Abstract
Genetic variability within Septoria carvi isolates obtained from various organs of caraway cultivated in south-eastern and central Poland was studied using the RAPD-PCR technique. The tests were performed using randomly selected primers. The DNA profiles obtained using four primers proved useful in determining genetic variability among the genotypes of Septoria carvi isolates. The present study characterized the differences in the nucleotide sequence within the internal transcribed spacer region of rDNA (ITS1, 5.8S, ITS2) of selected S. carvi isolates and reference strains of Septoria spp. Moreover, eight isolates were sequenced for three loci: actin, calmodulin and translation elongation factor 1-alpha, and the obtained sequences were compared with the sequences of Septoria reference strains affecting other plants of the family Apiaceae. Phylogenetic analysis showed distinct differences of the tested isolates, which allowed to treat them Septoria carvi species affecting the above-ground organs of caraway Carum carvi L. This study is the first report on the genetic characteristics of the species S. carvi.
Similar content being viewed by others
Introduction
Fungi of the genus Septoria (teleomorph Mycosphaerella) belong to the widespread pathogens of crops and wild plants in all regions of the world (Farr et al. 1995; Verkley et al. 2004, 2013; Marcinkowska 2012; Zalewska et al. 2015). Septoria spp. cause spots and symptoms on the leaves, leaf petioles, stems and fruits. On the basis of the morphological features observed in the host plant, almost 3000 species have been determined to date. Ongoing research in this important group of pathogens is aimed to revise the classification of this genus (Verkley et al. 2004, 2013; Marcinkowska 2012; Quaedvlieg et al. 2013). Septoria cytisi Desm. is a typical species of the genus Septoria. This species causes septoriose of Laburnum anagyroides and some other species of plants from the family Fabaceae (Farr et al. 1995; Muthumary 1999; Waterwereld 2009). The taxonomy of Septoria species is still confusing and depends mainly on the host plant. Anamorphs are very difficult to distinguish based only on morphological features. Currently, Septoria is identified mainly by pycnidial conidiomata, holoblastic conidiogenesis, hyaline, smooth-walled conidiogenous cells with sympodial proliferation as well as filiform, hyaline and smooth-walled multi-septate conidia (Sutton 1980; Constantinescu 1984; Farr 1992; Verkley et al. 2004). On the other hand, species with apparently non-proliferating, phialidic conidiogenous cells and of acervuloid to pycnidial structures bearing spores are also included in this taxa (Sutton 1980; Verkley et al. 2004). Among Septoria spp., S. cari (I.E. Brezhnev), S. umbelliferarum Kalchbr. and S. carvi Syd. are recognized as the cause of dying leaves and umbels of caraway cultivated in European countries (Sydow 1932; Pidopličko 1978; Ondŕej 1983; Teterevnikova-Babayan 1987; Farr et al. 1995; Odstrčilovà et al. 2002; Machowicz-Stefaniak and Zalewska 2004, 2008; Mazur and Nawrocki 2004; Bedlan 2005). In 2004, the species S. carvi was considered to be the major causal agent of caraway yield reduction in Austria. This fungus has been present in Poland since 2001 and its occurrence increases during periods of warm and humid weather (Machowicz-Stefaniak and Zalewska 2008; Zalewska 2012, 2013).
Knowledge of the phylogenetic relationship of the genus Septoria is still fragmentary. There are no reference isolates of S. carvi in the available databases. Sexual status of this species is unknown and only a small number of species produce teleomorphs, which are classified to the genus Mycosphaerella Johanson (Dothideales) (Verkley et al. 2004). Mycological studies carried out at the Department of Plant Protection indicated variation in the morphological features and growth conditions of Septoria carvi compared to other Septoria species colonizing various plants of the family Apiaceae (Machowicz-Stefaniak et al. 2008; Zalewska 2008, 2012, 2013; Verkley et al. 2013; Quaedvlieg et al. 2013). Based on our preliminary research, it is difficult to determine the affiliation of the studied isolates, especially by the analysis of growth, development and morphological structures of S. carvi in different culture conditions (Zalewska 2012). Thus it is necessary to apply different molecular tools.
The aim of the present study was to determine the genetic variability of Septoria carvi isolates obtained from caraway cultivated in various regions of Poland using RAPD-PCR (Random Amplified Polymorphic DNA), and to compare S. carvi rDNA sequences of four various loci with other Septoria spp. colonizing plants of the family Apiaceae available in the database. The purpose of our study was also to investigate whether S. carvi is a new pathogenic species of caraway or perhaps it is one of the described species colonizing other plants of the family Apiaceae. Moreover, we intended to submit the sequences of the studied isolates to the database.
