Abstract
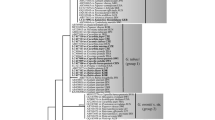
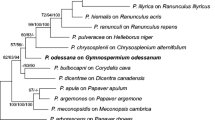
Species in the genera Hyaloperonospora and Peronospora (Oomycota, Peronosporaceae) are obligate biotrophic pathogens responsible for destructive downy mildew diseases of ornamental horticulture plants worldwide. Throughout the past decade, newly emergent downy mildew diseases have become increasingly prevalent, but often the identity of the pathogen is unknown or poorly defined. In this study we set out to identify the downy mildew pathogens infecting three widely grown ornamental plants: Agastache spp. (hyssops), Tarenaya hassleriana (syn. = Cleome hassleriana; spider-flower) and Monarda didyma (bee balm or bergamot). Phylogenetic analysis of the internal transcribed spacer rDNA and cox2 revealed two new species, which are described here as Peronospora monardae infecting both Monarda didyma and A. mexicana, and Hyaloperonospora daughtreyae infecting T. hassleriana. One sample of Peronospora collected from Agastache sp. ‘Bolero’ did not cluster with P. monardae, but its relationship to other species in the clade was not sufficiently resolved for taxonomic determination. Phylogenetic analysis also supported the combination of Peronospora iberidis, a pathogen of Iberis spp. (candytuft), into Hyaloperonospora. To assess intrasample ITS sequence variation from P. monardae, data were generated using high-throughput amplicon sequencing (HTAS). Multiple haplotypes were identified from the P. monardae samples, with less than 1% intrasample variation observed. From a core set of haplotypes shared across the HTAS the variant frequencies could not be completely explained by heterozygosity alone.








Similar content being viewed by others
Data availability
The dataset(s) supporting the conclusions of this article are available in the [repository name] repository, [unique persistent identifier and hyperlink to dataset(s) in http:// format]. Voucher specimens are available through the U.S. National Fungus Collections (BPI).
References
Beirn, L. A., Hempfling, J. W., Schmid, C. J., Murphey, J. A., Clarke, B. B., & Crouch, J. A. (2017). Differences among soil-inhabiting microbial communities in Poa annua turf throughout the growing season. Crop Science, 57, S262–S273.
Belbahri, L., Calmin, G., Pawlowski, J., & Lefort, F. (2005). Phylogenetic analysis and real time PCR detection of a presumably undescribed Peronospora species on sweet basil and sage. Mycological Research, 109, 12765–11287.
Bik, H. M., Fournier, D., Sung, W., Bergeron, R. D., & Thomas, W. K. (2013). Intra-genomic variation in the ribosomal repeats of nematodes. PLoS One, 8, e78230.
Brodin, J., Hedskog, C., Heddini, A., Benard, E., Neher, R. A., Mild, M., & Albert, J. (2015). Challenges with using primer IDs to improve accuracy of next generation sequencing. PLoS One, 10, e0119123.
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., et al. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods, 13, 581–583.
Choi, Y.-J., Hong, S. B., & Shin, H. D. (2003). Diversity of the Hyaloperonospora parasitica complex from core brassicaceous hosts based on ITS rDNA sequences. Mycological Research, 107, 1314–1322.
Choi, Y.-J., Shin, H.-D., & Thines, M. (2009). Two novel Peronospora species are associated with recent reports of downy mildew on sages. Mycological Research, 113, 13405–11350.
Choi Y-J, Mulenko W, Park JH, Shin HD (2012) First report of downy mildew of spider flower caused by a Hyaloperonospora sp. in Korea. Plant Disease 96: 4.
Choi, Y.-J., Klosterman, S. J., Kummer, V., Vogmayr, H. D., & Thines, M. (2015). Multi-locus tree and species tree approaches toward resolving a complex clade of downy mildews (Straminipila, Oomycota), including pathogens of beet and spinach. Molecular Phylogenetics and Evolution, 86, 245–234.
