Abstract
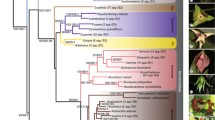
Cyttaria is a morphologically and biologically distinct genus comprising wood-inhabiting species that are biotrophic associates of trees in the genera Nothofagus sensu stricto and Lophozonia in southern South America, Australia, and New Zealand. The uniqueness of the fruit bodies and habitat of Cyttaria has led taxonomists to justify its placement in its own order, Cyttariales, in Leotiomycetes. A multilocus phylogenetic reconstruction of the class Leotiomycetes incorporating new sequence data from Cyttaria nigra shows Cyttariaceae to have an isolated position near the base of Helotiales, with weak support for a relationship with Polydesmia pruinosa (incertae sedis) and Chlorociboriaceae. Cyttariales is here proposed as a synonym of Helotiales.


source of the genome data for those that have had their genome sequenced (JGI joint genome initiative, MWLR Manaaki Whenua–Landcare Research, NCBI National Center for Biotechnology Information)
Similar content being viewed by others
Code availability
Not applicable.
References
Aveskamp MM, Verkley GJ, de Gruyter J, Murace MA, Perelló A, Woudenberg JHC, Groenewald JZ, Crous PW (2009) DNA phylogeny reveals polyphyly of Phoma section Peyronellaea and multiple taxonomic novelties. Mycologia 101:363–382. https://doi.org/10.3852/08-199
Baral HO, Haelewaters D, Pärtel K (2015) A new attempt to classify the families of the Helotiales. Poster presented at the Second International Workshop on Ascomycete Systematics, Amsterdam
Baral HO, Weber E, Marson G (2020) Monograph of Orbiliomycetes (Ascomycota) based on vital taxonomy. Part I National Museum of Natural History, Luxembourg
Chernomor O, von Haeseler A, Minh BQ (2016) Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol 65:997–1008. https://doi.org/10.1093/sysbio/syw037
Crisci JV, Gamundí IJ, Cabello MN (1988) A cladistic analysis of the genus Cyttaria (Fungi-Ascomycotina). Cladistics 4:279–290. https://doi.org/10.1111/j.1096-0031.1988.tb00475.x
Crous PW, Wingfield MJ, Burgess TI, Carnegie AJ, Hardy GESJ, Smith D, Summerell BA, Cano-Lira JF, Guarro J, Houbraken J, Lombard L, Martín MP, Sandoval-Denis M, Alexandrova AV, Barnes CW, Baseia IG, Bezerra JDP, Guarnaccia V, May TW, Hernández-Restrepo M, Stchigel AM, Miller AN, Ordoñez ME, Abreu VP, Accioly T, Agnello C, Colmán AA, Albuquerque CC, Alfredo DS, Alvarado P, Araújo-Magalhães GR, Arauzo S, Atkinson T, Barili A, Barreto RW, Bezerra JL, Cabral TS, Rodríguez FC, Cruz RHSF, Daniëls PP, da Silva BDB, de Almeida DAC, de Carvalho Júnior AA, Decock CA, Delgat L, Denman S, Dimitrov RA, Edwards J, Fedosova AG, Ferreira RJ, Firmino AL, Flores JA, García D, Gené J, Giraldo A, Góis JS, Gomes AAM, Gonçalves CM, Gouliamova DE, Groenewald M, Guéorguiev BV, Guevara-Suarez M, Gusmão LFP, Hosaka K, Hubka V, Huhndorf SM, Jadan M, Jurjević Ž, Kraak B, Kučera V, Kumar TKA, Kušan I, Lacerda SR, Lamlertthon S, Lisboa WS, Loizides M, Luangsa-ard JJ, Lysková P, Mac Cormack WP, Macedo DM, Machado AR, Malysheva EF, Marinho P, Matočec N, Meijer M, Mešić A, Mongkolsamrit S, Moreira KA, Morozova OV, Nair KU, Nakamura N, Noisripoom W, Olariaga I, Oliveira RJV, Paiva LM, Pawar P, Pereira OL, Peterson SW, Prieto M, Rodríguez-Andrade E, De Blas CR, Roy M, Santos ES, Sharma R, Silva GA, Souza-Motta CM, Takeuchi-Kaneko Y, Tanaka C, Thakur A, Smith MT, Tkalčec Z, Valenzuela-Lopez N, van der Kleij P, Verbeken A, Viana MG, Wang XW, Groenewald JZ et al (2017) Fungal planet description sheets: 625–715. Persoonia 39:270–467. https://doi.org/10.3767/persoonia.2017.39.11