Materials and methods
Sampling and isolation of S. carvi isolates
The material used for the study consisted of 54 isolates of S. carvi, obtained as a result of earlier research on the health of caraway in south-eastern and central regions of Poland. Among these isolates, 39 originated from the leaves and leaf petioles of caraway cultivated in Motycz near Lublin 51°14′21″N 22°22′46″E, while 15 other isolates from the same organs and additionally from the umbels of caraway cultivated in Trębanów near Kielce 50°51′06″N 21°29′10″E (Machowicz-Stefaniak and Zalewska 2008; Zalewska et al. 2015). Origin of isolates is shown in Table 1. Some of the aforementioned isolates were previously used to study morphological features and pathogenicity of this fungus towards different organs of caraway (Zalewska 2012, 2013). Single spore cultures of these isolates were grown on Difco™ Potato Dextrose Agar (PDA) medium in Petri dishes in a thermostat at a temperature 24 °C without access to light. The cultures were incubated for 2 weeks and 4 replications were made for each isolate (Table 2).
DNA extraction
Mycelium of S. carvi was sampled from the surface of the cultures grown on PDA medium and then transferred to an Eppendorf tube for DNA extraction. DNA isolation was performed using the CTAB method (Doyle and Doyle 1987) with some own modification. The obtained mycelium was transferred to an Eppendorf tube with a volume of 1.5 ml, and then frozen in liquid nitrogen. The frozen material was homogenized with a sterile pestle (SIGMA-Adrich). Then 600 ml of CTAB lysis buffer was added to each tube and incubated at a temperature of 65 °C for 2 h and subsequently centrifuged (10.000 rpm for 10 min). In the next step, 1.0 ml phenol/chloroform/alcohol mixture in the volume ratio of 25:24:1 was added to the supernatant and centrifuged (10.000 rpm for 8 min). DNA was precipitated with 40 μl of sodium acetate (5 M) and 400 μl of isopropanol. The obtained precipitate was washed with 70% ethanol, and then centrifuged at 14.000 rpm for 15 min and dried. Next, the supernatant was suspended in 50 μl of water for PCR (SIGMA-Adrich) supplemented with RNase A, and then incubated for 24 h at a temperature of 4 °C, 1 h at 37 °C, and 10 min at 65 °C. DNA concentration was first estimated on 1.5% agarose gel and compared with GeneRuler 100 bp DNA Ladder Plus (Thermo Scientific), and then estimated using a UV-Vis NanoDrop 2000c/2000 spectrophotometer (Thermo Scientific). DNA samples were diluted to a final concentration of 20 ng/μl and stored at -20 °C for further analysis.
Estimation of genetic variability of Septoria carvi by RAPD-PCR
The analysis of genetic diversity of S. carvi isolates were carried out based on the RAPD-PCR results. DNA amplification was performed according to the method of Williams et al. (1990) with some modifications. The reaction volume used was 20 μl and the reaction solution containing: 1 x buffer Taq (750 mM Tris HCl pH 8.8; 200 mM (NH4)2SO4; 0.1% Tween 20) (Thermo Scientific), 2.5 mM x MgCl2, 0,1 mM dNTP, 1.0 U polymerase Taq (Thermo Scientific), primer 0.2 μM, 20 ng/μl of genomic DNA and distilled water. Amplification was performed using a DNA Engine Dyad Thermal Cycler (Biorad), with the following program: initial denaturation for 3 min at 95 °C followed by 45 cycles consisting of 45 s of denaturation at 94 °C, 45 s of annealing at 37 °C, 45 s extension at 72 °C and a final extension at 72 °C for 10 min. Of 30 primers tested with five isolates of Septoria carvi, four that generated reproducible and detectable amplification products were selected for further experiments (Tab. 2). The amplification products were separated by electrophoresis using 1.5% agarose gel at 80 V for 1.5 h in 1 x TBE buffer containing 0.01% EtBr (bromidium ethidine). The products obtained were visualised under UV light and photographed.
