a population represents the sensitivity of that individual for the photoperiod. Average growth period variations between BC1 and BC2 population are represented in Suppl. Table S4. Some of the plants in BC1 were very late although they started their anthesis in September thus showing a similar behavior as that of Z. perennis. Plants with minimum days to reach at tasselling, silking, and anthesis stages were also spending more number of days than that of normal maize inbred lines do. The BC1 plants could be divided into perennial and annual types. The perennial plants were much slower in completing their growth stages as compared to annuals and their vegetative and flowering stages were also longer than annuals. The perennial plants start to wither at the end of December and in early March of the second year; they started to sprout from plant stubbles. Some families of BC2 generation were earlier than both BC1 and Mo17. These results showed that with increasing generations of backcross, growth periods of progenies transform gradually into cultivated corn.
Cytogenetics of BC1 and BC2
Chromosome segregation in BC1
Root-tip mitotic metaphase cells of 30 plants of BC1 were observed for chromosome counts; chromosome and frequency of chromosome number distribution in BC1 are shown in Suppl. Fig. S1. The results revealed an irregular distribution of mitotic chromosomes. The highest chromosome distribution frequency was 43–48, which was found in 19 plants, 5 plants contained less than 40 chromosomes, while plants having chromosome number 54, 57, 74, 76, 84, and 86 were also observed (Fig. 4). The theoretical chromosome value of BC1 plants was 48 but this chromosome number was observed in only two plants. Among a group of 19 plants, 8 plants possessed 45, 3 possessed 44, and 3 possessed 46 chromosomes. The possible reason of this random distribution is that the MTP parent was a new synthetic allopolyploid that contained chromosomes of three parental species and might cause meiotic disorders, leading to the production of univalent and multivalent chromosomes. Moreover, a large range of female meiotic gametes possessed changed a chromosome number upon fertilization. The mitotic chromosomes of BC1 were further analyzed by mcGISH to validate species-specific chromosome distribution. On the basis of chromosome distribution, the BC1 population was categorized into four different classes: (1) BC n+ n1 contained 15–16 chromosomes from T. dactyloides, 9–11 from Z. perennis and 18–22 from maize, (2) BC 2 n1 (2n = 74 = 34Tr + 20Zp + 20Mz) developed directly without reduction of female gametes (apomictic), (3) BC 2 n+ n1 (2n = 86 = 36Tr + 20Zp + 30Mz) was formed by the fertilization of an unreduced MTP female gamete and normal male gamete of maize, (4) BC NR1 was an irregular type that contained the same distribution of T. dactyloides and Z. perennis chromosomes like BC n+ n1 , but the distribution of maize chromosomes was random (Suppl. Table S5; Fig. 4). Chromosome translocations between maize and Z. perennis, maize and T. dactyloides, and T. dactyloides and Z. perennis chromosomes were also detected (Fig. 4e–h; Suppl. Table S5).
Somatic chromosome constitutions in BC1,a–d Somatic chromosome number in BC1-30 (2n = 44), BC1-9 (2n = 45), BC1-26 (2n = 74) and BC1-6 (2n = 86), respectively. e–h Chromosome constitutions in mitotic cells of root tips of BC1-31, BC1-26, BC1-6, BC1-5, respectively. Blue, red, and green represent chromosomes from T. dactyloides, Z. perennis, and maize, respectively. The yellow arrows point to translocations between T. dactyloides and maize chromosomes, while white arrows show translocations between maize and Z. perennis chromosomes. Bar 10 μm
Meiotic behavior in pollen mother cells (PMCs) of BC1 plants
The plants containing 2n = 45 (2n = 45 = 15Tr + 11Zp + 19Mz) chromosomes were selected to analyze meiosis in pollen mother cells. A relatively lower chromosome pairing and many univalent and multivalent chromosomes were found at diakinesis and laggard chromosomes were also present. The laggard chromosome could be seen at both sides and away from equatorial plates, and they might not be able to attach to spindle fibers at metaphase I and II (Fig. 5b, e). A great number of laggard chromosomes was also present at anaphase I and II (Fig. 5c, f). These laggard chromosomes might cause an unequal distribution of chromosomes at both poles during telophase I and II (Fig. 5d, h). Some chromosomes could also be seen that remain outside the nuclear membrane after late telophase II (Fig. 5i). These free outlying chromosomes were probably lost, and could not pass to the next generation thus leading to a decrease in the number of chromosomes in next generations.