Constantinescu, O., & Fateji, J. (2002). Peronospora-like fungi (Chromista, Peronosporales) parasitic on Brassicaceae and related hosts. Nova Hedwigia, 74, 2915–2338.
Crandall, S. G., Rahman, A., Quesada-Ocampo, L. M., Martin, F. M., Bilodeau, G. J., & Miles, T. D. (2018). Advances in diagnostics of downy mildews: Lessons learned from other oomycetes and future challenges. Plant Disease, 102, 2655–2275.
Darriba, D., Taboada, G. L., Doallo, R., & Posada, D. (2012). jModelTest 2: More models, new heuristics and parallel computing. Nature Methods, 9, 772.
Development Core Team, R. (2015). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
Etienne, R. S., Morlon, H., & Lambert, A. (2014). Estimating the duration of speciation from phylogenies. Evolution, 68, 24305–22440.
Farr, D. F., & Rossman, A. Y. (2019). Fungal databases, U.S. National Fungus Collections, ARS. USDA. Retrieved September 9, 2019, from https://nt.ars-grin.gov/fungaldatabases/.
Fletcher, K., Gil, J., Bertier, L. D., Kenefick, A., Wood, K. J., Zhang, L., Reyes-Chin-Wo, S., Cavanaugh, K., Tsuchida, C., Wong, J., & Michelmore, R. (2019). Genomic signatures of heterokaryosis in the oomycete pathogen Bremia lactucae. Nature Communications, 10, 2645.
Gabler, J., Hagedorn, G., & Braun, U. (2012). Taxonomy and phylogenetic placement of the downy mildew Personospora saturejae-horyensis. Mycotaxon, 121, 455–463.
Gäumann, E. A. (1918). Über die formen der Peronospora parasitica (Pers.) Fries. ein beitrag zur speziesfrage bei den parasitischen pilzen. Beih Bot Zentralblatt., 35, 395–533.
Gäumann, E. A. (1927). Mykologische mitteilungen III. Annales Mycologici, 25, 1675–1177.
Gawad, C., Koh, W., & Quake, S. R. (2016). Single-cell genome sequencing: Current state of the science. Nature Reviews Genetics, 17, 175–188.
Göker, M., Voglmayr, H., Blazquez, G. G., & Oberwinkler, F. (2009). Species delimitation in downy mildews: The case of Hyaloperonospora in the light of nuclear ribosomal ITS and LSU sequences. Mycological Research, 113, 3085–3325.
Gong, J., Dong, J., Liu, X., & Massana, R. (2013). Extremely high copy numbers and polymorphisms of the rDNA operon estimated from single cell analysis of oligotrich and peritrich ciliates. Protist, 164, 369–379.
Hudspeth, D. S. S., Nadler, S. A., & Hudspeth, M. E. S. (2000). A cox2 molecular phylogeny of the Peronosporomycetes. Mycologia, 92, 674–684.
Huson, D. H., & Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution, 23, 2545–2267.
Kates, H. R., Johnson, M. G., Gardner, E. M., Zerega, N. J. C., & Wickett, N. J. (2018). Allele phasing has minimal impact on phylogenetic reconstruction from targeted nuclear gene sequences in a case study of Artocarpus. American Journal of Botany, 105, 4045–4416.
Katoh K, Rozewicki J, Yamada K D (2017) MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefs in Bioinformatics bbx108.
Kebschull, J. M., & Zador, A. M. (2015). Sources of PCR-induced distortions in high-throughput sequencing datasets. Nucleic Acids Research, 43, e143.
Knaus, B., Tabima, J., Shakya, S., Judelson, H., & Grünwald, N. (2019). Genome-wide increased copy number is associated with emergence of super-fit clones of the Irish potato famine pathogen Phytophthora infestans. BioRxiv. https://doi.org/10.1101/633701.
Kumar, S., Stetcher, G., & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874.
