Abstract
Alternaria is a pathogenic and allergenic fungus affecting 400 plant species and 334 million people globally. This study aimed at assessing the diversity of Alternaria species in airborne samples collected from closely located (7 km apart) and heterogeneous sites (rural, urban and unmanaged grassland) in Worcester and Lakeside, the UK. A secondary objective was to examine how the ITS1 subregion varies from ITS2 in Alternaria species diversity and composition. Airborne spores were collected using Burkard 7-day and multi-vial Cyclone samplers for the period 5 July 2016–9 October 2019. Air samples from the Cyclone were amplified using the ITS1and ITS2 subregions and sequenced using Illumina MiSeq platform whereas those from the Burkard sampler were identified and quantified using optical microscopy. Optical microscopy and eDNA revealed a high abundance of Alternaria in the rural, urban and unmanaged sites. ITS1 and ITS2 detected five and seven different Alternaria species at the three sampling sites, respectively. A. dactylidicola, A. metachromatica and A. infectoria were the most abundant. The rural, urban and unmanaged grassland sites had similar diversity (PERMANOVA) of the species due to similarity in land use and proximity of the sites. Overall, the study showed that heterogeneous and neighbouring sites with similar land uses can have similar Alternaria species. It also demonstrated that an eDNA approach can complement the classical optical microscopy method in providing more precise information on fungal species diversity in an environment for targeted management. Similar studies can be replicated for other allergenic and pathogenic fungi.
Similar content being viewed by others
1 Introduction
Alternaria is a saprophytic, endophytic and pathogenic fungus with many species ubiquitous in nature (Rotem, 1994). Precisely 275 Alternaria species, having specific or wide host range, are pathogenic to nearly 400 plant species including those in the Poaceae, Cucurbitaceae, Brassicaceae and Solanaceae families (Lee et al., 2015; Meena et al., 2017b; Seifert & Gams, 2011; Simmons, 2007; Skjøth et al., 2016). Crop yield losses of up to 80% have been attributed to Alternaria diseases in several years of production (Maude & Humpherson-Jones, 1980; Nowicki et al., 2012). Alternaria is also an allergenic fungus affecting up to 70% of mould-allergic patients (Sanchez & Bush, 2001). About 37 Alternaria species are known to cause allergy and other respiratory tract disorders such as alveolitis, rhinitis, bronchitis and eczema (Hong et al., 2005; Meena et al., 2017a; Seifert & Gams, 2011; Skjøth et al., 2016). Inhalation of allergenic Alternaria spores triggers severe and potentially fatal asthma in sensitised individuals, most especially children (Gabriel et al., 2015; Sanchez & Bush, 2001). Often, the forecast advice to allergy sufferers and asthmatics for prevention is based on the daily Alternaria spp. spore concentration in the air in general (Grinn-Gofroń et al., 2019). This approach does not provide information on the particular species that are most abundant in the air thus leading to allergic individuals and asthmatics taking measures including medications based on basic forecast information.
Understanding fungal species diversity, e.g. in Alternaria spp. in an area helps to provide more targeted information on spore forecast and for targeted management of fungal diseases. However, aerobiological studies mostly use the classical microscopic method that relies on morphological features of the most abundant and recognisable fungal spores to provide spore data at the genus level (Banchi et al., 2018; Pashley et al., 2012; Sharma et al., 2015). The development of DNA metabarcoding alongside high-throughput sequencing (HTS) technology has enabled an increased number of studies in airborne plant diversity (Hebert et al., 2003; Joly & Peuch, 2012). However, few of such studies exist on airborne fungal diversity (Banchi et al., 2018). Moreover, some of the metabarcoding fungal diversity studies focus mostly on the genera level leaving out species diversity (Banchi et al., 2020a, 2020b; Rosa et al., 2020a, b Fort et al., 2016; Tordoni et al., 2021) which could otherwise provide cues on specific fungal species relevant for the prevention of allergy and pathological diseases.
In this study, we aimed to assess the diversity, richness, composition and abundance of Alternaria species in airborne samples collected from rural, urban and unmanaged sites that are located approximately 7 km apart in Worcester and Lakeside Container/Lakeside Circle, the UK. This was addressed by examining the hypothesis that there is a low diversity and high abundance of Alternaria species in nearby sites with similar land uses. Airborne eDNA and HTS fungal diversity studies focussing on both urban and rural sites are very limited (Bowers et al., 2013; Leppänen et al., 2018; Lin et al., 2018; Wady et al., 2004) and yet cases of allergy and asthma are high in such sites (Mitakakis et al., 2001; von Mutius, 2008). Moreover, there are no such eDNA and HTS studies of fungal diversity in unmanaged sites, e.g. grassland. Therefore, we sequenced the eDNA from the air samples using both the internal transcribed spacer (ITS1 and ITS2) subregions and the Illumina MiSeq platform.
2 Materials and methods
2.1 Spore sampling
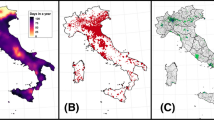
Alternaria spores were sampled using a Hirst-type Burkard 7-day volumetric spore trap (Hirst, 1952) and a Burkard automatic multi-vial Cyclone sampler (Burkard Manufacturing, UK). Samples were collected at St John’s campus (hereafter Worcester) and Lakeside campus (hereafter Lakeside Container and Lakeside Circle) of the University of Worcester (Fig. 1). Air samples were collected during the main Alternaria season at Worcester and Lakeside Container from 5 July 2016 to 9 October 2019 (Table S1). Sampling heights were set following the recommendations on the height of pollen/spore sampling (Galán et al., 2014). The Burkard 7-day and Cyclone samplers at Worcester were placed 10 m above ground level (AGL) on the rooftop of the Edward Elgar (EE) building (52.1970 N, − 2.2421 E, e.g. Sadyś et al., 2014) to capture spores at a regional scale. The Burkard 7-day and Cyclone samplers collected air at a rate of 10 L/min and 16.5 L/min, respectively (Hirst, 1952) and the flow rate of the samplers was checked weekly. The airborne particles were deposited by impaction on a tape coated with a thin film of petroleum jelly/wax mixture. The samplers at Worcester were unloaded weekly at 9:00 am while those at Lakeside Container and Lakeside Circle were emptied at 14:00 pm and their spore data every 2-h was later matched with those at Worcester. Worcester is an urban area surrounded by agricultural areas comprising permanent orchards for fruit and cider production (Sadyś et al., 2014), crops under rotation (Sadyś et al., 2015), grasslands and pasture within the public parks (Sadyś et al., 2016) and small woodlands (Skjøth et al., 2015). The nearest crop fields to the trap in Worcester were half a km away in a westerly direction (Apangu et al., 2020).
Spore sampling and meteorological station sites of Lakeside Container, Lakeside Circle and Worcester. Also indicated on the map are the (a) Lake near Lakeside Container and (b) Lake near Lakeside Circle and River Severn (dark blue spiralling feature on left map). Lakeside and St Johns maps on the right are enlargements of their corresponding map sections on the left
The Burkard and Cyclone samplers at Lakeside Container (52.2544 N, − 2.2537 E) were placed on a container at 4 m AGL. The Lakeside Container sampling site was in a rural environment and comprised mixed arable crop fields, permanent pastures, animal paddocks and patches of trees. The immediate vicinity of the sampler had no source of Alternaria spores and it was surrounded by buildings in the south, non-vegetated areas such as hard standing, roadways, and a man-made lake in the west, pine trees in the north and other trees and large areas of amenity grassland in the east. The area outside the vicinity of the sampler was comprised of crop fields under rotation and woodlands.
The third set of Burkard and Cyclone samplers collected spores at Lakeside Circle (52.2516 N, − 2.2535 E; Fig. 1) from 12 July 2018 to 9 October 2019 (Table S1). Historically, the area was an arable farm with annual rotations until after the 2016 crop harvest when it became amenity grassland. A circle, centrally on the field and with a diameter of 50 m was created in the grassland in 2018. The major grass and herb species growing in the circular grassland included Festuca rubra, Lolium perenne and Trifolium repens. Grasses such as Festuca are known hosts of Alternaria spp. (Awad, 2005; Wilson et al., 2014). Nitrogen, phosphorus, potassium (NPK) fertiliser mixture was applied to the grassland in the circle at the start of the observation period in spring 2018 to support root establishment and vigorous growth of the grasses (Leuschner et al., 2013). Thereafter, the grass circle remained unmanaged to enable natural production and release of fungal spores including Alternaria. A pair of Burkard 7-day and Cyclone samplers were placed 2.5 m AGL on a mast located in the centre of the circle. The grassland area outside the circle was mowed regularly (~ every 3 weeks) to ensure a green and well-growing grass patch without large amounts of decomposing plant material outside the circle. Lakeside Container and Lakeside Circle were approximately 310 m apart and both were approximately 7 km away from Worcester. The landscape outside the established grassland comprised mixed arable farms of rotational crop fields, permanent pastures and animal paddocks.
2.2 Microscopic Alternaria spp. spore identification
During slide preparation, the tape was cut into 7 pieces (each piece 48 mm long), each corresponding to one of the seven days in a week, and mounted on a microscopic slide (Sadyś et al., 2014). Slides were prepared according to a standard procedure used for more than 50 years in England and other European countries (Adams-Groom et al., 2002; BAF, 1995; Kasprzyk, 2008; Skjøth et al., 2008, 2015). Alternaria spores were identified to genus level and counted using the 12 transverse method at 400× magnification, an approach used for fungal spore monitoring in Worcester, UK (Apangu et al., 2020), Denmark (Skjøth et al., 2012) and Hungary (Paldy et al., 2014). The daily (24 h) mean Alternaria spore concentrations were expressed as spores/m3 of air by multiplying the microscopic spore counts with previously calculated correction factors (Lacey & Allit, 1995).
2.3 Cultivation of A. alternata conidia
A. alternata spores were cultivated in the Lab to be used in a mock community. Conidia of A. alternata procured from Fisher Scientific UK were cultured on potato dextrose agar for 23 days at 23 °C, similar to Smith et al. (2012). To harvest spores, sterile water (10 mL) was added to the culture petri dish and spore suspensions were obtained by gently scraping the surface of the culture using a sterile L-shaped spreader. Five mL of the spore suspension was drawn into a clean and sterile 50 mL centrifuge tube. The mycelial extract was recovered after 5 min of centrifugation at 2500 rpm. The supernatant was discarded and the pellet was transferred into a clean 2 mL microcentrifuge tube and resuspended in 1 mL sterile water.
2.4 DNA extraction from air samples and culture material
The daily air samples were pooled every seven consecutive days of sampling according to a pre-planned sampling date arrangement (Table S1), similar to Brennan et al. (2019). DNA was isolated from the air samples and the A. alternata culture material using a commercial protocol (Fast DNA spin kit for soil; MP Biomedicals), similar to previous studies (Chen et al., 2020; Degois et al., 2019; Ettenauer et al., 2012; Fröhlich-Nowoisky et al., 2012). The sample for 28/07/2018 at Lakeside circle, which comprised water, typically caused by heavy rain episodes or fog, was excluded from extraction. The concentration of DNA in the samples was quantified using a Nanodrop 2000c spectrophotometer instrument (Fisher Scientific, UK), similar to previous studies (Degois et al., 2019; Ettenauer et al., 2012; Shokere et al., 2009). The DNA was stored at − 20 °C for subsequent analyses.
2.5 Mock community
A large collection of pollen and spores of different plant and fungal species were originally acquired as clean reference samples from commercial companies and stored in a refrigerator at 4 °C in the Charles Darwin laboratory of the University of Worcester. Aiming at containing both allergenic pollen and allergenic and pathogenic spores, small amounts from the clean reference samples were pooled to form a mock community. The mock community sample consisted of Cladosporium sp., Alternaria sp., Dactylis glomerata, Lolium perenne, Artemisia vulgaris, Quercus ilex, Quercus robur, Alnus glutinosa, Betula pendula, Corylus avenella, Phleum pratense, Urtica dioica, Platanus × hispanica and A. alternata, similar to previous metabarcoding studies on fungal spores (Aguayo et al., 2018; Heeger et al., 2018; Pauvert et al., 2019). The species such as L. perenne, U. dioica and Cladosporium sp. were included because of their overlapping seasonality with Alternaria while A. glutinosa and C. avenella were added due to their seasonal differences with Alternaria. DNA was extracted from the mock community sample using the Fast DNA spit kit protocol, as for the air samples above. The mock community DNA was then stored at − 20 °C for downstream analyses.