The data were analysed and presented as a matrix on the basis of the presence or absence of bands – specific product - that were treated as a single feature. Genetic similarity between all isolates was estimated according to the formula by Nei and Li (1979). Matrix of genetic similarity index (SI index) was created based on the polymorphism of RAPD products: SI = 2Nxy/(Nx + Ny) where Nxy is the number of shared fragments for two genotypes X,Y, while Nx and Ny are the numbers of fragments obtained in X,Y genotypes respectively.
The obtained data were used to construct dendrograms using the UPGMA (Unweighted Pair Group Method with Arithmetic mean) method implemented in a free online program (Garcia-Valle et al. 1999; Loncaric et al. 2009; http://genomes.urv.cat/UPGMA/).
Sequence analysis of ITS region, actin, calmodulin, translation elongation factor
Based on the RAPD-PCR results, eight isolates of S. carvi, representing various subgroups of genetic similarity, originated from plants cultivated in both regions of Poland, i.e. KML 93, KTL 188, KTB 355, KLM 359, KMO 391, KML 1806 KML 1833 and KML 1860, were selected for further study. These isolates were very similar in terms of morphological characteristics and showed very high aggressiveness towards various organs of caraway (Zalewska 2012, 2013). Phylogenetic analyses were based on the differences in nucleotide sequences of the PCR-amplified fragments of the rDNA ITS region (ITS1, 5.8 rDNA gene, ITS2), and three loci: actin (Act), calmodulin (Cal) and translation elongation factor 1-alpha (EF 1-α). Two universal primers were used for each reaction: ITS1 and ITS4, ACT–512F and ACT2Rd, CAL-235F and CAL-2Rd and EF1-728F and EF2, respectively (Table 3). The amplification reactions volume was 25 μl and contained: 20 ng/μl of template DNA, 1 x Buffer Taq (750 mM Tris HCl pH 8.8; 200 mM (NH4)2SO4, 0.1% Tween 20), 60 μM MgCl2, 100 μM dNTP mix, 0.2 μM of each primer and 1.0 U polymerase (Thermo Scientific). Amplification was performed using a DNA Engine Dyad Thermal Cycler (Biorad), with the following program: an initial denaturation for 5 min at 95 °C, followed by 40 cycles at a denaturation temperature of 95 °C for 45 s, primer annealing at a temperature of 52 °C for 50 s, primer extension at 72 °C for 90 s and a final extension step at 72 °C for 5 min. PCR products with the addition of fluorescent dye were separated electrophoretically in 1.5% agarose gel at 80 V for 1.5 h in 1 x TBE buffer containing 0.01% EtBr and visualized under UV light. After checking and determining the size of the resulting PCR products, they were subjected to purification using an agarose gel of low melting point temperature (Sambrook and Russel 2001). The resulting fragments were sequenced using PCR primers and a Big Dye® Terminator Cycle Sequencing Kit V. 3.1 of Applied Biosystems (Life Technologies). Sequencing reaction products were separated by a capillary sequencer (3730XL DNA Analyzer). The products of the PCR reaction were sequenced by the company Genomed S.A. Poland. The obtained nucleotide sequences of Septoria carvi isolates were compared with reference strains of other Septoria spp. inhabiting plants from the family Apiaceae, downloaded from the GenBank at the National Centre for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) (Table 4) (Verkley et al. 2013). The nucleotide sequences were analysed with ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2) software and compared with sequences collected in the NCBI GenBank databases using BLAST software (http://www.ncbi.nlm.nih.gov/BLAST/; (Altschul et al. 1997).
The phylogenetic analysis of the studied native isolates of S. carvi was carried out in several steps using the Phylogeny.fr program (http://www.phylogeny.fr/). Sequences were aligned to each other using the MUSCLE (v 3.7), program and the sequence regions containing gaps or mismatched ends with the Gblocks (v 0.91b) program. A phylogenetic tree was constructed by using the PhyML bootstrap (v 3.0) with an HKY85 algorithm of the maximum likelihood of substitution model and presented graphically using TreeDyn (v 198.3) gamma shape parameter 88.402.
Results
A total of 67 DNA fragments were obtained on the basis of the RAPD reaction. The number of amplicons generated using a single primer ranged from 13 to 22. On average, 16.75 bands were produced by a single primer (Table 2). As a result of the RAPD-PCR reaction, 60 polymorphic products were obtained and the number of banding patterns scored for each primer ranged from 12 to 19. On average, 15 polymorphic banding patterns were generated by a single primer. Specific products were also obtained from three primers. In total, seven specific banding patterns were obtained and band number ranged from 1 to 3, depending on the isolate (Table 2). The matrix of Dice’s indicated genetic similarity between the isolates ranging from 0.254 to 0.607, whereas the average similarity of all isolates was 0.480. Cluster analysis using the UPGMA method indicated the presence of two main groups of isolates (Table 1, Fig. 1). The first group consisted of isolates obtained from central and south-eastern regions of Poland, while the second group contained mainly isolates from the south-eastern region of Poland (Table 1).