Crous PW, Wingfield MJ, Burgess TI, Hardy GESJ, Gené J, Guarro J, Baseia IG, García D, Gusmão LFP, Souza-Motta CM, Thangavel R, Adamčík S, Barili A, Barnes CW, Bezerra JDP, Bordallo JJ, Cano-Lira JF, de Oliveira RJV, Ercole E, Hubka V, Iturrieta-González I, Kubátová A, Martín MP, Moreau PA, Morte A, Ordoñez ME, Rodríguez A, Stchigel AM, Vizzini A, Abdollahzadeh J, Abreu VP, Adamčíková K, Albuquerque GMR, Alexandrova AV, Álvarez Duarte E, Armstrong-Cho C, Banniza S, Barbosa RN, Bellanger JM, Bezerra JL, Cabral TS, Caboň M, Caicedo E, Cantillo T, Carnegie AJ, Carmo LT, Castañeda-Ruiz RF, Clement CR, Čmoková A, Conceição LB, Cruz RHSF, Damm U, da Silva BDB, da Silva GA, da Silva RMF, Santiago ALCMdA, de Oliveira LF, de Souza CAF, Déniel F, Dima B, Dong G, Edwards J, Félix CR, Fournier J, Gibertoni TB, Hosaka K, Iturriaga T, Jadan M, Jany JL, Jurjević Ž, Kolařík M, Kušan I, Landell MF, Leite Cordeiro TR, Lima DX, Loizides M, Luo S, Machado AR, Madrid H, Magalhães OMC, Marinho P, Matočec N, Mešić A, Miller AN, Morozova OV, Neves RP, Nonaka K, Nováková A, Oberlies NH, Oliveira-Filho JRC, Oliveira TGL, Papp V, Pereira OL, Perrone G, Peterson SW, Pham THG, Raja HA (2018) Fungal planet description sheets: 716–784. Persoonia 40:240–393. https://doi.org/10.3767/persoonia.2018.40.10
Crous PW, Wingfield MJ, Chooi Y-H, Gilchrist CLM, Lacey E, Pitt JI, Roets F, Swart WJ, Cano-Lira JF, Valenzuela-Lopez N, Hubka V, Shivas RG, Stchigel AM, Holdom DG, Jurjević Ž, Kachalkin AV, Lebel T, Lock C, Martín MP, Tan YP, Tomashevskaya MA, Vitelli JS, Baseia IG, Bhatt VK, Brandrud TE, De Souza JT, Dima B, Lacey HJ, Lombard L, Johnston PR, Morte A, Papp V, Rodríguez A, Rodríguez-Andrade E, Semwal KC, Tegart L, Abad ZG, Akulov A, Alvarado P, Alves A, Andrade JP, Arenas F, Asenjo C, Ballarà J, Barrett MD, Berná LM, Berraf-Tebbal A, Bianchinotti MV, Bransgrove K, Burgess TI, Carmo FS, Chávez R, Čmoková A, Dearnaley JDW, de Azevedo SALCM, Denman F-N, S, Douglas B, Dovana F, Eichmeier A, Esteve-Raventós F, Farid A, Fedosova AG, Ferisin G, Ferreira RJ, Ferrer A, Figueiredo CN, Figueiredo YF, Reinoso-Fuentealba CG, Garrido-Benavent I, Cañete-Gibas CF, Gil-Durán C, Glushakova AM, Gonçalves MFM, González M, Gorczak M, Gorton C, Guard FE, Guarnizo AL, Guarro J, Gutiérrez M, Hamal P, Hien LT, Hocking AD, Houbraken J, Hunter GC, Inácio CA, Jourdan M, Kapitonov VI, Kelly L, Khanh TN, Kisło K, Kiss L, Kiyashko A, Kolařík M, Kruse J, Kubátová A, Kučera V, Kučerová I, Kušan I, Lee HB, Levicán G, Lewis A, Liem NV, Liimatainen K, Lim HJ, Lyons MN, Maciá-Vicente JG, Magaña-Dueñas V, Mahiques R, Malysheva EF, Marbach PAS, Marinho P, Matočec N, McTaggart AR, Mešić A, Morin L, Muñoz-Mohedano JM, Navarro-Ródenas A, Nicolli CP, Oliveira RL, Otsing E, Ovrebo CL, Pankratov TA, Paños A, Paz-Conde A, Pérez-Sierra A, Phosri C, Pintos Á, Pošta A, Prencipe S, Rubio E, Saitta A, Sales LS, Sanhueza L, Shuttleworth LA, Smith J, Smith ME, Spadaro D, Spetik M, Sochor M, Sochorová Z, Sousa JO, Suwannasai N, Tedersoo L, Thanh HM, Thao LD, Tkalčec Z, Vaghefi N, Venzhik AS, Verbeken A, Vizzini A, Voyron S, Wainhouse M, Whalley AJS, Wrzosek M, Zapata M, Zeil-Rolfe I, Groenewald JZ, (2020) Fungal Planet description sheets: 1042–1111. Persoonia 44:301–459. https://doi.org/10.3767/persoonia.2020.44.11
Darlington PJ (1965) Biogeography of the southern end of the world; distribution and history of far-southern life and land, with an assessment of continental drift. Harvard University Press, Cambridge, MA
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Ekanayaka AH, Hyde KD, Gentekaki E, McKenzie EHC, Zhao Q, Bulgakov TS, Camporesi E (2019) Preliminary Classification of Leotiomycetes Mycosphere 10:310–489. https://doi.org/10.5943/mycosphere/10/1/7
Gamundí IJ (1971) Las Cyttariales sudamericanas (Fungi-Ascomycetes). Darwiniana 16:461–510