2.6 Preparation of air samples and mock community for metabarcoding
For each site, the 7-day pooled samples were re-pooled for each sampling season to form a composite sample (Table S2). Ten re-pooled air samples and the mock community were used in Illumina sequencing. To prevent any contamination, microcentrifuge tubes were autoclaved before re-pooling the DNA of the air samples. Four µL of DNA were drawn from each weekly pooled air sample per season and added to the sterile microcentrifuge tube to form a composite sample. Meanwhile, 50 µL of the total DNA of the mock community was used for sequencing. The Nanodrop 2000c spectrophotometer was used for measuring the concentration of DNA in each sample. DNA of each sample and mock community was suspended in TE buffer [a mixture of 1 mM Ethylenediaminetetraacetic acid (EDTA) and 10 mM Tris(Hydroxymethyl)] to prevent DNA degradation and stored at 4 °C before shipment to Eurofins Genomics for ITS1/ITS2 amplification and sequencing, as recommended by Clasen et al. (2020). DES (DNase/Pyrogen-free water; 50 µL) was used as a negative control in the sequencing process.
2.7 PCR amplification and amplicon sequencing
Eurofins Genomics, Europe Sequencing GmbH, Konstanz, Germany synthesised the primers, performed the PCR assays and amplicon sequencing of the air samples and the mock community, similar to previous studies (Naveed et al., 2016; Nicolaisen et al., 2017; Rossmann et al., 2021; Senés-Guerrero & Schüßler, 2016). The PCR assay conditions were similar to Nicolaisen et al. (2017). The ITS is one of the most widely sequenced DNA regions in fungi and is the universally accepted genetic barcode for fungi (Abrego et al., 2018; Schoch et al., 2012). Therefore, we amplified 300 bp each of ITS1 and ITS2 regions using the universal primer pairs fwd-5′-GGAAGTAAAAGTCGTAACAAGG-3′; rev-5′-GCTGCGTTCTTCATCGATGC-3′ and fwd-5′-GCATCGATGAAGAACGCAGC-3′; rev-5′-TCCTCCGCTTATTGATATGC-3′, respectively, targeting specifically fungi (White et al., 1990). Each PCR reaction contained 1 µl of DNA template of air samples and mock community, 1 U of Taq DNA recombinant polymerase (Promega Corporation, Madison, WI, USA), 1 × PCR reaction buffer, 1.5 mM MgCl2, 1 mM of each primer and 0.2 mM dNTPs in a final volume of 25 µL. A GeneAmp PCR system 9700 thermal cycler (Fisher Scientific) was used for amplification. DNA was denatured at 94 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, 48 °C for 30 s, 72 °C for 1 min and elongation at 72 °C for 10 min. The amount of amplicon was visually estimated after gel electrophoresis. A visible smear of PCR products at approximately 340–760 bp was removed and purified using a QIAquick Gel Extraction Kit (QIAGEN, GmbH, Hilden, Germany).
2.8 Bioinformatics analysis
Illumina MiSeq-sequenced paired-end fastq sequences of fungal spores in air samples and the mock community that were amplified using ITS1 and ITS2 barcodes were demultiplexed and barcodes removed by Eurofins Genomics, similar to previous studies (Naveed et al., 2016; Nicolaisen et al., 2017; Rossmann et al., 2021; Senés-Guerrero & Schüßler, 2016). Primers were removed from demultiplexed sequences using the Cutadapt program (Martin, 2011). Reads were quality filtered, denoised, truncated and merged using the clustering-free Divisive Amplicon Denoising Algorithm (DADA2) v.1.14 (Callahan et al., 2016), similar to previous studies (Aguayo et al., 2018; Banchi et al., 2018; Banchi et al., 2020a; Kumari et al., 2016; Mbareche et al., 2020; Nicolaisen et al., 2017; Nilsson et al., 2019b; Schiro et al., 2019; Tordoni et al., 2021). The forward and reverse reads were truncated at 260 bp and 200 bp, respectively, to exclude lower quality reads while retaining only reads with fewer than two expected errors. After inference of sequence variation, reads were merged as described in the DADA2 tutorial. Chimeras were identified and removed, and the resulting amplicon sequences were used in subsequent analyses (Callahan et al., 2017). The “assignTaxonomy” function in the DADA2 pipeline was used for assigning amplicon sequence variants (ASVs) to specific sequences in the UNITE fungal database v.8.2 2020-02-04 (Abarenkov et al., 2020b), similar to previous studies (Banchi et al., 2018; da Silva et al., 2021; de Souza et al., 2021; Dyda et al., 2019; Ogaki et al., 2021). UNITE fungal databases, with 35,077 and 71,723 representative sequences, were used for taxonomic assignment of the ITS1 and ITS2 sequences, respectively. The UNITE eukaryotes database v.8.2 2020-02-04 (Abarenkov et al., 2020a) was used for taxonomic assignment in the mock community, similar to previous studies (Heeger et al., 2018; Tedersoo et al., 2018). Similarly, UNITE eukaryotes databases, with 91,074 and 183,678 representative sequences were used for taxonomic assignment of ITS1 and ITS2 sequences, respectively.
R packages “Phyloseq” v.1.3.0 (McMurdie & Holmes, 2013) and “ggplot2” v.3.3.2 alongside package “Biostrings” v.2.54.0 were used for downstream analysis of the ASV output from DADA2. The resulting taxonomic table was combined with the OTU table, matrix of ASV sequences and sample metadata. The relative abundance of fungal phyla detected using ITS1 and ITS2 barcodes at each site was visualised using bar charts. The data for the Alternaria genus and its species were extracted from the total fungal community data. ASVs specific to Alternaria species in the air samples and fungal and plant species in the mock community were assigned to the taxonomic groups and bar plots were constructed to visualise their relative abundance. Shannon and Simpson indices (α-diversity) combined with abundance were used to measure the richness and diversity of Alternaria species within the air samples (Spellerberg & Fedor, 2003). Mann–Whitney U test was performed to assess the significance of the α-diversity measures in the air samples using the R package “DESeq2” v.1.26.0, similar to Mbareche et al. (2020), Yang et al. (2018) and Archer et al. (2020). Beta diversity and species composition were assessed using the Bray–Curtis dissimilarity metric whereas UniFrac distance was used to measure phylogenetic community distance in the air samples (Mbareche et al., 2018). Weighted UniFrac and Unweighted UniFrac distances were used to evaluate the phylogenetic distances based on species abundance and presence or absence of the species in each sample, respectively (d’Entremont et al., 2020; Mbareche et al., 2018).
Principal Coordinates Analysis (PCoA) was performed using packages “plyr” v.1.8.6 alongside “ggplot2” to visualise the relationships. The PCoA was separated according to ITS barcodes and the environment of sampling (rural, urban and unmanaged) to examine the clusters. The clusters observed in the PCoA were statistically validated with a permutational analysis of variance (PERMANOVA) test at 999 permutations, similar to previous studies (Banchi et al., 2020a, 2020b; Fort et al., 2016; Tordoni et al., 2021). PERMANOVA was performed using the R package “vegan” v.2.5-6 with the function “adonis” (Oksanen et al., 2019). A p value of ≤ 0.05 was considered statistically significant for the Mann–Whitney U and PERMANOVA tests. The Mann–Whitney U test p values were adjusted using the Holm method.
The UNITE database, e.g. v.8.2 2020-02-04 dedicated to bioinformatics pipelines comprises sequences that are annotated, are released within a specific period of time and remain static, whereas the web-based version of the same database is an interactive and up-to-date version whose sequences continuously undergo taxonomic re-annotation to reflect the most recent nomenclatural and taxonomic changes and their associated metadata (Nilsson et al., 2019a). Furthermore, errors that accumulate during sample preparation and sequencing cannot be completely filtered during bioinformatics pipeline annotation of sequences using the static UNITE databases (Anslan et al., 2018; Nilsson et al., 2019b). Therefore, to minimise errors and improve taxonomic affiliations, sequences of all the ASVs of ITS1 and ITS2 were BLAST searched in the web-based UNITE reference database (https://unite.ut.ee/), similar to de Vere et al. (2012), Nilsson et al. (2016) and Anslan et al. (2018). To improve taxonomic specificity, similar to Be et al. (2015), BLAST search results with “Alternaria” in the taxon were further analysed to extract their species hypotheses while excluding those without “Alternaria” in their taxonomic name. Bioinformatics analyses were performed in R software v.3.6.3 (R Core Team, 2020).
2.9 Meteorological data
Two Campbell Scientific meteorological stations were established at Worcester and Lakeside Circle to provide half-hourly meteorological data for the period July 2017–October 2019. The meteorological stations were co-located with the Burkard 7-day and Cyclone samplers at Worcester and Lakeside Circle. The station at Lakeside Circle provided meteorological data for both itself and the Lakeside Container since they are closely located (310 m). There was a gap in meteorological data from January 2016 to July 2017 before the acquisition of the Campbell Scientific meteorological instruments. The gap in data was filled with hourly meteorological data obtained from a nearby (20 km away) UK Met station (Pershore Weather Station; MET Office, UK). This was after verifying that some of the Pershore weather data had a high correlation with Lakeside and Worcester weather data, similar to the approach of Skjøth et al. (2016).
The half-hourly and hourly weather data were independently averaged to provide daily meteorological data for each meteorological station to match the daily observation of Alternaria spores. Selected meteorological parameters including air temperature, pressure, relative humidity, solar radiation, precipitation, wind speed, wind direction, leaf wetness and dew point were extracted for analysis. Spearman’s rank correlation test was performed between the daily Alternaria spore concentrations and their corresponding meteorological variables, similar to Grinn-Gofroń et al. (2019) and Olsen et al. (2019).
3 Results
3.1 Taxonomy, relative abundance and diversity of Alternaria species in air samples
Ten composite air samples with DNA each representing the sampling period of 5 July 2016–09 October 2019 collected at Worcester, Lakeside Container and Lakeside Circle were sequenced. Sequencing, using ITS1 and ITS2 primers, resulted in a total of 926,319 and 644,016 reads after quality filtering, respectively (Table S3). Each sample contained between 56,100 and 116,955 reads for ITS1 and between 42,587 and 76,342 reads for ITS2. Reads were assembled into a total of 12,725 and 5369 ASVs for ITS1 and ITS2, respectively. A total of 114 (out of 1200 sequences) and 126 (out of 2795 sequences) chimeras were identified in ITS1 and ITS2 barcodes, respectively, and removed from the sequences as stipulated for PCR artefacts in the DADA2 guidelines.
3.2 Shared and unique taxa and their abundance
ITS1 and ITS2 barcodes were notably similar in composition and richness of fungal phyla, i.e. all the phyla detected in ITS1 were also found in ITS2 (Fig. 2a, b). Ascomycota and Basidiomycota were the most abundant phyla in all the sites. Alternaria sequences were detected in both ITS1 and ITS2 barcodes and they were correctly identified as Fungi (kingdom), Ascomycota (phylum), Dothideomycetes (class), Pleosporales (order), Pleosporaceae (family) and Alternaria (genus). The ITS1 barcode identified A. brassicae, A. dactylidicola, A. metachromatica, A. armoraciae and A. tenuissima species in the air samples (Fig. 2c). Meanwhile, the ITS2 barcode identified A. argyranthemi, A. infectoria, A. eichhorniae, A. mimicula, A. molesta, A. armoraciae and A. rosae species (Fig. 2d).
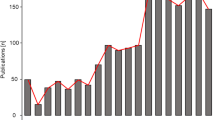
At the genus level, both the optical microscopy (Fig. 3a–d) and metabarcoding (using ITS1 and ITS2 barcodes in the DADA2 bioinformatics pipeline and UNITE v.8.2 2020-02-04; Fig. 4a, b) approaches corroborated the high abundance of Alternaria during the different years of observation. Both methods showed that Worcester had the highest frequency of Alternaria in 2016. Meanwhile, Lakeside Container and Lakeside Circle dominated in 2017 and 2019, respectively. However, the approaches differed in 2018 during which the optical microscopy detected the highest frequency of Alternaria at Worcester while metabarcoding (using both ITS1 and ITS2 barcodes in the DADA2 bioinformatics pipeline and UNITE v.8.2 2020-02-04) found the maximum at Lakeside Container. High spore concentrations (> 100 spores/m3) were observed in July and August of every season. At the species level, according to the ITS1 barcode, A. dactylidicola and A. metachromatica and for ITS2, A. infectoria, were the most abundant Alternaria species in the air of Lakeside Container, Lakeside Circle and Worcester. ITS1 barcode showed that A. dactylidicola was most abundant at Worcester in 2018 and Lakeside Container in 2019. According to ITS2, A. infectoria was the most abundant species at all the sampling sites and years of observation. There was a greater abundance of A. infectoria at both Worcester and Lakeside Circle in 2019 compared to 2018.