Chromosome distribution of BC2
The BC2 plants were divided into four groups on the basis of chromosome distribution pattern in BC1. The first group was grown from seeds of ten BC n+ n1 plants (2n = 44–46), the second group was a descendent of one plant of BC 2 n1 (2n = 76), the third group was produced by growing seeds of one plant of BC 2 n+ n1 (2n = 84) and the fourth was obtained from seeds of four plants from BC NR1 [two plants (2n = 37), one plant (2n = 54) and one plant (2n = 57)]. The “2n = 37” type seeds from two plants could give rise to only four weaker plants that could not provide good mitotic chromosome spreads. Only three plants were karyotyped from the BC BC12 n+ n2 group that showed the chromosome number 42, 47 and 49, respectively.
Chromosome distribution of BC BC1 n+ n2
A total of 64 plants from BC BC1 n+ n2 was karyotyped, in which the chromosome distribution range was 21–39 (Suppl. Fig. S2). Among them 42.19% plants contained 26–30 chromosomes, followed by 35.94% plants with 21–25 chromosomes, 21.87% plants with 30 chromosomes and one plant (1.56%) with 21 chromosomes (Suppl. Fig. S2). It could be observed that after two backcross generations, the MTP dropped its basic chromosome number below 30 and some plants returned to around 20 chromosomes. The mcGISH analysis of 19 BC BC1 n+ n2 plants confirmed that T. dactyloides chromosomes were in the range of 0–8, five plants (26.32%) completely lost T. dactyloides chromosomes, while nine plants (47.37%) contained only one T. dactyloides chromosome (Suppl. Table S6). The one plant of BC2 40-1 (2n = 21) containing one chromosome from T. dactyloides and 20 chromosomes from maize was an additional line of T. dactyloides—maize (Fig. 6a). Chromosome distribution range from Z. perennis genome was 0–6, while the most prevalent number was 3. It was found in 52.63% of the plants and one plant a complete loss of Z. perennis chromosomes (Suppl. Table S6), and maize chromosomes were in the range of 18–26 (Suppl. Table S6). Among 19 analyzed plants, a total of 17 plants (89.47%) possessed points of translocation. A total of 1–5 translocations was present in one plant genome with an average of 2.65 (Fig. 6a–f; Suppl. Table S6).
Chromosome constitutions of BC BC1 n+ n2 and BC BC12 n2 . a–f Chromosome constitutions in mitotic cells of root tips of BC2-40-1, BC2-25-12, BC2-40-5, BC2-40-6, BC2-25-19, BC2-25-12, respectively, which belong to BC BC1 n+ n2 . g–l GISH results of BC2-16-14, BC2-16-8, BC2-16-1, BC2-16-18, BC2-16-23 and BC2-16-31, respectively, from the BC BC12 n2 group. The yellow arrow shows translocation between T. dactyloides and maize, while the white arrows show translocation between maize and Z. perennis. Bar 10 μm
Chromosome distribution of BC BC12 n2
Mitotic chromosome observations of 21 plants revealed the presence of 39–81 chromosomes, while the most abundant range was 39–45 found in 57.14% plants, followed by 67–81 chromosomes in 42.86% of plants (Suppl. Fig. S3). Both groups of BC2 showed the tendency of chromosome subtraction compared to BC1. The chromosome number and distribution in BC 2 n1 were similar to MTP. Therefore, their backcross progenies should possess the same pattern of chromosome distribution, yet results were dissimilar. BC1 had a wider range of chromosome distribution and 2n = 45 plants were more abundant, while BC2 showed a narrow range of chromosome distribution and 2n = 41 plants were most prevalent. The plants of BC BC12 n2 were being divided into two main groups: first, plants containing 2n = 39–45 and second, plants containing 2n = 67–81; the former was developed by normal reduction of female gametes while the latter was developed by fertilization of unreduced female gametes (apomictic reproduction). The mcGISH results showed that the chromosomal distribution among non-apomictic plants in T. dactyloides, Z. perennis, and maize chromosomes was 11–15, 7–12, and 17–22, respectively, while the chromosomal distribution among the apomictic plant group of T. dactyloides, Z. perennis, and maize chromosomes was 29–32, 15–20, and 19–30, respectively (Suppl. Table S7). A total of 17 analyzed plants contained eight translocations; six between Z. perennis and maize and two between T. dactyloides and Z. perennis (Fig. 6g, h, l; Suppl. Table S7).