Haelewaters D, Filippova NV, Baral H-O (2018) A new species of Stamnaria (Leotiomycetes, Helotiales) from Western Siberia. MycoKeys 32:49–63. https://doi.org/10.3897/mycokeys.32.23277
Hernández-Restrepo M, Gené J, Castañeda-Ruiz RF, Mena-Portales J, Crous PW, Guarro J (2017) Phylogeny of saprobic microfungi from Southern Europe. Stud Mycol 86:53–97. https://doi.org/10.1016/j.simyco.2017.05.002
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS (2018) UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522. https://doi.org/10.1093/molbev/msx281
Hopple JS (1994) Phylogenetic investigations in the genus Coprinus based on morphological and molecular characters. PhD dissertation. Duke University, Durham
Hosoya T (2009) Enumeration of remarkable Japanese discomycetes (3): first records of three inoperculate helotialean discomycetes in Japan. Bull Natl Mus Nat Sci, Ser B 35:113–121
Jaklitsch W, Baral H-O, Lücking R, Lumbsch HT, Frey W (2016) Syllabus of plant families: A. Engler’s syllabus der Pflanzenfamilien part 1/2. Borntraeger, Stuttgart
Johnston PR, Baschien C (2020) Tricladiaceae fam. nov. (Helotiales, Leotiomycetes). Fungal Syst Evol 6:233–242. https://doi.org/10.3114/fuse.2020.06.10
Johnston PR, Quijada L, Smith CA, Baral H-O, Hosoya T, Baschien C, Pärtel K, Zhuang W-Y, Haelewaters D, Park D, Carl S, López-Giráldez F, Wang Z, Townsend JP (2019) A multigene phylogeny toward a new phylogenetic classification of Leotiomycetes. IMA Fungus 10:1. https://doi.org/10.1186/s43008-019-0002-x
Kalyaanamoorthy K, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. https://doi.org/10.1038/nmeth.4285
Karakehian JM, Quijada L, Friebes G, Tanney JB, Pfister DH (2019) Placement of Triblidiaceae in Rhytismatales and comments on unique ascospore morphologies in Leotiomycetes (Fungi, Ascomycota). MycoKeys 54:99–133. https://doi.org/10.3897/mycokeys.54.35697
Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008) Ainsworth and Bisby’s dictionary of the fungi. CAB International, Wallingford
Knapp M, Stöckler K, Havell D, Delsuc F, Sebastiani F, Lockhart PJ (2005) Relaxed molecular clock provides evidence for long-distance dispersal of Nothofagus (southern beech). Plos Biol 3:e14. https://doi.org/10.1371/journal.pbio.0030014
Korf RP (1983) Cyttaria (Cyttariales): Coevolution with Nothofagus, and evolutionary relationship to the Boedijnopezizeae (Pezizales, Sarcoscyphaceae). Aust J Bot Suppl Ser 13:77–87
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Landvik S, Eriksson OE (1994) Relationships of Tuber, Elaphomyces, and Cyttaria (Ascomycotina), inferred from 18S rDNA studies. In: Hawksworth DL (ed) Ascomycetes systematics: problems and perspectives in the nineties. Springer, Boston, pp 225–231. https://doi.org/10.1007/978-1-4757-9290-4_19
Liu YJ, Whelen S, Hall BD (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol 16:1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
Matheny PB (2005) Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol Phylogenet Evol 35:1–20. https://doi.org/10.1016/j.ympev.2004.11.014
Matheny PB, Liu YJ, Ammirati JF, Hall BD (2002) Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am J Bot 89:688–698. https://doi.org/10.3732/ajb.89.4.688
Mengoni TP (1986) El aparato apical del asco de Cyttaria harioti (Ascomycetes-Cyttariales) con microscopía fotónica y electronica. Bol Soc Argent Bot 24:393–401
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inferences of large phylogenetic trees. Proc Gateway Comp Environ Workshop 14:1–8. https://doi.org/10.1109/GCE.2010.5676129
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol 32:268–274. https://doi.org/10.1093/molbev/msu300
Pärtel K, Baral H-O, Tamm H, Põldmaa K (2017) Evidence for the polyphyly of Encoelia and Encoelioideae with reconsideration of respective families in Leotiomycetes. Fungal Divers 82:183–219. https://doi.org/10.1007/s13225-016-0370-0