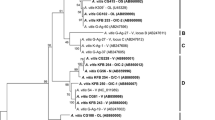
One characteristic band of approximately 580 bp was obtained for all 8 native S. carvi isolates, as a result of PCR reaction with the ITS1 and ITS4 primer set (Fig. 2). The analysis of the nucleotide sequence of ITS1 and ITS2 regions of S. carvi isolates showed 100% identity over a length of 418 nucleotides, i.e. 85% of the total length. Single substitution for isolate KLM 1806 was observed only in the ITS2 region. For this reason, only one of these amplicons (KML 93) was used in further studies as a representative of 8 native isolates. Sequence comparison of these isolates showed 98–100% homology with DNA sequences of the reference cultures along 83% of the total length (in all cases). Of all the sequences used in phylogenetic analyses, S. carvi displayed the greatest homology with S. petroselini (100%). In the case of other species, a lower sequence identity was observed: 99% to S. apiicola and S. sii, and 98% to S. aegopodina (Table 5). Some mutations were observed in the sequence of the ITS region in these isolates, namely these were: one transversion and 15 transitions in seven reference isolates representing S. apiicola, S. sii and S. aegopodina species. ITS dendrogram, based on the genetic distance using the Phylogeny program indicated segregation of 17 Septoria sequences in two main clusters: strains of S. aegopodina and S. sii were grouped into cluster 1. Isolates of S. carvi and reference strains of S. apiicola and S. petroselini were grouped in cluster 2 (Fig. 3). Two reference strains of S. apiicola constituted a distinct subgroup in this cluster. Isolates of S. carvi and S. petroselini were grouped in one subgroup (Fig. 3).
Electrophoresis of PCR products of native 8 isolates of S. carvi with actin primers showed the presence of a characteristic band of 650 bp in each isolate (Fig. 4a). The identity of isolate sequences was 97% in 24% of the sequence of the reference strains of S. sii and 93% in 22% of the sequence of S. aegopodina, S. apiicola and S. petroselini strains (Table 5). A comparison of S. carvi isolates and S. aegopodina actin sequences showed differences for four of eight isolates tested, i.e. isolates KML 93, KML 355, KMO 391 and KML 1860. The insertion of thymine was observed at position 11of the sequence of aforementioned isolates. Thymine insertion was also observed for the isolates KML 359 and KLM 1806 at position 15. Furthermore, in the sequence of isolates KTL 188 and KML 1833, thymine insertion was observed at position 5, cytosine also at position 5, adenine at position 3 and guanine at position 4 of the sequence compared to the sequence of S. aegopodina reference strains. In the sequence of S. carvi, the lack of a nucleotide at position 75 was observed. In addition, the occurrence of 13 transitions and 19 transversions was detected. A phylogenetic tree, based on the actin sequence of eight isolates of S. carvi and other selected Septoria reference strains generated using the Phylogeny.fr analysis, indicated the segregation of all isolates into two main clusters. The first cluster was divided into two sub-clusters, the first one grouped reference strains of S. sii and S. aegopodina, while the second sub-claster included strains of S. apiicola in an individual sub-group, and strains of S. petroselini, which were grouped in a separate sub-group and isolates of S. carvi in the second sub-group (Fig. 5a). Isolates of S. carvi in the two sub-groups and isolates KTL188 and KML1806 constituted two single separate sub-groups (Fig. 5a).