Daily mean Alternaria spp. spore concentrations at genus level during the sampling periods of (a) 30 Jun–28 Sep 2016, (b) 29 Jun–13 Sep 2017, (c) 12 Jul–19 Sep 2018 and (d) 05 Jul–09 Oct 2019 at Lakeside Container (red lines), Lakeside Circle (green lines) and Worcester (blue lines). Note: no spore sampling at Lakeside Circle in 2016 and 2017 and therefore Alternaria spore data for Lakeside Circle are not represented in the time series for 2016 and 2017
BLAST search identified 66.67% of the Alternaria species that were detected with the DADA2 bioinformatics pipeline. The BLAST search and threshold distance analysis of species hypothesis found A. metachromatica, A. brassicae, A. infectoria, A. eichhorniae A. sonchi, A. planifunda, A. carotiincultae, A. abundans and other unidentified Alternaria species in ITS1 sequences [(Table S4a (i) and (ii)]. A. infectoria was the most abundant species (69.6%) detected by the BLAST search.
For ITS2 sequences, the BLAST search and species hypothesis found A. infectoria, A. armoraciae, A. eichhorniae, A. rosae and A. mimicula. Other species detected by the search included A. alternata, A. metachromatica, A. abundans, A. linicola, A. brassicae, A. triticina, A. solani, A. brassicicola and other unidentified Alternaria species [(Table S4b (i) & (ii)]. Alternaria sp. was the most abundant (27.3%), followed by A. infectoria (9.1%) and A. solani (9.1%) in the BLAST search. Noteworthy, other uncommon Alternaria species including A. oregonensis, A. venezuelensis, A. tropica, A. multirostrata, A. cichorii, A. tumida, A. photistica, etc. were detected in the BLAST search.
3.3 Alternaria species richness and diversity
Alpha diversity was analysed using Shannon and Simpson diversity indices (Table 1). Statistical analysis (Mann–Whitney U test) showed significant (Shannon: p = 0.01437 and Simpson: p = 0.01418) difference in Alternaria species richness and diversity in the air samples. In general, the α-diversity showed high diversity and richness of Alternaria species at the observation sites. Both Shannon and Simpson indices of ITS1 and ITS2 barcodes showed that Worcester and Lakeside Container were the most and least diverse sites, respectively. Overall, ITS1 and ITS2 barcodes detected five and seven Alternaria species, respectively, and Shannon and Simpson indices were highly correlated.
Beta diversity was assessed using the Bray–Curtis dissimilarity metric, Weighted UniFrac and Unweighted UniFrac distances and visualised using PCoA plots. The maximum percentage variation of β-diversity for ITS1 and ITS2 barcodes was explained by Principal Coordinate (PC1; Fig. 5a, b). Bray–Curtis, weighted UniFrac and unweighted UniFrac metrics showed that air samples from Lakeside Container grouped with those from Worcester and Lakeside Circle. PERMANOVA analysis showed that the observed clusters were not significant (Table 2).
Principal Coordinates Analysis (PCoA) plots of air samples collected from Worcester (urban; blue), Lakeside Container (rural; green) and the unmanaged grassland of Lakeside Circle (natural; red) for the period 2016–2019, generated from ASV sequences of ITS1 (a) and ITS2 (b) and represented by (i) Bray–Curtis dissimilarity matrix (ii) weighted UniFrac and (iii) unweighted UniFrac distances. The points on the PCoA plots represent the β-diversity of Alternaria species at both the location of each sample and the whole sampling period (2016–2019)
3.4 Taxonomy and abundance of mock community individuals
Overall, the plants (except Corylus) and the fungi (Alternaria and Cladosporium) in the mock community were correctly identified from kingdom, phylum, class, order and family to genus level (Table S5; Fig. S1a–d). Dactylis, Phleum, Cladosporium and Alternaria were the most abundant genera. At the species level, D. glomerata (with both ITS1 and ITS2), P. pratense (with ITS1), Q. ilex and Q. robur (both with ITS2) were correctly identified using the UNITE eukaryotes database in the mock community (Fig. S2a and b). Among the plant species, D. glomerata and P. pratense were the most abundant in the mock community individuals (Fig. S2a and b). Among the fungal species, A. tenuissima, C. delicatulum and C. tenuissimum were the most abundant species detected (Fig. S2c & d).
However, there were some identification inaccuracies at the species level (Fig. S2a–d). For instance, A. glutinosa was identified as A. firma (with UNITE eukaryotes database and ITS1) and A. fauriei (with UNITE eukaryotes database and ITS2), Alternaria sp. and A. alternata were identified as A. tenuissima (with both UNITE eukaryotes and UNITE fungal databases and ITS1) and A. eichhorniae (with UNITE eukaryotes database and ITS2), Cladosporium sp. was identified as C. delicatulum (with both UNITE eukaryotes and fungal databases and ITS1) and C. tenuissimum (with both UNITE eukaryotes and fungal databases and ITS2), L. perenne was identified as L. temulentum (with UNITE eukaryotes database and ITS1 and ITS2). A. vulgaris, B. pendula, C. avenella, U. dioica, P. x hispanica were not detected in the mock community.
Other unexpected genera were detected in low abundances in the mock community and these included Aniselyton, Brassica, Chenopodium, Festuca, Laportea, Fraxinus and Mycocentrospora, Aureobasidium, Botrytis, Calloria, Camarosporium, Coniozyma, Filobasidium, Golovinomyces, Itersonilia, Lichtheimia, Muriphaeosphaeria, Phaecoccomyces, Pringsheimia, Sporobolomyces, Trebouxia and Vishniacozyma among others. Similarly, other unexpected species were detected in the mock community and these included Deutzia gracilis, Europiella artemisiae, Eschscholzia californica, Cochliobolus victoriae and Aureobasidium pullulans among others.
3.5 Effect of meteorological variables on daily Alternaria spp. spore concentrations
Spearman’s correlation test showed that several meteorological variables significantly correlated with the daily Alternaria spp. spore concentrations at the different sampling sites and seasons (Table 3). Temperature, relative humidity, precipitation and solar radiation were all strongly correlated with the daily spore concentration. Meanwhile, pressure, leaf wetness, wind speed and direction correlated weakly to moderately with the daily Alternaria spore concentration. The effect of the individual meteorological variables on the daily spore concentrations varied with observation site and year.
4 Discussion
4.1 Diversity and composition of Alternaria species in the rural, urban and unmanaged grassland habitats
The optical microscopy and metabarcoding approach showed a high spore concentration and relative abundance of Alternaria, respectively, for the sampling sites and in the different years of observation (Figs. 3a–d and 4a, b). Metabarcoding results showed a diversity of Alternaria species at Worcester and Lakeside (Fig. 2c, d). The species detected (with ITS1) included A. dactylidicola, A. metachromatica, A. brassicae, A. armoraciae and A. tenuissima and those with ITS2 included A. infectoria, A. armoraciae, A. eichhorniae, A. mimicula, A. molesta and A. rosae. BLAST search [Table S4a (i) and b (i)] and species hypotheses [Table S4a (ii) and b (ii)] confirmed 66.67% of the above species detected with the bioinformatics pipeline including A. alternata. Meanwhile, the BLAST search did not detect 33.33% of the Alternaria species because they could not meet the filtering criteria (similarity score = 93–100%, E-value = 0.001) and hence were considered putative artefacts or non-Alternaria species (Anslan et al., 2018). This is the first study to reveal the diversity and richness of Alternaria species in three heterogeneously diverse biogeographic locations that are ~ 7 km apart. Previous studies had found a high abundance (but not diversity) of Alternaria spp. in Worcester (O’Connor et al., 2014; Sadyś et al., 2015; Skjøth et al., 2016). Fungal diversity in an area depends on the pedology, land use, vegetation and climatic conditions of the area to provide substrate and conducive environments for the fungi to grow and multiply (Peay & Bruns, 2014). The Shannon and Simpson indices showed a significant difference in the diversity of Alternaria species within the samples. However, Bray–Curtis dissimilarity, Weighted UniFrac and Unweighted UniFrac distance metrics showed low variation in diversity of Alternaria species between the sites (Fig. 5a, b). This suggests similarity in Alternaria species at the three sites and this could be attributed to the fact that the unmanaged (Lakeside Circle), rural (Lakeside Container) and urban (Worcester) sites share similar vegetation (grass species) and agricultural landscapes since they are only ~ 7 km apart (Apangu et al., 2020; Brennan et al., 2019). Furthermore, the similarity in species diversity could also be attributed to the fact that the airborne Alternaria spores undergo dilution and mixing in the lower boundary layer before they are captured at the traps thus contributing to the genetic mixing, species diversity and the final concentration (Fröhlich-Nowoisky et al., 2016; Nicolaisen et al., 2017; Sicard et al., 2016). Moreover, the PCoA showed that the Alternaria spp. at the 3 study sites clustered together (Fig. 5a, b), suggesting that the sites share similar Alternaria species. Similar studies also found low fungal diversity, richness and composition in airborne samples collected from nearby (< 10 km apart) sites (Abrego et al., 2018; Dannemiller et al., 2014). Contrarily, Mhuireach et al. (2016), with more closely located samplers (50 m apart), reported high variations in abundance and diversity of bacterial and fungal communities. Such fine-scale (< 50 m) variations in species diversity and abundance of bioaerosols can be attributed to weather, season, land use, land cover and distance from bioaerosol sources (Mhuireach et al., 2016; Rathnayake et al., 2016; Rolph et al., 2018). Conversely and expectedly, studies with distantly (> 50 km apart) located sites reported a high fungal diversity due to the effect and interactions of site-specific factors, e.g. vegetation type, changing atmospheric conditions and shifts in microbial sources (Banchi et al., 2018; Bowers et al., 2013; Fröhlich-Nowoisky et al., 2012). Nonetheless, sometimes locations (even though distantly apart) may not have strong effects or require interaction with other variables to influence the diversity of fungal communities. For instance, Nicolaisen et al. (2017) found similar results to ours in that sampling location alone did not considerably contribute to airborne fungal diversity except between seasons and their interactions. Our study only presents the diversity and abundance of Alternaria species in the summer and early autumn and found no significant effect of season or their interaction with sampling site on Alternaria spp. diversity (Table 2). This is attributed to the fact that Worcester and Lakeside are neighbouring sites that likely have a synchronised Alternaria spore season in the middle of summer, as was previously reported in Central-Northern Europe (Skjøth et al., 2016). However, Banchi et al. (2020a) and Kumari et al. (2016) examined the influence of location, season and their interactions on fungal diversity and found that their interactions (especially during summer) greatly influenced fungal diversity. Meanwhile, Mbareche et al. (2020) considered only location and found that samples collected from sites of compost, biomethanisation and dairy farms significantly influenced fungal diversity in those areas. Overall, this study suggests that regardless of an urban, rural or unmanaged location, areas that border each other and have similar land use and land cover types tend to have low Alternaria species diversity and similar species composition. However, since we only investigated the Alternaria genus, further studies are needed to examine fungal diversity for a whole fungal community in such areas to draw a firm conclusion.
A. dactylidicola and A. metachromatica were among the Alternaria species detected in the air samples. A. dactylidicola and A. metachromatica are species observed for the first time in Worcester and Lakeside, the UK and they are relatively new species. Simmons (1994) first morphologically described A. metachromatica as a pathogenic species and together with A. infectoria belongs to the Alternaria section Infectoriae (Woudenberg et al., 2013). A. dactylidicola is a grass-inhabiting species that was first found to associate with the leaves, roots and dead stems of the grass D. glomerata in Italy and it is phylogenetically linked with Alternaria cesenica (Thambugala et al., 2017). Lolium, Holcus and Dactylis are the most abundant grass genera in Worcester (including Lakeside) and are harvested as feeds for livestock (Brennan et al., 2019; Frisk et al., 2021). Harvesting of grasses for hay is a common practice among livestock farmers and starts from June to September and peaks in July (Jefferson, 2005). Several studies have shown that cutting/mowing of grasses either for hay or in public parks releases a considerable amount of Alternaria spores into the atmosphere (Astray et al., 2010; Comtois et al., 1995; Corden et al., 2003; Irga & Torpy, 2016; Kilic et al., 2010; Mitakakis et al., 2001). Moreover, increased Alternaria spore concentrations in public urban parks were observed in July and August (Kasprzyk et al., 2021), a period when high Alternaria spp. spore concentrations were recorded in Worcester and Lakeside (Fig. 3) and when the parks are intensively utilised. Apart from grass mowing for hay, Worcester urban green areas such as grasslands, pastureland, public parks, the racecourse and the cricket ground are potential sources and were previously associated with Alternaria spores in the city (Sadyś et al., 2016). In Worcester, the grass species, D. glomerata, flowers in June–August (Brennan et al., 2019; Frisk et al., 2021), which coincides with the period of high Alternaria spp. spore concentrations in Worcester (Apangu et al., 2020). The Alternaria in the grasses can also actively or passively release many spores into the atmosphere when weather conditions are optimal (Crandall & Gilbert, 2017). Several meteorological parameters including temperature, humidity, rain, wind speed and direction significantly correlated with Alternaria spp. spore concentrations at the three sites (Table 3). Therefore, the high abundance of A. dactylidicola, A. metachromatica and A. infectoria in all the study sites could be attributed to the favourable weather conditions and the abundance of Dactylis grass in Worcester and Lakeside since it is a major host to the species (Brennan et al., 2019; Sadyś et al., 2015). Przemieniecki et al. (2019) found that endophytic fungi, including Alternaria, were capable of colonising multiple grass taxa as was observed with A. alternata inhabiting both L. perenne and P. pratense. Previously, Alternaria sp. was also found in the leaves and roots of the grass Holcus lanatus (Márquez et al., 2010). We hypothesise that A. dactylidicola can also colonise Lolium and Holcus grasses since those grass genera are abundant in Worcester (including Lakeside) (Brennan et al., 2019).
