Abstract
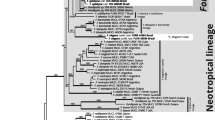
DNA sequences of Capitotricha bicolor from Quercus, Fagus sylvatica, Alnus alnobetula, and Nothofagus, and C. rubi from Rubus idaeus were obtained from apothecia to establish whether specimens from different hosts belong to separate species. The obtained ITS1–5.8S–ITS2 rDNA sequences were examined with Bayesian and parsimony phylogenetic analyses. Intra- and interspecific variation was also investigated based on molecular distances in the ITS region. The phylogenetic analyses supported the specific distinctness of Capitotricha rubi and the Capitotricha from Nothofagus, but also suggest specific distinctness between samples from Quercus, Fagus, and Alnus. The interspecific distances were larger than intraspecific distances for all examined units. The smallest distance was found between the “Alnus alnobetula” and “Fagus sylvatica” units. Two new sequences of Brunnipila are published. Capitotricha, Lachnum, and Erioscyphella are compared to each other based on hair and excipulum characteristics.




Similar content being viewed by others
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Baral H-O (1987) Lugol’s solution/IKI versus Melzer’s reagent: hemiamyloidity, a universal feature of the ascus wall. Mycotaxon 29:399–450
Baral H-O, Krieglsteiner GJ (1985) Bausteine zu einer Askomyzeten-Flora der BR Deutschland: In Süddeutschland gefundene Inoperculate Discomyzeten mit taxonomischen, ökologischen und chorologischen Hinweisen. Beih Zeitschr Mykol 6:1–160
Baral H-O, Garcia G, Bogale M, O’Hara MJ, Untereiner WA (2012) Colipila, a new genus in the Helotiales. Mycol Prog 11:201–214. https://doi.org/10.1007/s11557-011-0742-5
Bengtsson-Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A, De Wit P, Sánchez-García M, Ebersberger I, de Sousa F, Amend A, Jumpponen A, Unterseher M, Kristiansson E, Abarenkov K, Bertrand YJK, Sanli K, Eriksson KM, Vik U, Veldre V, Nilsson RH (2013) Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol 4:914–919. https://doi.org/10.1111/2041-210X.12073
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2017) GenBank. Nucleic Acids Res 45(D1):D37–D42. https://doi.org/10.1093/nar/gkw1070
Buchan A, Newell SY, Moreta JL, Moran MA (2002) Analysis of internal transcribed spacer (ITS) regions of rRNA genes in fungal communities in a southeastern U.S. salt marsh. Microb Ecol 43:329–340. https://doi.org/10.1007/s00248-001-1062-0
Cantrell SA, Hanlin RT (1997) Phylogenetic relationships in the family Hyaloscyphaceae inferred from sequences of ITS regions, 5.8S ribosomal DNA and morphological characters. Mycologia 89:745–755. https://doi.org/10.2307/3761131
Chlebická M (2009) Biodiversity of Lachnaceae on leaves of Acer, Quercus and Vaccinium. Mykol Listy Supplementum 2009:72–73
Farris JS, Albert VA, Källersjö M, Lipscomb D, Kluge AG (1996) Parsimony jackknifing outperforms neighbor-joining. Cladistics 12:99–124. https://doi.org/10.1111/j.1096-0031.1996.tb00196.x
Galtier N, Gouy M, Gautier C (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics 12:543–548
Gernandt DS, Camacho FJ, Stone JK (1997) Meria laricis, an anamorph of Rhabdocline. Mycologia 89:735–744. https://doi.org/10.2307/3761130
Goloboff PA, Farris JS, Nixon KC (2008) TNT, a free program for phylogenetic analysis. Cladistics 24:774–786. https://doi.org/10.1111/j.1096-0031.2008.00217.x
Guatimosim E, Schwartsburd PB, Crous PW, Barreto RW (2016) Novel fungi from an ancient niche: lachnoid and chalara-like fungi on ferns. Mycol Progress 15:1239–1267. https://doi.org/10.1007/s11557-016-1232-6
Han JG, Hosoya T, Sung GH, Shin HD (2014) Phylogenetic reassessment of Hyaloscyphaceae sensu lato (Helotiales, Leotiomycetes) based on multigene analyses. Fungal Biol 118:150–167. https://doi.org/10.1016/j.funbio.2013.11.004
Hein B (1980) Raster-elektronenmikroskopische Untersuchungen an Haaren von Hyaloscyphaceae (Ascomycetes, Helotiales). Nova Hedw 32:31–62
Hosoya T, Sasagawa R, Hosaka K, Sung GH, Hirayama Y, Yamaguchi K, Toyama K, Kakishima M (2010) Molecular phylogenetic studies of Lachnum and its allies based on the Japanese material. Mycoscience 51:170–181. https://doi.org/10.1007/s10267-009-0023-1