Lee, J. S., Lee, H. B., Shin, H.-D., & Choi, Y.-J. (2017). Diversity, phylogeny and host-specialization of Hyaloperonospora species in Korea. Mycobiology, 43, 1395–1149.
Lerch, A., Koepfli, C., Hofmann, N. E., Messerli, C., Wilcox, S., Kattenberg, J. H., Betuela, I., O’Connor, L., Mueller, I., & Felger, I. (2017). Development of amplicon deep sequencing markers and data analysis pipeline for genotyping multi-clonal malaria infections. BMC Genomics, 18, 864.
Li, Y., Shen, H., Zhou, Q., Qian, K., van der Lee, T., & Huang, S. (2016). Changing ploidy as a strategy: The Irish potato famine pathogen shifts ploidy in relation to its sexuality. Molecular Plant Microbe Interactions, 30, 45–52.
Lindner, D., & Banik, M. (2011). Intragenomic variation in the ITS rDNA region obscures phylogenetic relationships and inflates estimates of operational taxonomic units in genus Laetiporus. Mycologia, 103, 7315–7740.
Lyon, R., Correll, J., Feng, C., Bluhm, B., Shrestha, S., Shi, A., & Lamour, K. (2016). Population structure of Peronospora effusa in the southwestern United States. PLoS One, 11, e0148385.
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal, 17, 10–12.
Moncalvo, L., Wang, H., & Hseu, R. (1995). Phylogenetic relationships in Ganoderma inferred from the internal transcribed spacers and 25S ribosomal DNA sequences. Mycologia, 87, 223–238.
Oliver, A., Brown, S., Callaham, M., & Jumpponen, A. (2015). Polymerase matters: Non-proofreading enzymes inflate fungal community richness by up to 15%. Fungal Ecology, 15, 865–889.
Patchell, M., Roalson, E. H., & Hall, J. C. (2014). Resolved phylogeny of Cleomaceae based on all three genomes. Taxon, 63, 315–328.
Philippe, H., Brinkmann, H., Lavrov, D. V., Littlewood, D. T. J., Manuel, M., Wörheide, G., & Baurain, D. (2011). Resolving difficult phylogenetic questions: Why more sequences are not enough. PLoS Biology, 9, e1000602.
Rambaut, A. (2014). FigTree, v1.4.3 http://tree.bio.ed.ac.uk/software/figtree/.
Rivera Y, Salgado-Salazar C, Creswell TC, Ruhl G, Crouch JA (2016a) First report of downy mildew caused by Peronospora sp. on Agastache sp. in the United States. 100: 1249.
Rivera, Y., Salgado-Salazar, C., Gulya, T., & Crouch, J. A. (2016b). Newly emerged populations of Plasmopara halstedii infecting rudbeckia exhibit unique genotypic profiles and are distinct from sunflower-infecting isolates. Phytopathology, 106, 752–761.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M. A., & Huelsenbeck, J. P. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542.
Rottstock, T., Joshi, J., Kummer, V., & Fischer, M. (2014). Higher plant diversity promotes higher diversity of fungal pathogens, while it decreases pathogen infection per plant. Ecology, 95, 1907–1917.
Salgado-Salazar, C., Rane, K., & Crouch, J. A. (2017). First report of Hyaloperonospora sp. associated with downy mildew disease of Iberis sempervirens in the United States. Plant Disease, 101, 1058.
Salgado-Salazar, C., LeBlanc, N., Ismaiel, E., Rivera, Y., Warfield, C. Y., & Crouch, J. A. (2018a). Genetic variation of the pathogen causing impatiens downy mildew pre-dating and including 21stcentury epidemics on Impatiens walleriana. Plant Disease, 102, 2411–2420.
Salgado-Salazar, C., Shishkoff, N., Daughtrey, M., Palmer, C. L., & Crouch, J. A. (2018b). Downy mildew: A serious disease threat to rose health worldwide. Plant Disease, 102, 18735–11882.