A. dactylidicola is a saprobe (Thambugala et al., 2017), suggesting that it is active in decomposition within unmanaged grasslands and nutrient recycling in terrestrial ecosystems. Whereas pollen from D. glomerata, the main host of A. dactylidicola, is highly allergenic (D’Amato et al., 1991; Frisk et al., 2021), it is unknown whether A. dactylidicola also has such allergenic attributes since it is a newly discovered species. Neither is there any study on the pathological properties of A. dactylidicola. Future studies should elucidate the pathogenic and allergenic capabilities of A. dactylidicola. The high abundance of A. dactylidicola in the different sampling sites and years could also be related to differences in local meteorological conditions as it was found that local weather, e.g. temperature, relative humidity, rain, solar radiation, etc. observed at Worcester, Lakeside Container and Lakeside Circle all correlated with the daily Alternaria spore concentrations. Recently, Grinn-Gofroń et al. (2018) and Tordoni et al. (2021) observed a strong effect of the local weather and climate on fungal sporulation and the eventual release of the spores into the atmosphere.
The abundance of A. metachromatica in Worcester is linked to its host, oilseed rape (Brassica napus) (Al-lami et al., 2020). Oilseed rape is abundantly grown around Worcester and Lakeside and high concentrations of Alternaria spp. spores in Worcester were associated with its harvesting (Apangu et al., 2020). A. metachromatica is a multi-host pathogenic fungus as it was found to cause leaf spot on tomatoes (Lycopersicon esculentum) (Bashir et al., 2014) and infected the pasture plant, spotted knapweed (Centaurea stoebe), which is native to Europe (Broennimann et al., 2014). Related to A. metachromatica are A. solani, A. alternata, A. infectoria, A. arborescens, A. tenuissima, A. mimicula (Bessadat et al., 2017; Ramezani et al., 2019) and A. tomatophila (Rodrigues et al., 2010) that cause early blight in tomatoes and potatoes.
A. metachromatica belongs to the A. infectoria species group that is associated with many grass taxa in the family Poaceae, including wheat, barley, oat and rye (Andersen et al., 2002). Wheat, barley and oat are widely grown in Worcestershire and their combined harvesting was associated with high concentrations of Alternaria spp. spores in the area (Apangu et al., 2020). It is, therefore, likely that the harvesting of wheat, barley, oat and ryegrass (for hay) could have contributed to the high abundance of A. metachromatica in Worcester and Lakeside.
Alternaria spp. data of 2018 and 2019 analysed from Lakeside Circle strongly indicated that Alternaria spores were being released from Lakeside Circle when the grasses were still green and not expected to host Alternaria, suggesting that the litter from previous seasons was hosting Alternaria. Daily spore data analysis also showed that Alternaria spp. spore concentrations at Worcester and Lakeside Circle sites increased drastically from 2018 to 2019 (Fig. 3c, d). Metabarcoding results showed that there was a gradual increase in the abundance of A. dactylidicola, A. metachromatica and A. infectoria at Worcester and Lakeside Circle from 2018 to 2019 (Fig. 2c). Therefore, the increases in the abundance of A. dactylidicola, A. metachromatica and A. infectoria could also be attributed to the accumulation (2017–2019) of litter at the unmanaged site (Lakeside Circle) and other unmanaged sites surrounding Worcester and Lakeside, which provide a habitable environment for the species above. Similarly, de Vries et al. (2007) found that fungal biomass increased with the age of vegetation and they attributed it to the accumulation of organic matter content and minimal disturbance from agronomic practices, e.g. tillage, which encouraged the growth of the mycelial network. The current study presents the abundance and diversity of Alternaria species for the whole sampling period. Future studies should investigate species diversity and abundance at much higher temporal resolutions, e.g. weekly or three days, similar to Brennan et al. (2019), to produce precise and near real-time information for allergy sufferers, crop pathologists and medical practitioners.
Other Alternaria species detected by the ITS1 barcode included A. armoraciae, A. brassicae and A. tenuissima. A. armoraciae and A. tenuissima belong to the sections Chalastospora and Alternata, respectively, meanwhile the section of A. brassicae is yet undefined (Woudenberg et al., 2013). Like A. metachromatica, A. armoraciae and A. brassicae are species that infect plants in the Brassicaceae family, e.g. Brassica oleracea (da Cruz Cabral et al., 2016) and B. napus (Al-lami et al., 2020). Meanwhile, like A. dactylidicola, A. tenuissima infects crops and grasses in the Poaceae family including wheat and Dactylis (Dang et al., 2015; Thambugala et al., 2017). Other species detected by the ITS2 barcode included A. argyranthemi, A. armoraciae, A. eichhorniae, A. mimicula, A. molesta and A. rosae (Fig. 2d). BLAST search and threshold distance of species hypotheses also confirmed that A. alternata, A. armoraciae, A. eichhorniae, A. rosae and A. mimicula were among the Alternaria species in the air of Worcester, Lakeside Container and Lakeside Circle (Table S4b ii). A. alternata and A. eichhorniae belong to the section Alternata while A. mimicula and A. molesta belong to sections Brassicicola and Phragmosporae, respectively, and A. argyranthemi remains undefined (Woudenberg et al., 2013). The BLAST search also detected A. solani in the air samples. A. solani causes early blight in tomatoes and potatoes and therefore could have originated from the intensive horticultural farms and allotments (Agriculture & Horticulture Development Board, 2015; Apangu et al., 2020). Although the intensive farms manage the early blight through fungicide applications at earlier growth stages, such applications do not affect Alternaria sporulation at maturing and senescence stage of the plants (Skjøth et al., 2012). A. eichhorniae was detected at all the three sites and in all years of observation but was most abundant at Worcester in 2018. A. eichhorniae infects water hyacinth (Eichhornia crassipes) (Shabana et al., 2001) and parasitises other species of water hyacinth such as E. azurea, E. diversifolia, E. heterosperma, E. natans which are phylogenetically related (Cook, 1998; Pellegrini et al., 2018). Although E. crassipes is native to the Amazon basin, it was introduced as an ornamental plant (and grown in ponds) in more than 50 countries including the UK before it was banned by the European Union in 2016 (Patoka et al., 2016). Cases of allergy related to Alternaria and other allergens were previously reported in florist shops in Worcester (Emberlin et al., 2004). The detection of A. eichhorniae in the air samples suggests that E. crassipes weed still exists in ponds, lakes and rivers around Worcester and Lakeside and A. eichhorniae spores were possibly passively dispersed by the wind from such sources. Lakeside sampling site has two nearby Lakes (Fig. 1) and several ponds while River Severn (Fig. 1) flows through Worcester, which can all harbour E. crassipes, the host of A. eichhorniae. Studies have shown that thousands of invasive Eichhornia species including E. crassipes have been introduced to freshwater ecosystems worldwide (Johansson et al., 2018; Scriver et al., 2015). Another possibility could be that A. eichhorniae spores were transported from other remote sources to the UK, as seen with soybean rust spores being transported for over 1000 km in the USA (Isard et al., 2005, 2007). This also demonstrates the robustness of the DADA2 pipeline in detecting rare species of fungi (Nearing et al., 2018). A. mimicula causes early blight in tomato plants (Lycopersicon esculentum) (Ramezani et al., 2019; Woudenberg et al., 2013). A. molesta causes skin lesions on Harbour porpoise (Phocoena phocoena), a marine mammal that lives in coastal areas and river estuaries (Mamgain et al., 2013; Woudenberg et al., 2013). P. phocoena, being a protected cetacean among endangered species in the UK and EU, are abundant in British waters (Roberts et al., 2019). Magyar et al. (2016) explained that, through the “spiralling” process, suspended fungal propagules in flowing waters can be transported some distance (200–1000 m) before settling on leaves or other substrates at riverbanks or streambanks and eventually passively dispersed into the air. Fungal asexual spores of aquatic hyphomycetes have been found to use such dispersal pathways (Duarte et al., 2012; Magyar et al., 2016; Thomas et al., 1991). The River Severn and Bristol Channel are the possible habitats of P. phocoena nearest to Worcester and Lakeside. It is, therefore, possible that P. phocoena could have shed A. molesta spores into the river, which are later deposited at the riverbanks by spring bore tides and eventually passively dispersed into the air and detected at Worcester and Lakeside. Apart from seawater and seawater animals, e.g. P. phocoena, strains of A. molesta have also been found in soils and seawater plants but not land plants (Lawrence et al., 2016). A. rosae is a species in the section Pseudoalternaria (Lawrence et al., 2016). It causes black head mould in wheat and barley and infects sweet briar (Rosa rubiginosa) rose plants (Poursafar et al., 2018). A. rosae is also associated with locoweeds Astragalus variabilis and Sphaerophysa salsula that are commonly found in grazing grasslands and are toxic to animals (Lu et al., 2017). Overall, A. infectoria, A. metachromatica, A. brassicae, A. eichhorniae, A. mimicula, A. rosae, A. armoraciae and A. alternata were the main pathogenic Alternaria species detected in the air of Worcester, Lakeside Container and Lakeside Circle. A. infectoria and A. metachromatica were the most abundant pathogenic species.
4.2 Allergenic Alternaria species
The allergenicity of Alternaria spores to sensitised individuals is well documented (D’Amato et al., 1997). High cases of prescriptions for allergic rhinitis and asthma-related hospital admissions were previously reported from the population living in Worcester (including Lakeside) in the past (Emberlin & Lewis, 2006; Rowney et al., 2021; Watson et al., 1996). Phylogenetic analysis of Alternaria spores based on Alt a 1 allergen gene sequence shows that A. tenuissima is closely related with A. alternata while A. metachromatica is closely linked with A. infectoria and all are allergenic species (Hong et al., 2005). Other allergenic Alternaria species in this study include A. mimicula and A. argyranthemi (Hong et al., 2005). A. rosae causes cutaneous infection in humans (Liu et al., 2017). Meanwhile, no study has investigated the allergenic properties in A. dactylidicola, A. eichhorniae, A. molesta and A. armoraciae.
4.3 Similarities and differences between ITS1 and ITS2 barcodes
Although the ITS1 and ITS2 barcodes had similar fungal composition and richness at phylum level and UNITE database correctly identified all the fungi (and plants) in the air samples and mock community from kingdom to genus level (Table S5; Fig. S1a–d), there were notable variations at the species level (Table S5; Fig. S2a–d). For instance, A. alternata was incorrectly identified as A. tenuissima but correctly identified by BLAST search. Moreover, the ITS1 barcode differed from the ITS2 barcode in Alternaria species identification, apart from A. armoraciae. Such misidentifications and barcode differences are attributed to the quality of the reference databases and the biases and artefacts associated with the bioinformatics pipelines and these were also previously recognised (Abrego et al., 2018; Aguayo et al., 2021; Pauvert et al., 2019; Piper et al., 2019; Vasar et al., 2021). However, the BLAST search of the ASV sequences was performed to identify the exact taxonomy of the sequences and minimise the errors from the bioinformatics pipeline. The misidentifications in Alternaria spp. are also partly attributed to the several taxonomic re-descriptions of the 275 known Alternaria species, e.g. 32 new combinations, 16 new species and 10 old names resurrected, resulting in the addition of new species and transfer of some species to other genera, e.g. Prathoda (Simmons, 2007; Woudenberg et al., 2013). Alternaria morphospecies (300), indistinguishable using multi-gene phylogeny due to environmental conditions, are synonymised under A. alternata (He et al., 2021; Woudenberg et al. (2015), suggesting that some of the Alternaria species closely related with A. alternata could have been identified as other related species or as A. alternata. These findings suggest that taxonomic resolution of Alternaria species remains to be fully addressed for a more accurate molecular identification of Alternaria species in the future. de Vere et al. (2012) and Bulman et al. (2018) also recognised the uncertainty in species discrimination and emphasised that correct species identification of microorganisms is largely dependent on the quality of sequences in the reference databases.