Huhtinen S (1987) Three new species, and the histochemical delimitation of genera in the glassy-haired Hyaloscyphaceae. Mycotaxon 29:267–283
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Bio Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kornerup A, Wanscher JH (1981) Taschenlexikon der Farben. Muster-Schmidt, Zürich
Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. https://doi.org/10.1093/nar/gkw290
Miyoshi T, Ono Y, Shimizu S (2007) Occurrence of concave stem canker of citrus in Ehime prefecture and detection of the pathogenic fungus Lachnum abnorme by PCR. Japanese J Phytopathol 73:9–14
Nilsson RH, Ryberg M, Sjökvist E, Abarenkov K (2011) Rethinking taxon sampling in the light of environmental sequencing. Cladistics 27:197–203. https://doi.org/10.1111/j.1096-0031.2010.00336.x
Perić B, Baral H-O (2014) Erioscyphella curvispora spec. nov. from Montenegro. Mycol Monten 17:89–104
Phillips W, Plowright CB (1880) New and rare British fungi. Grevillea 8:97–109
Raitviir A (1970) Synopsis of the Hyaloscyphaceae. Scripta Mycol 1:1–115
Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. https://doi.org/10.1093/sysbio/sys029
Ryberg M, Kristiansson E, Sjökvist E, Nilsson RH (2009) An outlook on the fungal internal transcribed spacer sequences in GenBank and the introduction of a web-based tool for the exploration of fungal diversity. New Phytol 181:471–477. https://doi.org/10.1111/j.1469-8137.2008.02667.x
Slavík B (1988) Regionálně fytogeografické členění. In: Hejný S, Slavík B (eds) Květena České socialistické republiky 1. Academia, Praha, pp 103–121
Spooner BM (1987) Helotiales of Australasia: Geoglossaceae, Orbiliaceae, Sclerotiniaceae, Hyaloscyphaceae. Bibl Mycol 116:1–711
Suková M (2005) A revision of selected material of lignicolous species of Brunnipila, Capitotricha, Dasyscyphella and Neodasyscypha from the Czech Republic. Czech Mycol 57:139–172
Vrålstad T, Myhre E, Schumacher T (2002) Molecular diversity and phylogenetic affinities of symbiotic root-associated ascomycetes of the Helotiales in burnt and metal polluted habitats. New Phytol 155:131–148
White TJ, Bruns T, Lee SB, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, pp 315–322
Zhao P, Zhuang W-Y (2011) Evaluation of ITS region as a possible DNA barcode for the genus Lachnum (Helotiales). Mycosystema 30:932–937
Acknowledgements
The first author would like to thank Andrzej Chlebicki (W. Szafer Institute of Botany, Polish Academy of Sciences, Kraków) for organizing the expedition to Chile and field trips to Austria and the Carpathians (project no. 2 P04F 066 28, granted by the Ministry of Science and Information Society Technologies, Poland). Hans-Otto Baral kindly reviewed the manuscript and proposed many detailed corrections. Vladimír Melichar and Petr Lepší are acknowledged for consultation regarding Alnus alnobetula in the Czech Republic. Dr. Kamil Zágoršek is thanked for his technical help with the scanning electron microscopy. The present study was supported by the Ministry of Culture of the Czech Republic (DKRVO 2017/08, National Museum, 00023272), the Ministry of Education, Youth and Sports of the Czech Republic (project CZ.1.07/2.3.00/30.0003), and BIOCEV—Biotechnology and Biomedicine Center of the Academy of Sciences and Charles University (no. CZ.1.05/1.1.00/02.0109).
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editors: Teresa Iturriaga and Marc Stadler
This article is part of the “Special Issue on ascomycete systematics in honour of Richard P. Korf who died in August 2016”.
Rights and permissions
About this article
Cite this article
Šandová, M., Nilsson, R.H. & Kolařík, M. Relationships within Capitotricha bicolor (Lachnaceae, Ascomycota) as inferred from ITS rDNA sequences, including some notes on the Brunnipila and Erioscyphella clades. Mycol Progress 17, 89–101 (2018). https://doi.org/10.1007/s11557-017-1346-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-017-1346-5