Electrophoresis of PCR products (calmodulin primers) of 8 native isolates of S. carvi showed the presence of a characteristic band of 450 bp size (Fig. 4b). The comparison of isolate sequences with sequences of the reference strains demonstrated 99% identity of 77% of S. appicola sequence and 94% identity also of 77% of the sequence for S. aegopodina and S. sii reference strains. In contrast, 94% sequence identity of S. carvi isolates was observed in 73% of the length of S. petroselini sequence (Table 5). As regards the sequence differences between the isolates and S. apiicola, two transitions and two transversions were recorded while compared to the sequence of S. aegopodina, 13 transitions and five transversions occurred. Three additional tranversions were detected with respect to the CBS 102370 strain of S. sii. Three gaps, 10 transversions and five transitions were observed in the sequence of S. carvi isolates when compared to the corresponding sequence of S. petroselini. Phylogenetic tree of 8 isolates of S. carvi and other selected reference strains of Septoria generated using the Phylogeny.fr analysis of calmoduline sequence indicated segregation of all isolates into two main clusters (Fig. 5b). The first cluster contained strains of S. sii and S. aegopodina, which were divided into a single distinct sub-cluster (Fig. 5b). The second main cluster included all studied isolates of S. carvi divided into single sub-groups and a second sub-group with reference strains of S. petroselinii – and S. apiicola clustered into a separate single sub-groups (Fig. 5b).
Sequence analysis of PCR reaction of S. carvi isolates with selected EF 1-α primers showed the presence of a characteristic 600-bp band present in all isolates (Fig. 4c). A comparison of the sequence of our native isolates of S. carvi with the sequence of selected reference strains of other Septoria spp. demonstrated 99% identity along 81% of S. sii sequence length, 90% along 74% of S. apiicola sequence length and 89% identity of 75% of the S. petroselini sequence. Regarding the reference strains of S. aegopodina, the identity of S. carvi isolates was 82% in 80% of the sequence (Table 5). The comparison of the S. carvi isolate sequences to the sequence of S. sii reference strains showed the presence of two transitions and sequence containing adenine and guanine at position 120 in the CBS 1023 70 strain as well as adenine and guanine at position 145 in the reference strain sequences. The comparison of nucleotide sequences of S. carvi isolates with the sequences of S. apiicola strains demonstrated the presence of 12 transitions and 11 transversions. Moreover, the lack of purines or pyrimidines at positions 76, 77 and 110 was observed in the isolates` sequences, while insertions were observed at positions 88, 97 and 99. The comparison of S. petroselinii and S. carvi sequences with the sequences of the isolates revealed the presence of 16 transitions and 11 transversions. A phylogenetic tree of eight S. carvi isolates and other selected Septoria reference strains generated using the Phylogeny.fr analysis of the elongation factor EF 1-α sequence indicated segregation of all studied isolates into two clusters (Fig. 5c). One cluster included only two strains of S. sii, and the third strain of S. sii and other studied isolates were grouped into the second cluster. This cluster contained studied isolates of S. carvi - grouped into a single sub-group (Fig. 5c).
The obtained sequences of S. carvi isolates of have been submitted to the GenBank database (Table 6).
Discussion
Phylogenetic relationships of fungi of the genus Septoria have been the subject of an ongoing research of some Dutch researches (Quaedvlieg et al. 2013; Verkley et al. 2013). Mycologists constantly ask the following questions: “What are Septoria?” and “Which fungi belong to the genus Septoria?” The studies conducted by the above-mentioned authors showed that we can not clearly define the genus based solely on its morphology or host plants infected by these fungi. Investigating nucleotide sequence obtained in PCR reactions with various primers, allowed us to correctly identify fungal species and its affiliation to the genera. The study by Quaedvlieg et al. (2013) divided, the genus Mycosphaerella into 47 clades. Septoria is the first clade of the anamorphic stage of these fungi.
