Abstract
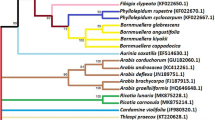
To investigate the phylogenetic relationships of Salvia euphratica sensu lato and its closely related species with a focus on the section Hymenosphace, we screened five different regions; one nuclear ribosomal DNA region (Internal Transcribed Spacer, ITS) and four chloroplast DNA regions [trnT-trnL intergenic spacer (IGS), trnL intron, trnL-trnF IGS and trnV intron]. Based on 19 sequences of 7 Salvia taxa produced in the study and different number of sequences obtained from GenBank, our results supported latest taxonomic treatments on Salvia pseudeuphratica and S. cerino-pruinosa as they are resurrected and accepted different species from S. euphratica. The results confirmed the latest phylogenetic findings as “the section Hymenosphace is a non-monophyletic group, originated thick textured, non-expanding ancestral group, and expanding calyces with widely diverging lips in fruiting stage evolved several times in parallel, not only in Salvia but also in the Iranian genus Zhumeria”. The species of the sect. Hymenosphace are mostly distributed in three different geographic regions [(1) Southwest Asia, Turkey, Russia and Iran, (2) Canary Islands, (3) Southern Africa] with different morphological characters. The results showed that ITS had the highest resolution power for discriminating studied taxa and the highest number of haplotypes was also observed in this region. The resolutions of the chloroplast regions were too low for taxa native to Turkey, but quite enough to discriminate species from the different clades whose sequences were obtained from database.



Similar content being viewed by others
References
Alziar G (1988–1993) Catalogue synonymique des Salvia L. du monde (Lamiaceae). I à VI. Biocosme Mésogéen, Nice 5:87–136; 6:79–115; 6: 163–204; 7:59–109; 9:413–497; 10:33–117
Artyukova EV, Gontcharov AA, Kozyrenko MM, Reunova GD, Zhuravlev YN (2005) Phylogenetic relationships of the far Eastern Araliaceae inferred from ITS sequences of nuclear rDNA. Russ J Genet 41:649–658
Baldwin BG (1992) Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the Compositae. Molec Phylogen Evol 1:3–16
Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ (1995) The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann Missouri Bot Gard 82:247–277
Behçet L, Avlamaz D (2009) A new record for Turkey: Salvia aristata Aucher ex Benth. (Lamiaceae). Turkish J Bot 33:61–63
Bentham G (1832–1836) Salvia. In: Bentham G (eds) Labiatarum genera et species: or, a description of the genera and species of plants of the order Labiatae with their general history, characters, affinities, and geographical distribution, J Ridgway and Sons, London, pp 190–312
Bentham G (1836) Salvia. Ann Sci Nat, Bot 2:40
Bentham G (1848) Labiatae. In: De Candolle A (ed) Prodromus systematis naturalis regni vegetabilis 12. Treuttel and Würtz, Paris, pp 262–358
Briquet J (1897) Labiatae. In: Engler A, Prantl K (eds) Die natürlichen Pflanzenfamilien nebst ihrer Gattungen und wichtigen Arten. Engelmann, Leipzig, pp 183–380
Celep F, Doğan M (2010) Salvia ekimiana (Lamiaceae), a new species from Turkey. Ann Bot Fenn 47:63–66
Celep F, Doğan M, Bagherpour S, Kahraman A (2009) A new variety of Salvia sericeotomentosa (Lamiaceae) from South Anatolia, Turkey. Novon 19:432–435
Celep F, Doğan M, Kahraman A (2010) Re-evaluated conservation status of Salvia L. (sage) in Turkey I: the Mediterranean and the Aegean geographic regions. Turkish J Bot 34:201–214
Celep F, Kahraman A, Doğan M (2011a) A new taxon of the genus Salvia L. (Lamiaceae) from Turkey. Pl Ecol Evol 144:111–114
Celep F, Kahraman A, Doğan M (2011b) Taxonomic notes for Salvia aucheri (Lamiaceae) from Southern Anatolia. Turkey Novon 21:34–35