Silvestro, D., & Michalak, I. (2012). raxmlGUI: A graphical front-end for RAxML. Organisms Diversity & Evolution, 12, 335–337.
Simon, U. K., & Weiss, M. (2008). Intragenomic variation of fungal ribosomal genes is higher than previously thought. Molecular Biology and Evolution, 25, 2251–2254.
Spring, O., Bachofer, M., Thines, M., Riethmüller, A., Göker, M., & Oberwinkler, F. (2006). Intraspecific relationship of Plasmopara halstedii isolates differing in pathogenicity and geographic origin based on ITS sequence data. European Journal of Plant Pathology, 114, 309–315.
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312–1313.
Thines, M. (2007). Characterization and phylogeny of repeated elements givin rise to exceptional length of ITS2 in several downy mildew genera (Peronosporaceae). Fungal Genetics and Biology, 44, 199–207.
Thines, M., & Choi, Y.-J. (2016). Evolution, diversity, and taxonomy of the Peronosporaceae, with focus on the genus Peronospora. Phytopathology, 106, 6–18.
Thines, M., & Kummer, V. (2013). Diversity and species boundaries in floricolous downy mildews. Mycological Progress, 12, 3215–3329.
Thines, M., Telle, S., Ploch, S., & Runge, F. (2009). Identity of the downy mildew pathogens of basil, coleus and sage with implications for quarantine measures. Mycological Research, 113, 5325–5540.
Voglmayr, H. (2003). Phylogenetic relationships of Peronospora and related genera based on nuclear ribosomal ITS sequences. Mycological Research, 107, 1132–1142.
Voglmayr, H., Choi, Y.-J., & Shin, H.-D. (2014). Multigene phylogeny, taxonomy and reclassification of Hyaloperonospora on Cardamine. Mycological Progress., 13, 1315–1144.
Wallace, E. C., Daughtrey, M. L., Rane, K., Salgado-Salazar, C., & Crouch, J. A. (2018a). First report of Peronospora sp. causing downy mildew disease on Geum sp. in Maryland and New York. Plant Disease, 102, 1463.
Wallace, E. C., Salgado-Salazar, C., Gregory, N., & Crouch, J. A. (2018b). Basidiophora delawarensis, a new downy mildew species infecting cultivated goldenrod (Solidago sphacelata) in the USA. Mycological Progress, 17, 1283–1291.
Yerkes, W. D., & Shaw, C. G. (1959). Taxonomy of the Peronospora species on Cruciferae and Chenopodiaceae. Phytopathology, 49, 4995–4507.
Acknowledgements
The authors wish to thank Sara May for providing samples included in this study.
Adherence to national and international regulations
Not applicable.
Funding
This work was funded by the U.S. Department of Agriculture (USDA), Agricultural Research Service (ARS) project 8042-22000-298-00-D and the USDA Animal and Plant Health Inspection Service (APHIS) Farm Bill 10007 program. CSS, NL and EW were participants in the ARS Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the USDA. ORISE is managed by ORAU under DOE contract number DEAC05-06OR23100. USDA-APHIS provided support in the form of stipend for one author (EW), but did not have any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of this author are articulated in the “author contributions” section. ORISE, DOE and ORAU did not have any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The U.S. Department of Agriculture (USDA) prohibits discrimination in all its programs and activities. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Contributions
CSS, NL, EW and MLD performed data generation and analysis. JAC conceived, supervised and administered the study and acquired funding. CSS, NL and JAC drafted and revised the original manuscript. All authors contributed to the interpretation of data, and read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Salgado-Salazar, C., LeBlanc, N., Wallace, E.C. et al. Peronospora monardae, Hyaloperonospora daughtreyae and H. iberidis: new species associated with downy mildew diseases affecting ornamental plants in the United States. Eur J Plant Pathol 157, 311–326 (2020). https://doi.org/10.1007/s10658-020-01989-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-020-01989-9