The study of genetic diversity of S. carvi isolates collected from the south-eastern and central regions of Poland using the RAPD-PCR method showed a high genetic diversity among them, despite very similar macro- and micro-morphological features. This fact could raise some doubts whether the studied isolates belonged to the same species. However, some authors have also observed a high variation within the populations of different fungal species. Zhou et al. (2005) found a wide variation of Phoma macrostoma isolates used for the biological control of weeds, similarly as did Tiwari et al. (2013) within the population of Phoma tropica isolates originated from different host plants growing under various environmental conditions. Likewise, Król (2002) observed genetic differentiation between the isolates of Phomopsis originated from different species of fruit trees, including few closely related genera from the same plant family. On the other hand, Baturo et al. (2004) showed little intraspecific variability among the isolates of Bipolaris sorokiniana from different regions of Brazil. Moreover, Balmas et al. (2005) found that Phoma tracheiphila isolates from Italy were genetically similar and created identical banding patterns with selected RAPD primers. Generally, it is believed that the RAPD-PCR technique is useful for detecting variability within many fungal species (Baturo et al. 2004; Sagar et al. 2011). Furthermore, the data obtained from the RAPD analysis may form the basis for further research using other molecular techniques (Baturo et al. 2004; Salem et al. 2007). The analyses of ITS1 and ITS2 regions of S. carvi isolates revealed their high similarity. Therefore, our data suggest that these isolates belong to one fungal species. The comparison of ITS1 and ITS2 sequences of native isolates of Septoria species to the sequences of various species (reference strains) occurring on different plants from the Apiaceae family showed 98, 99 or even 100% similarity. However, other study indicated differences between the tested isolates and the species S. petroselini (Verkley et al. 2013). Nonetheless, some authors have noted that Septoria material collected even from the same location and host species, but under different environmental conditions or at different times in the same season, can differ considerably in the average conidial sizes, particularly the length (Verkley et al. 2013). Simultaneously, a molecular phylogenetic study of the same authors on Septoria species infecting Asteraceae and woody perennials demonstrated that species capable of infecting hosts of the same plant family did not always cluster in monophyletic groups. Furthermore, Verkley et al. (2013) reported that the ITS sequence of Septoria anthrisci was distinct from that of S. apiicola, but identical to that of S. petroselini, which suggested insufficient diversity within the rDNA regions and the need to use different molecular tools.
In the most recent phylogenetic studies of the genus Septoria, there was no information about the phylogenetic relationships of Septoria carvi, although this species is recognized as a dangerous pathogen of caraway grown in the Czech Republic, Poland, Austria, Bulgaria and Lithuania (Ondŕej 1983; Odstrčilovà et al. 2002; Machowicz-Stefaniak and Zalewska 2004, 2008; Mazur and Nawrocki 2004; Bedlan 2005; Mačkinaitė 2012).
It seems that the difficulties in arriving at a clear definition of the relationship between native isolates of Septoria result mainly from the lack of the reference isolate of S. carvi and its nucleotide sequence, and perhaps also from little variation in the sequences of the ITS region of the genus Septoria. This fact makes the comparison of the ITS sequence impossible (Quaedvlieg et al. 2013). Similar relationships were found in species of the genus Phoma. Badillo-Vargas et al. (2008) observed that the ITS region sequences of the isolates of Phoma putaminum and P. macrostoma var. incolorata were similar to the sequences of closely related species. In another study on Phoma exigua species variants, it was impossible to distinguish the variants based on the nucleotide sequence of the ITS regions, despite clear differences in the appearance of the cultures (Abeln et al. 2002). The conducted research using typical differentiating proteins for Septoria genera showed too low sequence similarity to the sequences of reference strains infesting plants of the family Apiaceae. This means that the fungus tested can be considered a separate species Septoria carvi Syd. affecting Carum carvi L. plants. The results confirmed the usefulness of ITS barcodes to extract the genus of Septoria.
The obtained results in the phylogenetic analysis of actin, calmodulin and translation elongation factor 1-α sequences showed the distinctness of the eight studied isolates of S. carvi from the sequences of other species of the genus Septoria infecting plants of the family Apiceae. All translation elongation factor (EF 1-α) sequences of S. carvi isolates were located on one branch of the phylogenetic tree. They did not differ between each other, as opposed to the nucleotide sequences of actin and calmodulin. Actin gene variation was found between isolates KTL 188 and KLM 1806 compared to other S. carvi isolates. Calmodulin sequences showed differences in all isolates tested. All sequences of S. carvi formed one common group with S. pertoselinii and S. apiicola. This fact indicated close relationship of these species, which was demonstrated by the sequence analysis of ITS regions. The obtained results showed differences between the studied barcodes and indicated that translation elongation factor 1-α constituted the best primer to identify S. carvi. The results of our study are consistent with a previous study conducted by Verkley et al. (2013).
References
Abeln, E. C. A., Stax, A. M., de Gruyter, J., & van der As, H. A. (2002). Genetic differentiation of Phoma exigua varieties by means of AFLP fingerprints. Mycological Research, 106, 419–427.
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., & Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25, 3389–3402.
Badillo-Vargas, I. S., Rivera-Vargas, L. I., & Calle-Bellido, J. (2008). Morphological, pathogenic and molecular characterization of Phoma spp. isolated from onion field soils in Puerto Rico. Journal of Agriculture of the University of Puerto Rico, 92(1–2), 73–86.