Chromosome distribution of BC BC1NR2
A root-tip mitotic analysis of 21 plants’ chromosomes of BC BC1NR2 group displayed a range of 30–41 chromosomes (Suppl. Fig. S4). The most prevalent range was 33 and 35 that accounted for 38.10% of plant population. The mcGISH results showed that T. dactyloides, Z. perennis, and maize chromosomes were in the range of 0–6, 5–9, and 21–24, respectively, and 25% plants showed translocation between Z. perennis and maize.
Transfer of homoeologous chromosomes in backcross progeny and translocations frequency
The chromosome loss rate in BC1 plants of T. dactyloides and Z. perennis was 48.08% and 42.69%, respectively, while that of T. dactyloides and Z. perennis in BC BC12 n2 was 39.87% and 32.06, respectively. The chromosome loss rate was relatively lower in BC BC12 n2 . It might be due to the fact that with increasing backcross generation, the non-homologous chromosomes gradually coordinate in one nucleus and chromosomes with lower homologies reduced in each backcross generation. The chromosome loss rate for T. dactyloides and Z. perennis in BC BC1 n+ n2 was 94.59% and 83.95%, respectively, which was much higher than in the other three breeding routes. Therefore, it is a fast route to recover maize and probably the best way to select addition or substitution materials in early generations (Suppl. Table S8). The loss of T. dactyloides chromosomes was higher than of Z. perennis in all analyzed plants.
Three kinds of translocations were observed which were between maize and Z. perennis (Mz–Zp), maize and T. dactyloides (Mz–Tr) and T. dactyloides and Z. perennis (Tr–Zp) at the rate of 91.51%, 4.72%, and 3.77%, respectively (Suppl. Table S9). The translocation frequency between maize and Z. perennis was the highest and genetic exchange of T. dactyloides with both species was the lowest (Suppl. Table S9). The chromosome translocation frequency of Mz–Zp type was higher in BC BC1 n+ n2 and BC BC1NR2 groups. Therefore, it would be the best route to obtain Mz–Zp translocation and to get rid of exogenous chromosomes speedily. The Mz–Tr translocations were higher in BC BC12 n2 group; thus, it is the better pathway to obtain translocations in between T. dactyloides and maize. On the basis of these findings we presented a schematic diagram for recovering maize by different breeding pathways (Fig. 7).
Simplified diagram of some of the pathways to recover maize from MTP × Mo17. The different shapes represent the range of chromosome numbers, where black circles, 2n + n pathway; black and white circles, 2n (apomixes) pathway; black circle and black oval, n + n pathway; black and white ovals, irregular pathways; white square, recovered maize. On the right side of the dotted line is the predicted path
Discussion
The synthesis of allopolyploid bridge species for genetic transfer
Distant hybridization is considered to be a promising breeding route for crop improvement (Belanger et al. 2003). By wide hybridization, the boundaries between cultivars are broken and the scope of genetic recombination is broadened. However, the reproductive isolation between species makes it difficult to achieve distant hybridization (Cheng et al. 2016). Allopolyploids can overcome the sterility of distant hybrids (Miller and Venable 2000). In previous studies, allopolyploids bridge plants have been used to overcome such barriers (Eubanks 2006). The T. dactyloides genome has a partial homology with the maize genome (Maguire 1962; Kindiger et al. 1995) and Z. perennis is also distinctly related to maize; therefore, direct gene flow between them is difficult (Fu et al. 2015; Cheng et al. 2016). MTP is a female fertile hybrid, can produce many materials with varying genetic constitutions by backcrossing, which can be utilized in improving maize. In addition, MTP enables the genomes of the three species to cohabitate in one cell. Thus, this way provides an opportunity for genomic exchange and recombination in between the three species.