5 Conclusion
The results revealed several Alternaria species detected in the air of Worcester and Lakeside. These included A. dactylidicola, A. metachromatica, A. brassicae, A. tenuissima and A. armoraciae from ITS1 barcoding. Meanwhile, ITS2 barcoding revealed A. infectoria, A. alternata, A. argyranthemi, A. armoraciae, A. molesta, A. mimicula, A. rosae and A. eichhorniae. Some of the species, e.g. A. dactylidicola, A. metachromatica, A. armoraciae, A. argyranthemi, A. armoraciae, A. molesta, A. mimicula, A. rosae and A. eichhorniae are uncommon species detected for the first time in Worcester and Lakeside. A. dactylidicola and A. metachromatica were highly abundant in Worcester and Lakeside. The BLAST search and species hypothesis confirmed 66.67% of the above species (including A. alternata). Whereas the three sites exhibited a significant species diversity at each site (Shannon and Simpson indices), there was a minimum difference (PERMANOVA) in species diversity between the sites due to the uniformity of habitats in the three sites, the proximity of the sites to each other and genetic mix of the airborne spores before they are captured at the traps.
References
Abarenkov, K., Zirk, A., Piirmann, T., Pöhönen, R., Ivanov, F., Nilsson, R. H. & Kõljalg, U. (2020b). UNITE general FASTA release for Fungi. Version 04.02.2020b. UNITE Community.
Abarenkov, K., Zirk, A., Piirmann, T., Pöhönen, R., Ivanov, F., Nilsson, R. H. & Kõljalg, U. (2020a). UNITE general FASTA release for Eukaryotes. Version 04.02.2020a. UNITE Community.
Abrego, N., Norros, V., Halme, P., Somervuo, P., Ali-Kovero, H., & Ovaskainen, O. (2018). Give me a sample of air and I will tell which species are found from your region: Molecular identification of fungi from airborne spore samples. Molecular Ecology Resources, 18(3), 511–524. https://doi.org/10.1111/1755-0998.12755
Adams-Groom, B., Emberlin, J., Corden, J., Millington, W., & Mullins, J. (2002). Predicting the start of the birch pollen season at London, Derby and Cardiff, United Kingdom, using a multiple regression model, based on data from 1987 to 1997. Aerobiologia, 18(2), 117–123. https://doi.org/10.1023/A:1020698023134
Agriculture and Horticulture Development Board. (2015). GB end-November stocks up 23% on previous season. https://potatoes.ahdb.org.uk/publications/gb-end-november-stocks-23-previous-season
Aguayo, J., Fourrier-Jeandel, C., Husson, C., & Loos, R. (2018). Assessment of Passive Traps Combined with High-Throughput. Applied and Environmental Microbiology, 84(11), 1–17. https://doi.org/10.1128/AEM.02637-17
Aguayo, J., Husson, C., Chancerel, E., Fabreguettes, O., Chandelier, A., Fourrier-Jeandel, C., Dupuy, N., Dutech, C., Ioos, R., Robin, C., Thibaudon, M., Marçais, B., & Desprez-Loustau, M. L. (2021). Combining permanent aerobiological networks and molecular analyses for large-scale surveillance of forest fungal pathogens: A proof-of-concept. Plant Pathology, 70(1), 181–194. https://doi.org/10.1111/ppa.13265
Al-lami, H. F. D., You, M. P., Mohammed, A. E., & Barbetti, M. J. (2020). Virulence variability across the Alternaria spp. population determines incidence and severity of Alternaria leaf spot on rapeseed. Plant Pathology, 69(3), 506–517. https://doi.org/10.1111/ppa.13135
Andersen, B., Krøger, E., & Roberts, R. G. (2002). Chemical and morphological segregation of Alternaria arborescens, A. infectoria and A. tenuissima species-groups. Mycological Research, 106(2), 170–182. https://doi.org/10.1017/S0953756201005263
Anslan, S., Nilsson, R. H., Wurzbacher, C., Baldrian, P., Tedersoo, L., & Bahram, M. (2018). Great differences in performance and outcome of high-throughput sequencing data analysis platforms for fungal metabarcoding. MycoKeys, 39, 29–40. https://doi.org/10.3897/mycokeys.39.28109
Apangu, G. P., Frisk, C. A., Adams-Groom, B., Satchwell, J., Pashley, C. H., & Skjøth, C. A. (2020). Air mass trajectories and land cover map reveal cereals and oilseed rape as major local sources of Alternaria spores in the Midlands, UK. Atmospheric Pollution Research, 11(9), 1668–1679. https://doi.org/10.1016/j.apr.2020.06.026
Archer, S. D. J., Lee, K. C., Caruso, T., King-Miaow, K., Harvey, M., Huang, D., Wainwright, B. J., & Pointing, S. B. (2020). Air mass source determines airborne microbial diversity at the ocean–atmosphere interface of the Great Barrier Reef marine ecosystem. ISME Journal, 14(3), 871–876. https://doi.org/10.1038/s41396-019-0555-0
Astray, G., Rodríguez-Rajo, F. J., Ferreiro-Lage, J. A., Fernández-González, M., Jato, V., & Mejuto, J. C. (2010). The use of artificial neural networks to forecast biological atmospheric allergens or pathogens only as Alternaria spores. Journal of Environmental Monitoring, 12(11), 2145. https://doi.org/10.1039/c0em00248h
Awad, A. H. A. (2005). Vegetation: A source of air fungal bio-contaminant. Aerobiologia, 21(1), 53–61. https://doi.org/10.1007/s10453-004-5878-1
BAF. (1995). Airborne pollens and spores: A guide to trapping and counting. Harpenden: BAF The British Aerobiology Federation.
Banchi, E., Ametrano, C. G., Greco, S., Stanković, D., Muggia, L., & Pallavicini, A. (2020a). PLANiTS: A curated sequence reference dataset for plant ITS DNA metabarcoding. Database, 2020, 1–9. https://doi.org/10.1093/database/baz155
Banchi, E., Ametrano, C. G., Stanković, D., Verardo, P., Moretti, O., Gabrielli, F., Lazzarin, S., Borney, M. F., Tassan, F., Tretiach, M., Pallavicini, A., & Muggia, L. (2018). DNA metabarcoding uncovers fungal diversity of mixed airborne samples in Italy. PLoS ONE, 13(3), 1–20. https://doi.org/10.1371/journal.pone.0194489
Banchi, E., Ametrano, C. G., Tordoni, E., Stanković, D., Ongaro, S., Tretiach, M., Pallavicini, A., Muggia, L., Verardo, P., Tassan, F., Trobiani, N., Moretti, O., Borney, M. F., & Lazzarin, S. (2020b). Environmental DNA assessment of airborne plant and fungal seasonal diversity. Science of the Total Environment, 738, 1–14. https://doi.org/10.1016/j.scitotenv.2020.140249
Bashir, U., Mushtaq, S., & Akhtar, N. (2014). First report of Alternaria metachromatica from Pakistan causing leaf spot of tomato. Pakistan Journal of Agricultural Sciences, 51(2), 315–318.
Be, N. A., Thissen, J. B., Fofanov, V. Y., Allen, J. E., Rojas, M., Golovko, G., Fofanov, Y., Koshinsky, H., & Jaing, C. J. (2015). Metagenomic analysis of the airborne environment in urban spaces. Microbial Ecology, 69(2), 346–355. https://doi.org/10.1007/s00248-014-0517-z
Bessadat, N., Berruyer, R., Hamon, B., Bataille-Simoneau, N., Benichou, S., Kihal, M., Henni, D. E., & Simoneau, P. (2017). Alternaria species associated with early blight epidemics on tomato and other Solanaceae crops in northwestern Algeria. European Journal of Plant Pathology, 148(1), 181–197. https://doi.org/10.1007/s10658-016-1081-9
Bowers, R. M., Clements, N., Emerson, J. B., Wiedinmyer, C., Hannigan, M. P., & Fierer, N. (2013). Seasonal variability in bacterial and fungal diversity of the near-surface atmosphere. Environmental Science and Technology, 47(21), 12097–12106. https://doi.org/10.1021/es402970s
Brennan, G. L., Potter, C., de Vere, N., Griffith, G. W., Skjøth, C. A., Osborne, N. J., Wheeler, B. W., McInnes, R. N., Clewlow, Y., Barber, A., Hanlon, H. M., Hegarty, M., Jones, L., Kurganskiy, A., Rowney, F. M., Armitage, C., Adams-Groom, B., Ford, C. R., Petch, G. M., The PollerGEN Consortium and Creer, S. (2019). Temperate airborne grass pollen defined by spatio-temporal shifts in community composition. Nature Ecology and Evolution, 3, 750–754. https://doi.org/10.1038/s41559-019-0849-7
Broennimann, O., Mráz, P., Petitpierre, B., Guisan, A., & Müller-Schärer, H. (2014). Contrasting spatio-temporal climatic niche dynamics during the Eastern and Western invasions of spotted knapweed in North America. Journal of Biogeography, 41(6), 1126–1136. https://doi.org/10.1111/jbi.12274
Bulman, S. R., McDougal, R. L., Hill, K., & Lear, G. (2018). Opportunities and limitations for DNA metabarcoding in Australasian plant-pathogen biosecurity. Australasian Plant Pathology, 47(5), 467–474. https://doi.org/10.1007/s13313-018-0579-3
Callahan, B. J., McMurdie, P. J., & Holmes, S. P. (2017). Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME Journal, 11(12), 2639–2643. https://doi.org/10.1038/ismej.2017.119
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., & Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods, 13(7), 581–583. https://doi.org/10.1038/nmeth.3869
Chen, L., Fang, K., Dong, X. F., Yang, A. L., Li, Y. X., & Zhang, H. B. (2020). Characterization of the fungal community in the canopy air of the invasive plant Ageratina adenophora and its potential to cause plant diseases. PLoS ONE, 15(3), 1–16. https://doi.org/10.1371/journal.pone.0230822
Clasen, L. A., Detheridge, A. P., Scullion, J., & Griffith, G. W. (2020). Soil stabilisation for DNA metabarcoding of plants and fungi. Implications for sampling at remote locations or via third-parties. Metabarcoding and Metagenomics, 4, 135–147. https://doi.org/10.3897/MBMG.4.58365
Comtois, P., Morand, S., Infante-Rivard, C., Gautrin, D., Vanderplass, O., & Malo, J. L. (1995). Exposure to spores during mowing: A comparative assessment of workers, parks and town. Aerobiologia, 11(2), 145–150. https://doi.org/10.1007/BF02738280
Cook, C. D. K. (1998). Pontederiaceae. In K. Kubitzki (Ed.), Flowering plants. Monocotyledons. The families and genera of vascular Plants (pp. 395–403). Berlin: Springer. https://doi.org/10.1007/978-3-662-03531-3_37
Corden, J. M., Millington, W. M., & Mullins, J. (2003). Long-term trends and regional variation in the aeroallergen Alternaria in Cardiff and Derby UK—are differences in climate and cereal production having an effect? Aerobiologia, 19, 191–199.
Crandall, S. G., & Gilbert, G. S. (2017). Meteorological factors associated with abundance of airborne fungal spores over natural vegetation. Atmospheric Environment, 162, 87–99. https://doi.org/10.1016/j.atmosenv.2017.05.018
D’Amato, G., Chatzigeorgiou, G., Corsico, R., Gioulekas, D., Jäger, L., Jäger, S., Kontou-Fili, K., Kouridakis, S., Liccardi, G., Meriggi, A., Palma-Carlos, A., Palma-Carlos, M. L., Pagan Aleman, A., Parmiani, S., Puccinelli, P., Russo, M., Spieksma, F. T., Torricelli, R., & Wüthrich, B. (1997). Evaluation of the prevalence of skin prick test positivity to Alternaria and Cladosporium in patients with suspected respiratory allergy. A European multicenter study promoted by the Subcommittee on Aerobiology and Environmental Aspects of Inhalant Allerg. Allergy, 52(7), 711–716.
