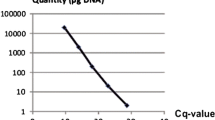
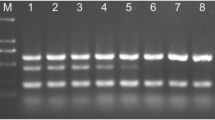
Abstract
Five primer/probe sets to identify the tomato wilt pathogen, Fusarium oxysporum f. sp. lycopersici (FOL), and its three races selectively were designed based on the rDNA-intergenic spacer and avirulence genes. Real-time PCR using genomic DNA from mycelia and soil DNA with the primer/probe sets allowed the successful identification of FOL and its races.

Similar content being viewed by others
References
Agrios GN (2005) Genetics of virulence in pathogens and of resistance in host plants. In: Agrios GN (ed) Plant pathology, 5th edn. Academic Press, New York, pp 139–161
Alexander LJ, Tucker CM (1945) Physiologic specialization in the tomato wilt fungus Fusarium oxysporum f. sp. lycopersici. J Agric Res 70:303–313
Arie T, Christiansen SK, Yoder OC, Turgeon BG (1997) Efficient cloning of ascomycete mating type genes by PCR amplification of the conserved MAT HMG box. Fungal Genet Biol 21:118–130
Balogun OS, Hirano Y, Teraoka T, Arie T (2008) PCR-based analysis of disease in tomato singly or mixed inoculated with Fusarium oxysporum f. sp. lycopersici races 1 and 2. Phytopathol Mediterr 47:50–60
Berry SD, Fargette M, Spaull VW, Morand S, Cadet P (2008) Detection and quantification of root-knot nematode (Meloidogyne javanica), lesion nematode (Pratylenchus zeae) dagger nematode (Xiphinema elongatum) parasites of sugarcane using real-time PCR. Mol Cell Probes 22:168–176
Clayton EE (1923) The relation of temperature to the Fusarium wilt of the tomato. Am J Bot 10:71–89
Cullen DW, Lees AK, Toth IK, Duncan JM (2001) Conventional PCR and real-time quantitative PCR detection of Helminthosporium solani in soil and on potato tubers. Eur J Plant Pathol 107:387–398
Flor HH (1956) The complementary genic systems in flax and flax rust. Adv Genet 8:29–54
Grattidge R, O’Brien RG (1982) Occurrence of a third race of Fusarium wilt of tomatoes in Queensland. Plant Dis 66:165–166
Hirano Y, Arie T (2006) PCR-based differentiation of Fusarium oxysporum ff. sp. lycopersici and radicis-lycopersici and races of F. oxysporum f. sp. lycopersici. J Gen Plant Pathol 72:273–283
Houterman PM, Cornelissen BJC, Rep M (2008) Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog 4:e1000061
Houterman PM, Ma L, van Ooijen G, de Vroomen MJ, Cornelissen BJC, Takken FLW, Rep M (2009) The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J 58:970–978
Huang J, Wu J, Li C, Xiao C, Wang G (2009) Specific and sensitive detection of Ralstonia solanacearum in soil with quantitative, real-time PCR assays. J Appl Microbiol 107:1729–1739
Ippolito A, Schena L, Nigro F, Ligorio VS, Yaseen T (2004) Real-time detection of Phytophthora nicotianae and P. citrophthora in citrus roots and soil. Eur J Plant Pathol 110:833–843
Kawabe M, Kobayashi Y, Okada G, Yamaguchi I, Teraoka T, Arie T (2005) Three evolutionary lineages of tomato wilt pathogen, Fusarium oxysporum f. sp. lycopersici, based on sequences of IGS, MAT1, and pg1, are each composed of isolates of a single mating type and a single or closely related vegetative compatibility group. J Gen Plant Pathol 71:263–272
Kistler HC (1997) Genetic diversity in the plant-pathogenic fungus Fusarium oxysporum. Phytopathology 87:474–479
Lievens B, Brouwer M, Vanachter ACRC, Cammue BPA, Thomma BPHJ (2006) Real-time PCR for detection and quantification of fungal and oomycete tomato pathogens in plant and soil samples. Plant Sci 171:155–165
Lievens B, Houterman PM, Rep M (2009) Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiol Lett 300:201–215
Morimoto S, Hoshino YT (2008) Methods for analysis of soil communities by PCR-DGGE (1) Bacterial and fungal communities (in Japanese). Soil Microorg 62:63–68
Okubara PA, Schroeder KL, Paulitz TC (2005) Real-time polymerase chain reaction: applications to studies on soilborne pathogens. Can J Plant Pathol 27:300–313
Ratti C, Budge G, Ward L, Clover G, Rubies-Autonell C, Henry C (2004) Detection and relative quantitation of Soil-borne cereal mosaic virus (SBCMV) and Polymyxa graminis in winter wheat using real-time PCR (TaqMan®). J Virol Methods 122:95–103
Saito H, Banno S, Kabe T, Urushibara T, Fujimura M (2007) Application of real-time PCR for quantitative detection of Verticillium longisporum in cabbage fields (abstract in Japanese). Ann Phytopathol Soc Jpn 73:213
van der Does HC, Lievens B, Claes L, Houterman PM, Cornelissen BJC, Rep M (2008) The presence of a virulence locus discriminates Fusarium oxysporum isolates causing tomato wilt from other isolates. Environ Microbiol 10:1475–1485
Wang Y, Zhang W, Wang Y, Zheng X (2006) Rapid and sensitive detection of Phytophthora sojae in soil and infected soybeans by species-specific polymerase chain reaction assays. Phytopathology 96:1315–1321
Acknowledgments
We thank Yuji Hosobuchi (Sakata Seed, Kimitsu, Japan), Hideaki Tateishi (Kureha, Iwaki, Japan), Taiji Miyake (Kureha), Ken Watanabe (Ibaraki Agricultural Center, Kasama, Japan), Suminaga Suwa (Gunma prefecture, Gunma, Japan), Yoshimiki Amemiya (Chiba University, Matsudo, Japan), and Fujio Kodama (previously at Hokkaido Central Agricultural Experiment Station, Naganuma, Japan) for providing fungal isolates. This study was partly supported by eDNA Project (Development of soil diversity analysis system with environmental DNA) of the Ministry of Agriculture, Forestry, and Fisheries (MAFF), and Grants-in-Aid (16405021, 18380030) from The Japan Society for the Promotion of Sciences (JSPS) for TA.
Author information
Authors and Affiliations
Corresponding author
Additional information
K. Inami and C. Yoshioka have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Inami, K., Yoshioka, C., Hirano, Y. et al. Real-time PCR for differential determination of the tomato wilt fungus, Fusarium oxysporum f. sp. lycopersici, and its races. J Gen Plant Pathol 76, 116–121 (2010). https://doi.org/10.1007/s10327-010-0224-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10327-010-0224-7