Modes of reproductions and pathways to recover maize
It is important to understand genetic relationships and reproductive systems of target plants to design and control a breeding experiment, especially to regulate exogenous chromosomes and genetic recombination. MTP exhibited four types of reproductive systems: (1) 2n + n type, that was developed by fertilization of an unreduced gamete of MTP with normal maize gamete, (2) 2n type, developed directly by apomixes, (3) n + n type, that was formed by a normal chromosome reduction process during meiosis and (4), an irregular type that was produced by fertilization of the normal haploid set from MTP and a different chromosome number from maize. The T. dactyloides (one of MTP parent) was an apomictic parent in which diploid genotypes reproduce sexually, while apomixes is expressed in polyploidy lineages (Burson et al. 1990; Grimanelli et al. 1998). The varying number of chromosomes in irregular types might be due to meiotic disorders at various stages. Therefore, it is speculated that irregular chromosome production might be due to the presence of more univalent and multivalent chromosomes at terminals during meiosis. Such meiotic disorders cause imbalance of chromosome segregation in gametes, but further meiotic studies are required to validate this hypothesis. The Z. perennis never exhibit the apomictic mode of reproduction. Therefore, 2n + n and 2n reproductive modes were inherited from T. dactyloides, but expression frequency (13.33%) of apomixes was much lower than in T. dactyloides itself. That might be due to the fact that the inheritance of apomictic expression deteriorated as much as the proportion of maize chromosome increased (Kindiger et al. 1996b).
Transfer of exogenous chromosomes or chromosomal segments in progeny material
The transfer of beneficial genes from distinct genomes has greatly improved agricultural crops (Tang et al. 2014; Kang et al. 2016). However, introduction of a whole chromosome or whole set of exogenous chromosomes in cultivating species may break the genetic balance of receptors along with the induction of many undesirable traits (Kole 2011). Therefore, choice of small fragments (translocations) that carry the target genes is the best way to transfer interesting genes into target species. The translocations generally have a better genetic coordination and stability than the additional and substitution lines (Qi et al. 2007). In the current study, 69.23% plants of BC1 depicted chromosomal translocation and 89.47% plants of BC BC1 n+ n2 also displayed chromosomal translocation. The frequency of translocations in BC1 was higher than in the female parent MTP, which might be due to the fact that loss of chromosomes with relatively lower homology increases the overall proportion of chromosomes with higher homologies in BC1. The higher percentage of chromosomal translocations in BC2 than in BC1 might be due to a gradual integration of parental chromosomes into the nucleus. A relatively higher Mz–ZP translocation rate (91.51%) revealed the closer genetic relationship between maize and Z. perennis (Takahashi et al. 1999; González et al. 2006) and lower translocation frequency between Tr–Zp than Tr–Mz suggested a higher genetic distance between T. dactyloides and Z. perennis genome compared to T. dactyloides and maize genomes.