Celep F, Kahraman A, Atalay Z, Doğan M (2014) Morphology, anatomy, palynology, mericarp and trichome micromorphology of the rediscovered Turkish endemic Salvia quezelii (Lamiaceae) and their taxonomic implications. Pl Syst Evol 300:1945–1958
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Molec Ecol 9:1657–1659
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gao T, Yao H, Song JY, Zhu YJ, Liu C, Chen SL (2010) Evaluating the feasibility of using candidate DNA barcodes in discriminating species of the large Asteraceae family. BMC Evol Biol 10:324
Harley RM, Atkins S, Budantsey AL, Cantino PD, Conn BJ, Grayer R, de Kok R, Krestovskaja T, Morales R, Paton AJ, Ryding O, Upson T (2004) Labiatae. In: Kadereit JW (ed) The families and genera of vascular plants, vol 7. Springer, Berlin Heidelberg, pp 167–275
Hedge IC (1965) Studies in the Flora of Afghanistan III: an account of Salvia. Notes RBG Edinburgh 26:408
Hedge IC (1972) Salvia L. In: Tutin TG, Heywood VH, Burges NA, Valentine DH, Walters SM, Webb DA (eds) Flora Europaea 3. Cambridge University Press, Cambridge, pp 188–192
Hedge IC (1974) A revision of Salvia in Africa including Madagascar and the Canary Islands. Notes Roy Bot Gard Edinburgh 33:1–121
Hedge IC (1982) In: Davis PH (ed), Flora of Turkey and the East Aegean Islands: Salvia L., vol 7. University of Edinburgh Press, Edinburgh
Hedge IC (1982) In: Rechinger KH (ed), Flora Iranica: Salvia L., Akademische Druck und Verlagsanstalt, Graz, 150:403–476
Hsiao C, Chatterton NJ, Asay KH, Jensen KB (1995) Phylogenetic relationships of the monogenomic species of the wheat tribe, Triticeae (Poaceae), inferred from nuclear rDNA (internal transcribed spacer) sequences. Genome 38:221–223
Kahraman A, Celep F, Doğan M (2009) Morphology, anatomy and palynology of Salvia indica L. (Labiatae). World Appl Sci J 6:289–296
Kahraman A, Celep F, Dogan M, Bagherpour S (2010a) A taxonomic revision of Salvia euphratica sensu lato and its closely related species (sect. Hymenosphace, Lamiaceae) by using multivariate analysis. Turkish J Bot 34:261–276
Kahraman A, Celep F, Dogan M (2010b) Anatomy, trichome morphology and palynology of Salvia chrysophylla Stapf (Lamiaceae). S African J Bot 76:187–195
Kahraman A, Celep F, Dogan M (2010c) Morphology, anatomy, palynology and nutlet micromorphology of Salvia macrochlamys (Labiatae) in Turkey. Biologia (Bratislava) 65:219–227
Kahraman A, Celep F, Doğan M, Guerin GR, Bagherpour S (2011) Mericarp morphology and its systematic implications for the genus Salvia L. Section Hymenosphace Benth. (Lamiaceae) in Turkey. Pl Syst Evol 292:33–39
Lahaye R, Savolainen V, Barraclough TG, Duthoit S, Maurin O, Gigot G, Pupulin F, Warner-Pineda J, Bogarín D, van der Bank M, ed. Janzen, Daniel H (2008) DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci USA 150:2923–2928
Martin E, Çetin Ö, Kahraman A, Celep F, Doğan M (2011) A cytomorphological study in some taxa of the genus Salvia L. (Lamiaceae). Caryologia 64:272–287
Metcalfe CR, Chalk L (1950) Anatomy of the Dicotyledons, vol I. Oxford University Press, Oxford
Metcalfe CR, Chalk L (1972) Anatomy of the Dicotyledons, vol II. Oxford University Press, Oxford
Nakipoğlu M (1993) Karyological studies on Salvia L. species of Turkey I. Salvia fruticosa Miller, Salvia tometosa Miller, Salvia officinalis L., Salvia smyrnaea Boiss. (Lamiaceae). Turkish J Bot 17:21–25
Pleines T, Jakob SS, Blattnerl FR (2009) Application of non coding DNA regions in intraspecific analyses. Pl Syst Evol 282:281–294
Pobedimova EG (1954) In: Schischkin BK (ed) Flora of the USSR: Salvia L. vol. 21:178–260. Translated from Russian, Israel Prog Sci Transl, Jerusalem
Posada D, Crandall KA (2001) Intraspecific gene genealogies: trees grafting into networks. Trends Ecol Evol 16:37–45