D’Amato, G., De Palma, R., Verga, A., Martucci, P., Liccardi, G., & Lobefalo, G. (1991). Antigenic activity of nonpollen parts (leaves and stems) of allergenic plants (Parietaria judaica and Dactylis glomerata). Annals of Allergy, Asthma & Immunology, 67, 421–424.
d’Entremont, T. W., Migicovsky, Z., López-Gutiérrez, J. C., & Walker, A. K. (2020). Saltmarsh rhizosphere fungal communities vary by sediment type and dominant plant species cover in Nova Scotia, Canada. Environmental Microbiology Reports, 00(00), 1–6. https://doi.org/10.1111/1758-2229.12904
da Cruz Cabral, L., Terminiello, L., Fernández Pinto, V., Fog Nielsen, K., & Patriarca, A. (2016). Natural occurrence of mycotoxins and toxigenic capacity of Alternaria strains from mouldy peppers. International Journal of Food Microbiology, 236, 155–160. https://doi.org/10.1016/j.ijfoodmicro.2016.08.005
da Silva, T. H., Câmara, P. E. A. S., Pinto, O. H. B., Carvalho-Silva, M., Oliveira, F. S., Convey, P., Rosa, C. A., & Rosa, L. H. (2021). Diversity of fungi present in permafrost in the South Shetland Islands, Maritime Antarctic. Microbial Ecology. https://doi.org/10.1007/s00248-021-01735-6
Dang, H. X., Pryor, B., Peever, T., & Lawrence, C. B. (2015). The Alternaria genomes database: A comprehensive resource for a fungal genus comprised of saprophytes, plant pathogens, and allergenic species. BMC Genomics, 16(1), 1–9. https://doi.org/10.1186/s12864-015-1430-7
Dannemiller, K. C., Mendell, M. J., Macher, J. M., Kumagai, K., Bradman, A., Holland, N., Harley, K., Eskenazi, B., & Peccia, J. (2014). Next-generation DNA sequencing reveals that low fungal diversity in house dust is associated with childhood asthma development. Indoor Air, 24, 236–247. https://doi.org/10.1111/ina.12072
de Souza, L. M. D., Ogaki, M. B., Câmara, P. E. A. S., Pinto, O. H. B., Convey, P., Carvalho-Silva, M., Rosa, C. A., & Rosa, L. H. (2021). Assessment of fungal diversity present in lakes of Maritime Antarctica using DNA metabarcoding: A temporal microcosm experiment. Extremophiles, 25(1), 77–84. https://doi.org/10.1007/s00792-020-01212-x
de Vere, N., Rich, T. C. G., Ford, C. R., Trinder, S. A., Long, C., Moore, C. W., Satterthwaite, D., Davies, H., Allainguillaume, J., Ronca, S., Tatarinova, T., Garbett, H., Walker, K., & Wilkinson, M. J. (2012). DNA barcoding the native flowering plants and conifers of wales. PLoS ONE, 7(6), 1–12. https://doi.org/10.1371/journal.pone.0037945
de Vries, F. T., Bloem, J., van Eekeren, N., Brusaard, L., & Hoffland, E. (2007). Fungal biomass in pastures increases with age and reduced N input. Soil Biology and Biochemistry, 39(7), 1620–1630. https://doi.org/10.1016/j.soilbio.2007.01.013
Degois, J., Simon, X., Bontemps, C., Leblond, P., & Duquenne, P. (2019). Characterization of experimental complex fungal bioaerosols: Impact of analytical method on fungal composition measurements. Aerosol Science and Technology, 53(2), 146–159. https://doi.org/10.1080/02786826.2018.1557320
Duarte, S., Seena, S., Bärlocher, F., Cássio, F., & Pascoal, C. (2012). Preliminary insights into the phylogeography of six aquatic hyphomycete species. PLoS ONE, 7(9), 1–7. https://doi.org/10.1371/journal.pone.0045289
Dyda, M., Pyzik, A., Wilkojc, E., Kwiatkowska-Kopka, B., & Sklodowska, A. (2019). Bacterial and fungal diversity inside the medieval building constructed with sandstone plates and lime mortar as an example of the microbial colonization of a nutrient-limited extreme environment (Wawel Royal Castle, Krakow, Poland). Microorganisms, 7(416), 1–17. https://doi.org/10.3390/microorganisms7100416
Emberlin, J., Adamsgroom, B., Treu, R., & Carswell, F. (2004). Airborne pollen and fungal spores in Florist shops in Worcester and in Bristol, UK: A potential problem for occupational health. Aerobiologia, 20, 153–160. https://doi.org/10.1023/B:AERO.0000032952.99736.f5
Emberlin, J. C., & Lewis, R. A. (2006). A double blind, placebo controlled trial of inert cellulose powder for the relief of symptoms of hay fever in adults. Current Medical Research and Opinion, 22(2), 275–285. https://doi.org/10.1185/030079906X80440
Ettenauer, J. D., Piñar, G., Lopandic, K., Spangl, B., Ellersdorfer, G., Voitl, C., & Sterflinger, K. (2012). Microbes on building materials—Evaluation of DNA extraction protocols as common basis for molecular analysis. Science of the Total Environment, 439, 44–53. https://doi.org/10.1016/j.scitotenv.2012.09.005
Fort, T., Robin, C., Capdevielle, X., Delière, L., & Vacher, C. (2016). Foliar fungal communities strongly differ between habitat patches in a landscape mosaic. PeerJ, 2016(11), 1–20. https://doi.org/10.7717/peerj.2656
Frisk, C. A., Adams-groom, B., & Skjøth, C. A. (2021). Stochastic flowering phenology in Dactylis glomerata populations described by Markov chain modelling. Aerobiologia, 2021, 1–16. https://doi.org/10.1007/s10453-020-09685-1
Fröhlich-Nowoisky, J., Burrows, S. M., Xie, Z., Engling, G., Solomon, P. A., Fraser, M. P., Mayol-Bracero, O. L., Artaxo, P., Begerow, D., Conrad, R., Andreae, M. O., Després, V. R., & Pöschl, U. (2012). Biogeography in the air: Fungal diversity over land and oceans. Biogeosciences, 9(3), 1125–1136. https://doi.org/10.5194/bg-9-1125-2012
Fröhlich-Nowoisky, J., Kampf, C. J., Weber, B., Huffman, J. A., Pöhlker, C., Andreae, M. O., Lang-Yona, N., Burrows, S. M., Gunthe, S. S., Elbert, W., Su, H., Hoor, P., Thines, E., Hoffmann, T., Després, V. R., & Pöschl, U. (2016). Bioaerosols in the Earth system: Climate, health, and ecosystem interactions. Atmospheric Research, 182, 346–376. https://doi.org/10.1016/j.atmosres.2016.07.018
Gabriel, M. F., Postigo, I., Gutiérrez-Rodríguez, A., Suñén, E., Tomaz, C. T., & Martínez, J. (2015). Development of a PCR-based tool for detecting immunologically relevant Alt a 1 and Alt a 1 homologue coding sequences. Medical Mycology, 53, 636–642. https://doi.org/10.1093/mmy/myv022
Galán, C., Smith, M., Thibaudon, M., Frenguelli, G., Oteros, J., Gehrig, R., Berger, U., Clot, B., & Brandao, R. (2014). Pollen monitoring: Minimum requirements and reproducibility of analysis. Aerobiologia, 30(4), 385–395. https://doi.org/10.1007/s10453-014-9335-5
Grinn-Gofroń, A., Bosiacka, B., Bednarz, A., & Wolski, T. (2018). A comparative study of hourly and daily relationships between selected meteorological parameters and airborne fungal spore composition. Aerobiologia, 34, 45–54. https://doi.org/10.1007/s10453-017-9493-3
Grinn-Gofroń, A., Nowosad, J., Bosiacka, B., Camacho, I., Pashley, C., Belmonte, J., De Linares, C., Ianovici, N., Manzano, J. M. M., Sadyś, M., Skjøth, C., Rodinkova, V., Tormo-Molina, R., Vokou, D., Fernández-Rodríguez, S., & Damialis, A. (2019). Airborne Alternaria and Cladosporium fungal spores in Europe: Forecasting possibilities and relationships with meteorological parameters. Science of the Total Environment, 653, 938–946. https://doi.org/10.1016/j.scitotenv.2018.10.419
He, L., Cheng, H., Zhao, L., Htun, A. A., Yu, Z. H., Deng, J. X., & Li, Q. L. (2021). Morphological and molecular identification of two new Alternaria species (Ascomycota, Pleosporaceae) in section Radicina from China. MycoKeys 78, 187–198. https://doi.org/10.3897/mycokeys.78.64853
Hebert, P. D. N., Cywinska, A., Ball, S. L., & DeWaard, J. R. (2003). Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences, 270(1512), 313–321. https://doi.org/10.1098/rspb.2002.2218
Heeger, F., Bourne, E. C., Baschien, C., Yurkov, A., Bunk, B., Spröer, C., Overmann, J., Mazzoni, C. J., & Monaghan, M. T. (2018). Long-read DNA metabarcoding of ribosomal RNA in the analysis of fungi from aquatic environments. Molecular Ecology Resources, 18(6), 1500–1514. https://doi.org/10.1111/1755-0998.12937
Hirst, J. M. (1952). An automatic volumetric spore trap. Annals of Applied Biology, 39(2), 257–265. https://doi.org/10.1111/j.1744-7348.1952.tb00904.x
Hong, S. G., Cramer, R. A., Lawrence, C. B., & Pryor, B. M. (2005). Alt a 1 allergen homologs from Alternaria and related taxa: Analysis of phylogenetic content and secondary structure. Fungal Genetics and Biology, 42(2), 119–129. https://doi.org/10.1016/j.fgb.2004.10.009
Irga, P. J., & Torpy, F. R. (2016). A survey of the aeromycota of Sydney and its correspondence with environmental conditions: Grass as a component of urban forestry could be a major determinant. Aerobiologia, 32(2), 171–185. https://doi.org/10.1007/s10453-015-9388-0
Isard, S. A., Gage, S. H., Comtois, P., & Russo, J. M. (2005). Principles of the atmospheric pathway for invasive species applied to soybean rust. BioScience, 55(10), 851–861. https://doi.org/10.1641/0006-3568(2005)055[0851:POTAPF]2.0.CO;2
Isard, S. A., Russo, J. M., & Ariatti, A. (2007). The integrated aerobiology modeling system applied to the spread of soybean rust into the Ohio River valley during September 2006. Aerobiologia, 23(4), 271–282. https://doi.org/10.1007/s10453-007-9073-z
Jefferson, R. G. (2005). The conservation management of upland hay meadows in Britain: A review. Grass and Forage Science, 60(4), 322–331. https://doi.org/10.1111/j.1365-2494.2005.00489.x
Johansson, M. L., Dufour, B. A., Wellband, K. W., Corkum, L. D., MacIsaac, H. J., & Heath, D. D. (2018). Human-mediated and natural dispersal of an invasive fish in the eastern Great Lakes. Heredity, 120(6), 533–546. https://doi.org/10.1038/s41437-017-0038-x
Joly, M., & Peuch, V. H. (2012). Objective classification of air quality monitoring sites over Europe. Atmospheric Environment, 47, 111–123. https://doi.org/10.1016/j.atmosenv.2011.11.025
Kasprzyk, I. (2008). Non-native Ambrosia pollen in the atmosphere of Rzesz (SE Poland); Evaluation of the effect of weather conditions on daily concentrations and starting dates of the pollen season. International Journal of Biometeorology, 52(5), 341–351. https://doi.org/10.1007/s00484-007-0129-0
Kasprzyk, I., Grinn-Gofroń, A., Ćwik, A., Kluska, K., Cariñanos, P., & Wójcik, T. (2021). Allergenic fungal spores in the air of urban parks. Aerobiologia, 37(1), 39–51. https://doi.org/10.1007/s10453-020-09671-7
Kilic, M., Ufuk Altintas, D., Yilmaz, M., Güneşer Kendirli, S., Bingöl Karakoc, G., Taskin, E., Ceter, T., & Pinar, N. M. (2010). The effects of meteorological factors and Alternaria spore concentrations on children sensitised to Alternaria. Allergologia Et Immunopathologia, 38(3), 122–128. https://doi.org/10.1016/j.aller.2009.09.006
Kumari, P., Woo, C., Yamamoto, N., & Choi, H. L. (2016). Variations in abundance, diversity and community composition of airborne fungi in swine houses across seasons. Scientific Reports, 6(37929), 1–11. https://doi.org/10.1038/srep37929
Lacey, J., & Allit, U. (1995). Airborne pollen and spores: A guide to trapping and counting. Harpenden: The British Aerobiology Federation.
Lee, H. B., Patriarca, A., & Magan, N. (2015). Alternaria in food: Ecophysiology, mycotoxin production and toxicology. Mycobiology, 43(2), 93–106. https://doi.org/10.5941/MYCO.2015.43.2.93.