Impacts of exogenous genetic material on plant phenotypes
The expression of exogenous genes in recipient species is of particular interest. The MTP showed four reproductive modes. Therefore, the number and the composition of chromosomes were different, which causes differences in genetic information and gene expressions among offspring leading to the phenotypic variations. Plants having chromosome numbers around 2n = 50 in BC1 produced fewer tillers, thicker stems and wider leaves because of the increased proportion of maize chromosomes in the cells. However, plants having chromosomes around/below 2n = 30 in BC2 showing some wild-type traits might result from the presence of a large proportion of foreign genes in the cells. Compared with the female parent MTP, the BC BC12 n2 had fewer tillers and leaves, lower plant height and thicker stems, although chromosomal compositions of both were the same but phenotypes were different. The possible explanation is that three parental genomes in the same nucleus might undergo multiple changes, such as repeated sequence loss, DNA methylation and gene silencing that alters gene-expression landscapes (Chen 2007; Gaeta et al. 2007; Matsushita et al. 2012). By backcrossing with maize, the proportion of exogenous chromosomes in progeny of plant cells decreased and the proportion of maize chromosomes increased that converted he vast majority of BC2 plants into cultivated corn-type, i.e., plants were without tillers, stout stalks, increased ear rows, and exposed grains.
Prospects of future generations in genetic research and breeding
Polyploidy and distinct hybridization are some of the important driving forces for plant evolution (Masterson 1994). Important crops such as wheat, rapeseed, cotton, and potato are polyploids. Crops that are considered diploid such as corn, soybean, and cabbage have also undergone polyploidization during evolutionary history, hence, they are paleoploids (Shoemaker et al. 1996; Lagercrantz and Lydiate 1996; Doebley et al. 1997). These natural polyploids underwent long-run chromosome evolution and a genomic diploidization process, during which well-adapted lineage was naturally selected. The newly synthesized allopolyploids are generally associated with genomic instabilities at early stages that cause variation in gene structure, organization, and expressions (Kashkush et al. 2002; He et al. 2003). We synthesized allopolyploids with different genetic compositions with clear genetic relationships. These allopolyploids would be suitable to explore the evolution of allopolyploids in early generation since most of available allopolyploids are primitive and are difficult to track evolutionary.
One of the most important roles of this allopolyploid pathway is to direct a barrier-free interspecies gene flow to get access to more genetic variations and exogenous gene utilization. A series of materials with different genetic constitutions was synthesized by backcrossing of allopolyploid bridge plants with maize. The BC2 had a chromosome composition of 2n = 1Tr + 20Mz, that was a corn—T. dactyloides addition line. There were also four substitution lines containing only three Z. perennis chromosomes, hence, with increasing progeny population, a greater number of T. dactyloides—maize and/or maize—Z. perennis addition or substitution lines can be obtained. Additionally, these materials are suitable for utilizing in crop breeding, gene mapping, comparative genomic research, and construction of exogenous DNA libraries. BC1 and BC2 generations had basic groups of introgressed lines. These introgressed lines may contain excellent beneficial genes that could be effective for maize improvement. Although the phenotypes of these plants are far away from the inbred lines of maize and although it is difficult to directly breed them for high-yielding grain hybrids, the route can be used as an intermediate material for new forage maize breeding.
Author contribution statement
MZI did experiment and wrote the manuscript. MC, YZ, YS, YL, WJ, HL, XW, LZ, assisted with data collection and analysis, and contributed to the manuscript. AA contributed to the manuscript writing. TR reviewed the manuscript and QT funded and supervised each stage of the project. All authors read and approved the final manuscript.