Reales A, Riviera D, Palazon JA, Obon C (2004) Numerical taxonomy study of Salvia sect. Salvia L. (Labiatae). Bot J Linn Soc 145:353–371
Rechinger KH (1952) Labiatae Novae Orientalis. Öst Bot Zeitschr 99:48–52
Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Amer J Bot 92:142–166
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of the three non-coding regions of chloroplast DNA. Pl Mol Biol 17:1105–1109
Takano A, Okada H (2011) Phylogenetic relationships among subgenera, species, and varieties of Japanese Salvia L. (Lamiaceae). J Pl Res 124:245–252
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molec Biol Evol 10:512–526
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 101:11030–11035
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA 5: molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molec Biol Evol 28:2731–2739
Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132:619–633
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Walker JB, Sytsma KJ (2007) Staminal Evolution in the Genus Salvia (Lamiaceae): molecular Phylogenetic Evidence for Multiple Origins of the Staminal Lever. Ann Bot (Oxford) 100:375–391
Walker JB, Sytsma KJ, Treutlein J, Wink M (2004) Salvia (Lamiaceae) is not monophyletic: implications for the systematic, radiation, and ecological specializations of Salvia and tribe Mentheae. Amer J Bot 91:1115–1125
Wang XR, Tsumura Y, Yoshimaru H, Nagasaka K, Szmidt AE (1999) Phylogenetic relationships of Eurasian pines (Pinus, Pinaceae) based on chloroplast rbcL, matK, rpl20-rps18 spacer, and tnV intron sequences. Amer J Bot 86:1742–1753
Wang M, Zhao HX, Wang L, Wang T, Yang RW, Wang XL, Zhou YH, Ding CB, Zhang L (2013) Potential use of DNA barcoding for the identification of Salvia based on cpDNA and nrDNA sequences. Gene 528:206–215
Will M, Claßen-Bockhoff R (2014) Why Africa matters: evolution of Old World Salvia (Lamiaceae) in Africa. Ann Bot 114:61–83
Wojciechowski MF (2005) Astragalus (Fabaceae): a molecular phylogenetic perspective. Brittonia 57:382–396
Wojciechowski MF, Sanderson MJ, Baldwin BG, Donoghue MJ (1993) Monophyly of aneuploid Astragalus (Fabaceae): evidence from nuclear ribosomal DNA internal transcribed spacer sequences. Amer J Bot 80:711–722
Xu H, Wang ZT, Cheng KT, Wu T, Gu LH, Hu ZB (2009) Comparison of rDNA ITS sequences and tanshinones between Salvia miltiorrhiza populations and Salvia species. Bot Stud (Taipei) 50:127–136
Yao H, Song J, Liu C, Luo K, Han J, Li Y, Pang X, Xu H, Zhu Y, Xiao P, Chen S (2010) Use of ITS2 Region as the Universal DNA Barcode for Plants and Animals. PLoS ONE 5:e13102. doi:10.1371/journal.pone.0013102
Zhang Y, Wang Y, Li D (2008) Analysis of ITS sequences of some medicinal plants and their related species in Salvia. Acta Pharm Sin 42:1309–1313
Acknowledgments
This study was financially supported in part by Yuzuncu Yil University (Scientific Research Project Foundation, 2013-FEN-B039), Van, Turkey and TUBITAK (Project Number: 104 T 450). Laboratory studies were carried out in Department of Biological Sciences, Middle East Technical University and Department of Molecular Biology and Genetics, Yüzüncü Yıl University. Comments by Prof. Dr. Pablo Vargas (Madrid, Spain) and an anonymous reviewer improved the manuscript.
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Pablo Vargas.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dizkirici, A., Celep, F., Kansu, C. et al. A molecular phylogeny of Salvia euphratica sensu lato (Salvia L., Lamiaceae) and its closely related species with a focus on the section Hymenosphace . Plant Syst Evol 301, 2313–2323 (2015). https://doi.org/10.1007/s00606-015-1230-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-015-1230-1