Lawrence, D. P., Rotondo, F., & Gannibal, P. B. (2016). Biodiversity and taxonomy of the pleomorphic genus Alternaria. Mycological Progress, 15(3), 1–22. https://doi.org/10.1007/s11557-015-1144-x
Leppänen, H. K., Täubel, M., Jayaprakash, B., Vepsäläinen, A., Pasanen, P., & Hyvärinen, A. (2018). Quantitative assessment of microbes from samples of indoor air and dust. Journal of Exposure Science and Environmental Epidemiology, 28(3), 231–241. https://doi.org/10.1038/jes.2017.24
Leuschner, C., Gebel, S., & Rose, L. (2013). Root trait responses of six temperate grassland species to intensive mowing and NPK fertilisation: A field study in a temperate grassland. Plant and Soil, 373, 687–698. https://doi.org/10.1007/s11104-013-1836-4
Lin, W. R., Wang, P. H., Tien, C. J., Chen, W. Y., Yu, Y. A., & Hsu, L. Y. (2018). Changes in airborne fungal flora along an urban to rural gradient. Journal of Aerosol Science, 116, 116–123. https://doi.org/10.1016/j.jaerosci.2017.11.010
Liu, A. W., Bateman, A. C., Greenbaum, A., Garvin, K., Clarridge, J., & Grim, J. (2017). Cutaneous phaeohyphomycosis in a hematopoietic stem cell transplant patient caused by Alternaria rosae: First case report. Transplant Infectious Disease, 19(3), 1–4. https://doi.org/10.1111/tid.12698
Lu, H., Haiyun, Q., Qiwu, Z., Zhenhui, R., Ruixu, X., Baoyu, Z., & Rebecca, C. (2017). Endogenous fungi isolated from three locoweed species from rangeland in western China. African Journal of Microbiology Research, 11(5), 155–170. https://doi.org/10.5897/ajmr2016.8392
Magyar, D., Vass, M. & Li, D.-W. (2016). Dispersal strategies of microfungi. In Li, D.-W (Eds.), Biology of microfungi (pp. 315–371). Springer. https://doi.org/10.1007/978-3-319-29137-6_14
Mamgain, A., Roychowdhury, R., & Tah, J. (2013). Review: Alternaria pathogenicity and its strategic controls. Research Journal of Biology, 1, 1–9.
Márquez, S. S., Bills, G. F., Acuña, L. D., & Zabalgogeazcoa, I. (2010). Endophytic mycobiota of leaves and roots of the grass Holcus lanatus. Fungal Diversity, 41, 115–123. https://doi.org/10.1007/s13225-009-0015-7
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet Journal, 17(1), 10–12. https://doi.org/10.14806/ej.17.1.200
Maude, R. B., & Humpherson-Jones, F. M. (1980). Studies on the seed-borne phases of dark leaf spot Alternaria brassicicola and grey leaf spot Alternaria brassicae of brassicas. Annals of Applied Biology, 55, 311–319. https://doi.org/10.1111/j.1744-7348.1980.tb04752.x
Mbareche, H., Veillette, M., Bilodeau, G. J., & Duchaine, C. (2018). Bioaerosol sampler choice should consider efficiency and ability of samplers to cover microbial diversity. Applied and Environmental Microbiology, 84(23), 1–22. https://doi.org/10.1128/AEM.01589-18
Mbareche, H., Veillette, M., Bilodeau, G., & Duchaine, C. (2020). Comparison of the performance of ITS1 and ITS2 as barcodes in amplicon-based sequencing of bioaerosols. PeerJ, 8(e8523), 1–36. https://doi.org/10.7717/peerj.8523
McMurdie, P. J., & Holmes, S. (2013). Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE, 8(4), e61217. https://doi.org/10.1371/journal.pone.0061217
Meena, M., Gupta, S. K., Swapnil, P., Zehra, A., Dubey, M. K., & Upadhyay, R. S. (2017a). Alternaria toxins: Potential virulence factors and genes related to pathogenesis. Frontiers in Microbiology, 8(1451), 1–14. https://doi.org/10.3389/fmicb.2017.01451
Meena, M., Swapnil, P., & Upadhyay, R. S. (2017b). Isolation, characterization and toxicological potential of Alternaria-mycotoxins (TeA, AOH and AME) in different Alternaria species from various regions of India. Scientific Reports, 7(1), 8777. https://doi.org/10.1038/s41598-017-09138-9
Mhuireach, G., Johnson, B. R., Altrichter, A. E., Ladau, J., Meadow, J. F., Pollard, K. S., & Green, J. L. (2016). Urban greenness influences airborne bacterial community composition. Science of the Total Environment, 571, 680–687. https://doi.org/10.1016/j.scitotenv.2016.07.037
Mitakakis, T. Z., Clift, A., & McGee, P. A. (2001). The effect of local cropping activities and weather on the airborne concentration of allergenic Alternaria spores in rural Australia. Grana, 40(4–5), 230–239. https://doi.org/10.1080/001731301317223268
Naveed, M., Herath, L., Moldrup, P., Arthur, E., Nicolaisen, M., Norgaard, T., Ferré, T. P. A., & de Jonge, L. W. (2016). Spatial variability of microbial richness and diversity and relationships with soil organic carbon, texture and structure across an agricultural field. Applied Soil Ecology, 103, 44–55. https://doi.org/10.1016/j.apsoil.2016.03.004
Nearing, J. T., Douglas, G. M., Comeau, A. M., & Langille, M. G. I. (2018). Denoising the Denoisers: An independent evaluation of microbiome sequence error- correction approaches. PeerJ, 2018(8), 1–22. https://doi.org/10.7717/peerj.5364
Nicolaisen, M., West, J. S., Sapkota, R., Canning, G. G. M., Schoen, C., & Justesen, A. F. (2017). Fungal communities including plant pathogens in near surface air are similar across northwestern Europe. Frontiers in Microbiology, 8(SEP), 1–11. https://doi.org/10.3389/fmicb.2017.01729
Nilsson, R. H., Anslan, S., Bahram, M., Wurzbacher, C., Baldrian, P., & Tedersoo, L. (2019a). Mycobiome diversity: High-throughput sequencing and identification of fungi. Nature Reviews Microbiology, 17(2), 95–109. https://doi.org/10.1038/s41579-018-0116-y
Nilsson, R. H., Larsson, K. H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., Kennedy, P., Picard, K., Glöckner, F. O., Tedersoo, L., Saar, I., Kõljalg, U., & Abarenkov, K. (2019b). The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research, 47(D1), D259–D264. https://doi.org/10.1093/nar/gky1022
Nilsson, R. H., Wurzbacher, C., Bahram, M., Coimbra, V. R. M., Larsson, E., Tedersoo, L., Eriksson, J., Ritter, C. D., Svantesson, S., Sánchez-García, M., Ryberg, M., Kristiansson, E., & Abarenkov, K. (2016). Top 50 most wanted fungi. MycoKeys, 12, 29–40. https://doi.org/10.3897/mycokeys.12.7553
Nowicki, M., Nowakowska, M., Niezgoda, A., & Kozik, E. (2012). Alternaria black spot of Crucifers: Symptoms, importance of disease, and perspectives of resistance breeding. Vegetable Crops Research Bulletin, 76(1), 5–19. https://doi.org/10.2478/v10032-012-0001-6
O’Connor, D. J., Sadyś, M., Skjøth, C. A., Healy, D. A., Kennedy, R., & Sodeau, J. R. (2014). Atmospheric concentrations of Alternaria, Cladosporium, Ganoderma and Didymella spores monitored in Cork (Ireland) and Worcester (England) during the summer of 2010. Aerobiologia, 30(4), 397–411. https://doi.org/10.1007/s10453-014-9337-3
Ogaki, M. B., Câmara, P. E. A. S., Pinto, O. H. B., Lirio, J. M., Coria, S. H., Vieira, R., Carvalho-Silva, M., Convey, P., Rosa, C. A., & Rosa, L. H. (2021). Diversity of fungal DNA in lake sediments on Vega Island, north-east Antarctic Peninsula assessed using DNA metabarcoding. Extremophiles, 25(3), 257–265. https://doi.org/10.1007/s00792-021-01226-z
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R. & O’Hara, R. B. (2019). Community Ecology Package: package “vegan” v2.5–6. In CRAN (pp. 1–298).
Olsen, Y., Gosewinkel, U. B., Skjøth, C. A., Hertel, O., Rasmussen, K., & Sigsgaard, T. (2019). Regional variation in airborne Alternaria spore concentrations in Denmark through 2012–2015 seasons: The influence of meteorology and grain harvesting. Aerobiologia, 35(3), 533–551. https://doi.org/10.1007/s10453-019-09587-x
Paldy, A., Bobvos, J., Fazekas, B., Manyoki, G., Malnasi, T., & Magyar, D. (2014). Characterisation of the pollen season by using climate specific pollen indicators. European Journal of Occupational and Environmental Medicine, 20, 199–214.
Pashley, C. H., Fairs, A., Free, R. C., & Wardlaw, A. J. (2012). DNA analysis of outdoor air reveals a high degree of fungal diversity, temporal variability, and genera not seen by spore morphology. Fungal Biology, 116, 214–224. https://doi.org/10.1016/j.funbio.2011.11.004
Patoka, J., Bláha, M., Kalous, L., Vrabec, V., Buřič, M., & Kouba, A. (2016). Potential pest transfer mediated by international ornamental plant trade. Scientific Reports, 6, 1–6. https://doi.org/10.1038/srep25896
Pauvert, C., Buée, M., Laval, V., Edel-Hermann, V., Fauchery, L., Gautier, A., Lesur, I., Vallance, J., & Vacher, C. (2019). Bioinformatics matters: The accuracy of plant and soil fungal community data is highly dependent on the metabarcoding pipeline. Fungal Ecology, 41, 23–33. https://doi.org/10.1016/j.funeco.2019.03.005
Peay, K. G., & Bruns, T. D. (2014). Spore dispersal of basidiomycete fungi at the landscape scale is driven by stochastic and deterministic processes and generates variability in plant-fungal interactions. New Phytologist, 204(1), 180–191. https://doi.org/10.1111/nph.12906
Pellegrini, M. O. O., Horn, C. N., & Almeida, R. F. (2018). Total evidence phylogeny of Pontederiaceae (Commelinales) sheds light on the necessity of its recircumscription and synopsis of Pontederia L. PhytoKeys, 83(108), 25–83. https://doi.org/10.3897/phytokeys.108.27652
Piper, A. M., Batovska, J., Cogan, N. O. I., Weiss, J., Cunningham, J. P., Rodoni, B. C., & Blacket, M. J. (2019). Prospects and challenges of implementing DNA metabarcoding for high-throughput insect surveillance. GigaScience, 8(8), 1–22. https://doi.org/10.1093/gigascience/giz092
Poursafar, A., Ghosta, Y., Orina, A. S., Gannibal, P. B., Javan-Nikkhah, M., & Lawrence, D. P. (2018). Taxonomic study on Alternaria sections Infectoriae and Pseudoalternaria associated with black (sooty) head mold of wheat and barley in Iran. Mycological Progress, 17(3), 343–356. https://doi.org/10.1007/s11557-017-1358-1
Przemieniecki, S. W., Damszel, M., Kurowski, T. P., Mastalerz, J., & Kotlarz, K. (2019). Identification, ecological evaluation and phylogenetic analysis of non-symbiotic endophytic fungi colonizing timothy grass and perennial ryegrass grown in adjacent plots. Grass and Forage Science, 74(1), 42–52. https://doi.org/10.1111/gfs.12404
R Core Team. (2020). The R foundation for statistical computing platform. https://www.r-project.org/
Ramezani, Y., Taheri, P., & Mamarabadi, M. (2019). Identification of Alternaria spp. associated with tomato early blight in Iran and investigating some of their virulence factors. Journal of Plant Pathology, 101(3), 647–659. https://doi.org/10.1007/s42161-019-00259-w
Rathnayake, C. M., Metwali, N., Baker, Z., Jayarathne, T., Kostle, P. A., Thorne, P. S., Shaughnessy, P. T. O., & Stone, E. A. (2016). Urban enhancement of PM10 bioaerosol tracers relative to background locations in the Midwestern United States. Journal of Geophysical Research: Atmospheres, 121, 5071–5089. https://doi.org/10.1002/2015JD024538.Received
Roberts, L., Collier, S., Law, S., & Gaion, A. (2019). The impact of marine vessels on the presence and behaviour of harbour porpoise (Phocoena phocoena) in the waters off Berry Head, Brixham (South West England). Ocean and Coastal Management, 179(104860), 1–7. https://doi.org/10.1016/j.ocecoaman.2019.104860
Rodrigues, T. T. M. S., Berbee, M. L., Simmons, E. G., Cardoso, C. R., Reis, A., Maffia, L. A., & Mizubuti, E. S. G. (2010). First report of Alternaria tomatophila and A. grandis causing early blight on tomato and potato in Brazil. New Disease Reports. https://doi.org/10.5197/j.2044-0588.2010.022.028
Rolph, C. A., Gwyther, C. L., Tyrrel, S. F., Nasir, Z. A., Drew, G. H., Jackson, S. K., Khera, S., Hayes, E. T., Williams, B., Bennett, A., Collins, S., Walsh, K., Kinnersley, R., & Gladding, T. L. (2018). Sources of airborne endotoxins in ambient air and exposure of nearby communities—A review. Atmosphere, 9(375), 1–21. https://doi.org/10.3390/atmos9100375
Rosa, L. H., Pinto, O. H. B., Convey, P., Carvalho-Silva, M., Rosa, C. A., & Câmara, P. E. A. S. (2020a). DNA metabarcoding to assess the diversity of airborne fungi present over Keller Peninsula, King George Island, Antarctica. Microbial Ecology. https://doi.org/10.1007/s00248-020-01627-1
Rosa, L. H., Pinto, O. H. B., Šantl-Temkiv, T., Convey, P., Carvalho-Silva, M., Rosa, C. A., & Câmara, P. E. A. S. (2020b). DNA metabarcoding of fungal diversity in air and snow of Livingston Island, South Shetland Islands, Antarctica. Scientific Reports, 10(1), 1–11. https://doi.org/10.1038/s41598-020-78630-6
Rossmann, S., Lysøe, E., Skogen, M., Talgø, V., & Brurberg, M. B. (2021). DNA metabarcoding reveals broad presence of plant pathogenic oomycetes in soil from internationally traded plants. Frontiers in Microbiology, 12(637068), 1–12. https://doi.org/10.3389/fmicb.2021.637068
Rotem, J. (1994). The genus alternaria: Biology, epidemiology and pathogenicity. Minnesota: American Phytopathological Society.