Balmas, V., Scherm, B., Ghignone, S., Salem, A. O. M., Cacciola, S. O., & Migheli, Q. (2005). Characterization of Phoma tracheiphila by RAPD-PCR, microsatellite-primed PCR and ITS rDNA sequencing and development of specific primers for in planta PCR detection. European Journal of Plant Pathology, 111, 235–247.
Baturo, A., Mehta, Y. R., & Sadowski, C. K. (2004). Identification of genetic variability in Bipolaris sorokiniana isolates from wheat in Brazil. Summa Phytopathologica, 30(4), 470–474.
Bedlan, G. (2005). Septoria carvi an Kümmel. Gemüse, 11, 25.
Carbone, I., & Kohn, L. M. (1999). A method for designing pimer sets for speciation studies in filamentous ascomycetes. Mycologia, 91, 553–556.
Constantinescu, O. (1984). Taxonomic revision of Septoria-Like Fungi parasitic on Betulaceae. Transactions of the British Mycological Society, 83, 383–398.
Doyle, J. L., & Doyle, J. J. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin., 19, 11–15.
Farr, D. F. (1992). Species of Septoria on the Fabaceae, subfamily Faboidae, tribe Genistate. Sydowia, 44, 13–31.
Farr, D. F., Bills, G. F., Chamuris, G. P., & Rossman, A. Y. (1995). Fungi on plants products in the United States. St. Paul.
Garcia-Valle, S., Palau, J., & Romeu, A. (1999). Horizontal gene transfer in glycosylic hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Molecular Biology and Evolution, 9, 1125–1134.
Groenewald, J. Z., Nakashima, C., Nishikawa, J., Shin, H.-D., Park, J.-H., Jama, A. N., Groenewald, M., Braun, U., & Crous, P. W. (2013). Species concepts in Cercospora: spotting the weeds among the roses. Studies in Mycology, 75, 115–170.
Król, E. (2002). Determination of genetic variability within Phomopsis spp. using RAPD method. Phytopathologia Polonica, 25, 35–46.
Loncaric, I., Oberlerchner, J. T., Heissenberger, B., & Moosbeckhofer, R. (2009). Phenotypic and genotypic diversity among strains of Aureobasidium pullulans in comparison with related species. Antonie Van Leeuwenhoek, 95(2), 165–178.
Machowicz-Stefaniak, Z., & Zalewska, E. (2004). The fungi threaten to some species of herbs from Apiaceae Family cultivated in south-East Poland. Folia Universitatis Agriculturae Stetinensis Agricultura, 239(95), 223–228.
Machowicz-Stefaniak, Z., & Zalewska, E. (2008). Biodiversity of fungi inhabiting various parts of caraway (Carum carvi L.). EJPAU. Horticulturae. 2008; 11(1)#21 http://www.ejpau.media.pl/volume11/issue1/art-21.html.
Machowicz-Stefaniak, Z., Zalewska, E., & Król, E. (2008). Biotic effect of caraway phyllosphere fungi on the pathogenic Septoria carvi Syd. Herba Polonica, 54(3), 70–80.
Mačkinaitė, R. (2012). Potential pathogens of common caraway (Carum carvi L.) seeds and search for measures suppressing their spread. Žemdirbystė = Agriculture, 99(2), 179–188.
Marcinkowska, J. (2012). Oznaczanie rodzajów grzybów sensu lato ważnych w fitopatologii: PWRiL Sp. Warszawa: Z o.o.
Mazur, S., & Nawrocki, J. (2004). Fungal diseases threat on caraway plantations in south region of Poland In: Proceeding of the XVI Slovak and Czech Plant Protection Conference. Acta Fytotechnica et Zootechnica, 7, 201–203.
Muthumary, J. (1999). First contribution to a morphology of Septoria species in India. Madras: Centre for advanced studies in Botany.
Nei, M., & Li, W. H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences of the United States of America, 76(10), 5269–5273.
O’Donnell, K., Kistler, H. C., Cigelnik, E., & Ploetz, R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America, 95, 2044–2049.
Odstrčilovà, L., Ondŕej, M., Kocourkovà, B., & Růžičkovà, G. (2002). Monitoring of incidence and determination of chemical protection. In: Proc 6th Conf EFPP. Praha. Plant Protect Sci., 38 (Special Issue 2), pp. 340–343.
Ondŕej, M. (1983). Wýskyt hub na kminu (Carum carvi L.) v ČSSR. Ochr Rostl., 19, 235–237.