Abbreviations
- mcGISH:
-
Multi-color Genomic in Situ-Hybridization
- MTP:
-
Maize, T. dactyloides, and Z. perennis hybrid
- Mz:
-
Maize, Zea mays L. ssp. mays
- Zp:
-
Zea perennis
- Tr:
-
Tripsacum dactyloides L
- Chr:
-
Chromosomes
References
Belanger FC, Plumley K, Day P, Meyer WA (2003) Interspecific hybridization as a potential method for improvement of species. Crop Sci 43(6):2172–2176. https://doi.org/10.2135/cropsci2003.2172
Belova I, Tarakanova TK, Abdyrahmanova E, Sokolov VA, Panikhin PA (2010) Chromosome control of apomixis in maize-gamagrass hybrids. Russ J Genet 46(9):1055–1057. https://doi.org/10.1134/S1022795410090103
Burson BL, Voigt PW, Sherman RA, Dewald CL (1990) Apomixis and sexuality in eastern gamagrass. Crop Sci 30(1):86–89. https://doi.org/10.2135/cropsci1990.0011183X003000010020x
Chen ZJ (2007) Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58(1):377–406. https://doi.org/10.1146/annurev.arplant.58.032806.103835
Cheng M, Zheng M, Yang S, Li Y, Dong X, Li J, Sun R, Li H, Zhou S, Wu Y (2016) The effect of different genome and cytoplasm on meiotic pairing in maize newly synthetic polyploids. Euphytica 207(3):593–603. https://doi.org/10.1007/s10681-015-1552-7
Comai L (2005) The advantages and disadvantages of being polyploid. Nat Rev Genet 6(11):836–846. https://doi.org/10.1038/nrg1711
Doebley J (1990) Molecular evidence for gene flow among Zea species. BioScience 40(6):443–448. https://doi.org/10.2307/1311391
Doebley JF, Iltis HH (1980) Taxonomy of Zea (Gramineae). I. A subgeneric classification with key to taxa. Am J Bot. https://doi.org/10.2307/2442441
Doebley J, Stec AO, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386(6624):485–488. https://doi.org/10.1038/386485a0
Duvick DN (1984) Genetic diversity in major farm crops on the farm and in reserve. Econ Bot 38:161–178. https://doi.org/10.1007/BF02858829
Emerson RA, Beadle GW (1930) A fertile tetraploid hybrid between Euchlaena perennis and Zea mays. Am Naturalist 64:190–192. https://doi.org/10.1086/280311
Eubanks MW (2006) A genetic bridge to utilize Tripsacum germplasm in maize improvement. Maydica 51(2):315–327
Farquharson LI (1955) Apomixis and polyembryony in Tripsacum dactyloides. Am J Bot 42(8):737. https://doi.org/10.1002/j.1537-2197.1955.tb10416.x
Fu J, Yang XY, Cheng MJ, Lü GH, Wang P, Wu YQ, Zheng MM, Zhou SF, Rong TZ, Tang QL (2015) Perennial aneuploidy as a potential material for gene introgression between maize and Zea perennis. J Integr Agr 14(5):839–846. https://doi.org/10.1016/S2095-3119(14)60874-1
Gaeta RT, Pires JC, Iniguezluy FL, Leon EJ, Osborn TC (2007) Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19(11):3403–3417. https://doi.org/10.1105/tpc.107.054346
Galinat WC (1973) Intergenomic mapping of maize, teosinte and Tripsacum. Evolution 27(4):644–655. https://doi.org/10.1111/j.1558-5646.1973.tb00712.x
González G, Comas C, Confalonieri V, Naranjo C, Poggio L (2006) Genomic affinities between maize and Zea perennis using classical and molecular cytogenetic methods (GISH–FISH). Chromosome Res 14(6):629–635. https://doi.org/10.1007/s10577-006-1072-3
Grimanelli D, Leblanc O, Espinosa E, Perotti E, De Leon DG, Savidan Y (1998) Mapping diplosporous apomixis in tetraploid Tripsacum: one gene or several genes? Heredity 80(1):33–39. https://doi.org/10.1046/j.1365-2540.1998.00263.x
Hajjar R, Hodgkin T (2007) The use of wild relatives in crop improvement: a survey of developments over the last 20 years. Euphytica 156(1–2):1–13. https://doi.org/10.1007/s10681-007-9363-0
Hancock WG, Lewis RS (2017) Heterosis, transmission genetics, and selection for increased growth rate in a N. tabacum × synthetic tobacco cross. Mol Breeding 37(4):53