Rowney, F. M., Brennan, G. L., Skjøth, C. A., Wheeler, B., Osborne, N. J., & Creer, S. (2021). Report Environmental DNA reveals links between abundance and composition of airborne grass pollen and respiratory health Environmental DNA reveals links between abundance and composition of airborne grass pollen and respiratory health. Current Biology, 31, 1–9. https://doi.org/10.1016/j.cub.2021.02.019
Sadyś, M., Adams-Groom, B., Herbert, R. J., & Kennedy, R. (2016). Comparisons of fungal spore distributions using air sampling at Worcester, England (2006–2010). Aerobiologia, 32(4), 619–634. https://doi.org/10.1007/s10453-016-9436-4
Sadyś, M., Skjøth, C. A., & Kennedy, R. (2014). Back-trajectories show export of airborne fungal spores (Ganoderma sp.) from forests to agricultural and urban areas in England. Atmospheric Environment, 84, 88–99. https://doi.org/10.1016/j.atmosenv.2013.11.015
Sadyś, M., Skjøth, C. A., & Kennedy, R. (2015). Determination of Alternaria spp. habitats using 7-day volumetric spore trap, Hybrid Single Particle Lagrangian Integrated Trajectory model and geographic information system. Urban Climate, 14, 429–440. https://doi.org/10.1016/j.uclim.2014.08.005
Sanchez, H., & Bush, R. K. (2001). A review of Alternaria alternata sensitivity. Revista Iberoamericana De Micologia, 18(2), 56–59.
Schiro, G., Colangeli, P., & Müller, M. E. H. (2019). A metabarcoding analysis of the mycobiome of wheat ears across a topographically heterogeneous field. Frontiers in Microbiology, 10(2095), 1–12. https://doi.org/10.3389/fmicb.2019.02095
Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. A., Chen, W., Bolchacova, E., Voigt, K., Crous, P. W., Miller, A. N., Wingfield, M. J., Aime, M. C., An, K. D., Bai, F. Y., Barreto, R. W., Begerow, D., Bergeron, M. J., Blackwell, M., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America, 109(16), 6241–6246. https://doi.org/10.1073/pnas.1117018109
Scriver, M., Marinich, A., Wilson, C., & Freeland, J. (2015). Development of species-specific environmental DNA (eDNA) markers for invasive aquatic plants. Aquatic Botany, 122, 27–31. https://doi.org/10.1016/j.aquabot.2015.01.003
Seifert, K. A., & Gams, W. (2011). The genera of Hyphomycetes—2011 update. Persoonia: Molecular Phylogeny and Evolution of Fungi, 27, 119–129. https://doi.org/10.3767/003158511X617435
Senés-Guerrero, C., & Schüßler, A. (2016). A conserved arbuscular mycorrhizal fungal core-species community colonizes potato roots in the Andes. Fungal Diversity, 77(1), 317–333. https://doi.org/10.1007/s13225-015-0328-7
Shabana, Y. M., Elwakil, M. A., & Charudattan, R. (2001). Effect of nutrition and physical factors on mycelial growth and production of pigments and nonchromatic UV-absorbing compounds of Alternaria eichhorniae. Journal of Phytopathology, 149(1), 21–27. https://doi.org/10.1046/j.1439-0434.2001.00564.x
Sharma, A., Clark, E., McGlothlin, J. D., & Mittal, S. K. (2015). Efficiency of Airborne Sample Analysis Platform (ASAP) bioaerosol sampler for pathogen detection. Frontiers in Microbiology, 6(512), 1–7. https://doi.org/10.3389/fmicb.2015.00512
Shokere, L. A., Holden, M. J., & Ronald Jenkins, G. (2009). Comparison of fluorometric and spectrophotometric DNA quantification for real-time quantitative PCR of degraded DNA. Food Control, 20(4), 391–401. https://doi.org/10.1016/j.foodcont.2008.07.009
Sicard, M., Izquierdo, R., Alarcón, M., Belmonte, J., Comerón, A., & Baldasano, J. M. (2016). Near-surface and columnar measurements with a micro pulse lidar of atmospheric pollen in Barcelona, Spain. Atmospheric Chemistry and Physics, 16(11), 6805–6821. https://doi.org/10.5194/acp-16-6805-2016
Simmons, E. G. (2007). Alternaria: An identification manual (vol. 6). CBS Biodiversity, CBS Fungal Biodiversity Centre.
Simmons, E. G. (1994). Alternaria themes and variations (106-111). Mycotaxon, 50, 409–427.
Skjøth, C. A., Baker, P., Sadyś, M., & Adams-Groom, B. (2015). Pollen from alder (Alnus sp.), birch (Betula sp.) and oak (Quercus sp.) in the UK originate from small woodlands. Urban Climate, 14, 414–428. https://doi.org/10.1016/j.uclim.2014.09.007
Skjøth, C. A., Damialis, A., Belmonte, J., De Linares, C., Fernández-Rodríguez, S., Grinn-Gofroń, A., Jędryczka, M., Kasprzyk, I., Magyar, D., Myszkowska, D., Oliver, G., Páldy, A., Pashley, C. H., Rasmussen, K., Satchwell, J., Thibaudon, M., Tormo-Molina, R., Vokou, D., Ziemianin, M., & Werner, M. (2016). Alternaria spores in the air across Europe: Abundance, seasonality and relationships with climate, meteorology and local environment. Aerobiologia, 32(1), 3–22. https://doi.org/10.1007/s10453-016-9426-6
Skjøth, C. A., Sommer, J., Brandt, J., Hvidberg, M., Geels, C., Hansen, K. M., Hertel, O., Frohn, L. M., & Christensen, J. H. (2008). Copenhagen—A significant source of birch (Betula) pollen? International Journal of Biometeorology, 52(6), 453–462. https://doi.org/10.1007/s00484-007-0139-y
Skjøth, C. A., Sommer, J., Frederiksen, L., & Gosewinkel Karlson, U. (2012). Crop harvest in Denmark and Central Europe contributes to the local load of airborne Alternaria spore concentrations in Copenhagen. Atmospheric Chemistry and Physics, 12(22), 11107–11123. https://doi.org/10.5194/acp-12-11107-2012
Smith, D. J., Jaffe, D. A., Birmele, M. N., Griffin, D. W., Schuerger, A. C., Hee, J., & Roberts, M. S. (2012). Free Tropospheric Transport of microorganisms from Asia to North America. Microbial Ecology, 64(4), 973–985. https://doi.org/10.1007/s00248-012-0088-9
Spellerberg, I. F., & Fedor, P. J. (2003). A tribute to Claude-Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the “Shannon-Wiener” Index. Global Ecology and Biogeography, 12(3), 177–179. https://doi.org/10.1046/j.1466-822X.2003.00015.x
Tedersoo, L., Tooming-Klunderud, A., & Anslan, S. (2018). PacBio metabarcoding of Fungi and other eukaryotes: Errors, biases and perspectives. New Phytologist, 217(3), 1370–1385. https://doi.org/10.1111/nph.14776
Thambugala, K. M., Wanasinghe, D. N., Phillips, A. J. L., Camporesi, E., Bulgakov, T. S., Phukhamsakda, C., Ariyawansa, H. A., Goonasekara, I. D., Phookamsak, R., Dissanayake, A., Tennakoon, D. S., Tibpromma, S., Chen, Y. Y., Liu, Z. Y., & Hyde, K. D. (2017). Mycosphere notes 1–50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere, 8(4), 697–796. https://doi.org/10.5943/MYCOSPHERE/8/4/13
Thomas, K., Chilvers, G. A., & Norris, R. H. (1991). A dynamic model of fungal spora in a freshwater stream. Mycological Research, 95(2), 184–188. https://doi.org/10.1016/S0953-7562(09)81009-5
Tordoni, E., Ametrano, C. G., Banchi, E., Ongaro, S., Pallavicini, A., Bacaro, G., & Muggia, L. (2021). Integrated eDNA metabarcoding and morphological analyses assess spatio-temporal patterns of airborne fungal spores. Ecological Indicators, 121(107032), 1–8. https://doi.org/10.1016/j.ecolind.2020.107032
Vasar, M., Davison, J., Neuenkamp, L., Sepp, S., Young, J. P. W., Moora, M., & Öpik, M. (2021). User-friendly bioinformatics pipeline gDAT (graphical downstream analysis tool) for analysing rDNA sequences. Molecular Ecology Resources, 21, 1380–1392. https://doi.org/10.1111/1755-0998.13340
von Mutius, E. (2008). Rhinitis as predictor of adult-onset asthma. The Lancet, 372(9643), 1012–1014. https://doi.org/10.1016/S0140-6736(08)61418-X
Wady, L., Shehabi, A., Szponar, B., Pehrson, C., Sheng, Y., & Larsson, L. (2004). Heterogeneity in microbial exposure in schools in Sweden, Poland and Jordan revealed by analysis of chemical markers. Journal of Exposure Analysis and Environmental Epidemiology, 14(4), 293–299. https://doi.org/10.1038/sj.jea.7500324
Watson, J. P., Cowen, P., & Lewis, R. A. (1996). The relationship between asthma admission rates, routes of admission, and socioeconomic deprivation. European Respiratory Journal, 9(10), 2087–2093. https://doi.org/10.1183/09031936.96.09102087
White, T. J., Bruns, T., Lee, S. & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics: PCR—protocols and applications—a laboratory manual. In PCR protocols: A guide to methods and applications (pp. 315–322). Academic Press, Inc.
Wilson, H. E., Carroll, G. C., Roy, B. A., & Kai Blaisdell, G. (2014). Tall fescue is a potential spillover reservoir host for Alternaria species. Mycologia, 106(1), 22–31. https://doi.org/10.3852/12-330
Woudenberg, J. H. C., Groenewald, J. Z., Binder, M., & Crous, P. W. (2013). Alternaria redefined. Studies in Mycology, 75, 171–212. https://doi.org/10.3114/sim0015
Woudenberg, J. H. C., Seidl, M. F., Groenewald, J. Z., de Vries, M., Stielow, J. B., Thomma, B. P. H. J., & Crous, P. W. (2015). Alternaria section Alternaria: Species, formae speciales or pathotypes? Studies in Mycology, 82, 1–21. https://doi.org/10.1016/j.simyco.2015.07.001
Yang, R. H., Su, J. H., Shang, J. J., Wu, Y. Y., Li, Y., Bao, D. P., & Yao, Y. J. (2018). Evaluation of the ribosomal DNA internal transcribed spacer (ITS), specifically ITS1 and ITS2, for the analysis of fungal diversity by deep sequencing. PLoS ONE, 13(10), 1–17. https://doi.org/10.1371/journal.pone.0206428
Acknowledgements
We acknowledge financial support from the European Commission through a Marie Curie Career Integration Grant (Project ID CIG630745 and Acronym SUPREME) awarded to Carsten A. Skjoth and co-financing the Ph.D. project awarded to Godfrey P. Apangu.
Author information
Authors and Affiliations
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Apangu, G.P., Frisk, C.A., Petch, G.M. et al. Environmental DNA reveals diversity and abundance of Alternaria species in neighbouring heterogeneous landscapes in Worcester, UK. Aerobiologia 38, 457–481 (2022). https://doi.org/10.1007/s10453-022-09760-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10453-022-09760-9