Pidopličko, H. M. (1978). The fungi parasites of culture plants, T 3. Fungi with pycnidia. Kijów: Naukowa Dumka.
Quaedvlieg, W., Groenewald, J. Z., de Jesús Yáñez-Morales, M., & Crous, P. W. (2012). DNA barcoding of Mycosphaerella species of quarantine importance to Europe. Persoonia, 29, 101–115.
Quaedvlieg, W., Verkley, G. J. M., Shin, H. D., Barreto, R. W., Algenas, A. C., Swart, W. J., Groenewald, J. Z., & Crous, P. W. (2013). Sizing up Septoria. Studies in Mycology, 75, 307–390.
Sagar, M. S. I., Meah, M. B., Rahman, M. M., & Ghose, A. K. (2011). Determination of genetic variations among different Trichoderma isolates using RAPD marker in Bangladesh. Journal of the Bangladesh Agricultural University, 9(1), 9–20.
Salem, H. H., Ali, B. A., Huang, T. H., Qin, A., Wang, X., & Xie, Q. (2007). Use of random amplified polymorphic DNA analysis for economically important food crops. Journal of Integrative Plant Biology, 49(12), 1670–1680.
Sambrook, J., & Russel, D. W. (2001). Molecular cloning: a laboratory manual (3rd ed.). New York: Cold Spring Laboratory.
Sutton, B. C. (1980). The Coelomycetes. Kew: Commonwealth Mycological Institute 696 p.
Sydow, H. (1932). Novae fungorum species – XXI. Annales Mycologici, 30, 114.
Teterevnikova-Babayan, D. N. (1987). Griby roda Septoria w SSR: the USSR Academy of Sciences’ Armenian Branch.
Tiwari, V. V., Gade, A. K., & Rai, M. (2013). A study of phylogenetic variations among Indian Phoma tropica species by RAPD-PCR and ITS-rDNA sequencing. Indian Journal of Biotechnology, 12, 187–194.
Verkley, G. J. M., Starink-Willemse, M., Iperen, A., & van Abeln, E. C. A. (2004). Phylogenetic analyses of Septoria species based on the ITS and LSU-D2 regions of nuclear ribosomal DNA. Mycologia, 96, 558–571.
Verkley, G. J. M., Quaedvlieg, W., Shin, H. D., & Crous, P. W. (2013). A new approach to species delimination in Septoria. Studies in Mycology, 75, 2013–2305.
Waterwereld (2009) Golden chain tree. http://www.waterwereld.nu/goudenregeneng.php#
White, T. J., Bruns, T., Lee, S., & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetic. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, & T. J. White (Eds.), PCR Protocols (pp. 315–322). San Diego: Academic Press.
Williams, J. G. K., Kubelik, A. R., Livak, K. J., Rafalski, J. A., & Tingey, S. V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research, 18(22), 6531–6535.
Zalewska, E. (2008). Occurrence and characterization of Septoria carvi Syd. (Coelomycetes, Sphaeropsidales). Herba Polonica, 54(1), 25–33.
Zalewska, E. (2012). Growth and sporulation of Septoria carvi Syd. In different culture conditions. Acta Scientiarum Polonorum Hortorum Cultus, 11(1), 93–107.
Zalewska, E. (2013). Pathogenicity of Septoria carvi Syd. Towards caraway Carum carvi L. (Apiaceae). Journal of Agricultural Science and Technology Part A, 3, 711–723.
Zalewska, E. D., Machowicz-Stefaniak, Z., & Król, E. (2015). Fungi colonizing caraway (Carum carvi L.) in different regions of cultivation. Acta Scientiarum Polonorum Hortorum Cultus, 14(6), 175–188.
Zhou, L., Bailey, K. L., Chen, C. Y., & Keri, M. (2005). Molecular and genetic analyses of geographic variation in isolates of Phoma macrostoma used for biological weed control. Mycologia, 6, 612–620.
Acknowledgments
We would like to kindly thank Professor Krzysztof Kowalczyk, the head of the Institute of Plant Genetics, Breeding and Biotechnology for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Zalewska, E.D., Król, E.D., Abramczyk, B.A. et al. Genetic variability within Septoria carvi Syd.- a pathogen of caraway Carum carvi L. Eur J Plant Pathol 148, 825–838 (2017). https://doi.org/10.1007/s10658-016-1139-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-016-1139-8