Hardin B (1994) Eastern gamagrass: corn’s comeback cousin. Agr Res 42(4):12
He P, Friebe B, Gill BS, Zhou J (2003) Allopolyploidy alters gene expression in the highly stable hexaploid wheat. Plant Mol Biol 52(2):401–414. https://doi.org/10.1023/A:1023965400532
Hussain SW, Williams WM (1997) Development of a fertile genetic bridge between Trifolium ambiguum M. Bieb. and T. repens L. Theor Appl Genet 95(4):678–690. https://doi.org/10.1007/s001220050612
Jiang ZL, Wu BH, Wang ZZ, Hu JL, Yuan J, Chen HL, Liu J, Zheng YL, Liu DC (2017) Enriching novel Glu-Ax alleles and significantly strengthening gluten properties of common wheat through wide hybridization with wild emmer. J Cereal Sci. https://doi.org/10.1016/j.jcs.2017.04.018
Kang HY, Zhang ZJ, Xu LL, Qi WL, Tang Y, Wang H, Zhu W, Li DY, Zeng J, Wang Y (2016) Characterization of wheat—Psathyrostachys huashanica small segment translocation line with enhanced kernels per spike and stripe rust resistance. Genome 59(4):221–229. https://doi.org/10.1139/gen-2015-0138
Kashkush K, Feldman M, Levy AA (2002) Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160(4):1651–1659
Kindiger B, Beckett JB (1990) Cytological evidence supporting a procedure for directing and enhancing pairing between maize and Tripsacum. Genome 33(4):495–500. https://doi.org/10.1139/g90-073
Kindiger B, Blakey C, Dewald C (1995) Sex reversal in maize x Tripsacum hybrids. Allelic non-complementation of ts2 and gsf1. Maydica (Italy)
Kindiger B, Bai D, Sokolov V (1996a) Assignment of a gene (s) conferring apomixis in Tripsacum to a chromosome arm: cytological and molecular evidence. Genome 39(6):1133–1141. https://doi.org/10.1139/g96-143
Kindiger B, Sokolov V, Khatypova IV (1996b) Evaluation of apomictic reproduction in a set of 39 chromosome maize—Tripsacum backcross hybrids. Crop Sci 36(5):1108–1113. https://doi.org/10.2135/cropsci1996.0011183X003600050006x
Kole C (2011) Wild crop relatives: genomic and breeding resources: Cereals, vol 1. Springer. https://link.springer.com/book/10.1007/978-3-642-14228-4
Lagercrantz U, Lydiate DJ (1996) Comparative genome mapping in Brassica. Genetics 144(4):1903–1910
Lamb JC, Birchler JA (2006) Retroelement genome painting: cytological visualization of retroelement expansions in the genera Zea and Tripsacum. Genetics 173(2):1007–1021. https://doi.org/10.1534/genetics.105.053165
Leblanc O, Grimanelli D, Hernandez-Rodriguez M, Galindo PA, Soriano-Martinez AM, Perotti E (2009) Seed development and inheritance studies in apomictic maize-Tripsacum hybrids reveal barriers for the transfer of apomixis into sexual crops. Int J Dev Biol 53(4):585. https://doi.org/10.1387/ijdb.082813ol
Li Q, Zhou Q, Mei J, Zhang Y, Li J, Li Z, Ge X, Xiong Z, Huang Y, Qian W (2014) Improvement of Brassica napus via interspecific hybridization between B. napus and B. oleracea. Mol Breeding 34(4):1955–1963. https://doi.org/10.1007/s11032-014-0153-9
Maazou A-RS, Qiu J, Mu J, Liu Z (2017) Utilization of wild relatives for maize (Zea mays L.) improvement. Afr J Plant Sci 11(5):105–113. https://doi.org/10.5897/AJPS2017.1521
Maguire MP (1962) Common loci in corn and Tripsacum. J Hered 53(2):87–88. https://doi.org/10.1093/oxfordjournals.jhered.a107130
Masterson J (1994) Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264(5157):421–423. https://doi.org/10.1126/science.264.5157.421
Matsushita SC, Tyagi AP, Thornton GM, Pires JC, Madlung A (2012) Allopolyploidization lays the foundation for evolution of distinct populations: evidence from analysis of synthetic Arabidopsis allohexaploids. Genetics 191(2):535–547. https://doi.org/10.1534/genetics.112.139295
Miller JS, Venable DL (2000) Polyploidy and the evolution of gender dimorphism in plants. Science 289(5488):2335–2338. https://doi.org/10.1126/science.289.5488.2335
Molina MDC, Garcia M, Chorzempa SE (2006) Meiotic study of Zea mays ssp. mays (2n = 40) x Tripsacum dactyloides (2 n = 72) hybrid and its progeny. Electron J Biotechnol. https://doi.org/10.4067/S0717-34582006000300020
Poggio L, Confalonieri V, Comas C, Cuadrado A, Jouve N, Naranjo CA (1999) Genomic in situ hybridization (GISH) of Tripsacum dactyloides and Zea mays ssp. mays with B chromosomes. Genome 42(4):687–691. https://doi.org/10.1139/g98-157
Qi LL, Friebe B, Zhang P, Gill BS (2007) Homoeologous recombination, chromosome engineering and crop improvement. Chromosome Res 15(1):3–19. https://doi.org/10.1007/s10577-006-1108-8
Ramos JM, Furuta T, Uehara K, Chihiro N, Angeles-Shim RB, Shim J, Brar DS, Ashikari M, Jena KK (2016) Development of chromosome segment substitution lines (CSSLs) of Oryza longistaminata. Euphytica 210(2):151–163. https://doi.org/10.1007/s10681-016-1685-3
Reagon M, Snow AA (2006) Cultivated Helianthus annuus (Asteraceae) volunteers as a genetic “bridge” to weedy sunflower populations in North America. Am J Bot 93(1):127–133. https://doi.org/10.3732/ajb.93.1.127
Schnable JC, Springer NM, Freeling M (2011) Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci USA 108(10):4069–4074. https://doi.org/10.1073/pnas.1101368108
Shakiba E, Eizenga GC (2014) Unraveling the secrets of rice wild species. In: Rice germplasm, genetics and improvement. InTech, pp 1–58. https://doi.org/10.5772/58393
Shaver D (1964) Perennialism in Zea. Genetics 50(3):393
Shoemaker RC, Polzin K, Labate J, Specht JE, Brummer EC, Olson T, Young ND, Concibido V, Wilcox J, Tamulonis JP (1996) Genome duplication in soybean (Glycine subgenus soja ). Genetics 144(1):329–338
Stebbins GL (1971) Chromosomal evolution in higher plants. Addison-Wesley, London
Takahashi C, Marshall J, Bennett M, Leitch I (1999) Genomic relationships between maize and its wild relatives. Genome 42(6):1201–1207. https://doi.org/10.1139/g99-074
Tang Q, Rong T, Song Y, Yang J, Pan G, Li W, Huang Y, Cao M (2005) Introgression of perennial teosinte genome into maize and identification of genomic in situ hybridization and microsatellite markers. Crop Sci 45(2):717–721. https://doi.org/10.2135/cropsci2005.0717
Tang X, Shi D, Xu J, Li Y, Li W, Ren Z, Fu T (2014) Molecular cytogenetic characteristics of a translocation line between common wheat and Thinopyrum intermedium with resistance to powdery mildew. Euphytica 197(2):201–210. https://doi.org/10.1007/s10681-013-1059-z
Acknowledgements
This work was supported by the National Program on Key Basic Research Project of China (973 Program, 2014CB138705), the National Natural Science Foundation of China (31371640) and Major Science and Technology Projects of Sichuan Province (2018).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Suppl. Fig. S1
Frequency of chromosome distribution in BC1 (DOCX 48 kb)
Suppl. Fig. S2
Frequency of chromosome distribution in BC BC1 n+ n2 (DOCX 44 kb)
Suppl. Fig. S3
Frequency of chromosome distribution in BC BC12 n2 (DOCX 36 kb)
Suppl. Fig. S4
Frequency of chromosome distribution in BC BC1NR2 (DOCX 38 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Iqbal, M.Z., Cheng, M., Su, Y. et al. Allopolyploidization facilitates gene flow and speciation among corn, Zea perennis and Tripsacum dactyloides. Planta 249, 1949–1962 (2019). https://doi.org/10.1007/s00425-019-03136-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-019-03136-z