Peterson KR, Pfister DH (2010) Phylogeny of Cyttaria inferred from nuclear and mitochondrial sequence and morphological data. Mycologia 102:1398–1416. https://doi.org/10.3852/10-046
Peterson KR, Pfister DH, Bell CD (2010) Cophylogeny and biogeography of the fungal parasite Cyttaria and its host Nothofagus, southern beech. Mycologia 102:1417–1425. https://doi.org/10.3852/10-048
Pino-Bodas R, Zhurbenko MP, Stenroos S (2017) Phylogenetic placement within Lecanoromycetes of lichenicolous fungi associated with Cladonia and some other genera. Persoonia 39:91–117. https://doi.org/10.3767/persoonia.2017.39.05
Prieto M, Schultz M, Olariaga I, Wedin M (2019) Lichinodium is a new lichenized lineage in the Leotiomycetes. Fungal Divers 94:23–39. https://doi.org/10.1007/s13225-018-0417-5
Quandt CA, Haelewaters D (2021) Phylogenetic advances in Leotiomycetes, an understudied clade of taxonomically and ecologically diverse fungi. In: Zaragoza Ó, Casadevall A (eds) Encyclopedia of Mycology, Vol 1. Elsevier, Oxford, pp. 284–294. https://doi.org/10.1016/B978-0-12-819990-9.00052-4
Quijada L, Tanney JB, Popov E, Johnston PR, Pfister DH (2020) Cones, needles and wood: Micraspis (Micraspidaceae, Micraspidales fam. et ord. nov.) speciation segregates by host plant tissues. Fungal Syst Evol 5:99–111. https://doi.org/10.3114/fuse.2020.05.05
Rehner SA, Buckley E (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97:84–98. https://doi.org/10.3852/mycologia.97.1.84
Santesson R (1945) Cyttaria, a genus of inoperculate discomycetes. Sven Bot Tidskr 39:319–345
Schmitt I, Crespo A, Divakar PK, Fankhauser JD, Herman-Sackett E, Kalb K, Nelsen MP, Nelson NA, Rivas-Plata E, Shimp AD, Widhelm T, Lumbsch HT (2009) New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia 23:35–40. https://doi.org/10.3767/003158509x470602
Schwessinger B, McDonald M (2017) High quality DNA from fungi for long read sequencing e.g. PacBio, Nanopore MinION. protocols.io. https://doi.org/10.17504/protocols.io.k6qczdw
Setoguchi H (2005) Co-evolution of Nothofagus plants and the ascomycete Cyttaria spp. with the Gondwanaland breakup. In: Sugiyama J (ed) Diversity and evolution of fungi, bacteria and viruses. Shokabo Publishing, Tokyo, pp 155–156
Spegazzini CL (1888) Fungi fuegiana. Bol Acad Nac Cienc Córdoba 11:135–311
Steenis CGGJ (1971) Nothofagus, key genus of plant geography, in time and space, living and fossil, ecology and phylogeny. Blumea 19:65–98
Stenroos S, Laukka T, Huhtinen S, Döbbeler P, Myllys L, Syrjänen K, Hyvönen J (2010) Multiple origins of symbioses between ascomycetes and bryophytes suggested by a five-gene phylogeny. Cladistics 26:281–300. https://doi.org/10.1111/j.1096-0031.2009.00284.x
Stiller JW, Hall BD (1997) The origin of red algae: implications for plastid evolution. Proc Natl Acad Sci USA 94:4520–4525. https://doi.org/10.1073/pnas.94.9.4520
Suija A, Ertz D, Lawrey JD, Dierderich P (2015) Multiple origin of the lichenicolous life habit in Helotiales, based on nuclear ribosomal sequences. Fungal Divers 70:55–72. https://doi.org/10.1007/s13225-014-0287-4
Tanney JB, Seifert KA (2020) Mollisiaceae: An overlooked lineage of diverse endophytes. Stud Mycol 95:293–380. https://doi.org/10.1016/j.simyco.2020.02.005
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. https://doi.org/10.1128/JB.172.8.4238-4246.1990
Wang Z, Johnston PR, Takamatsu S, Spatafora JW, Hibbett DS (2006) Toward a phylogenetic classification of the Leotiomycetes based on rDNA data. Mycologia 98:1065–1075. https://doi.org/10.3852/mycologia.98.6.1065
White TJ, Bruns TD, Lee SB, Taylor JW (1990) Analysis of phylogenetic relationships by amplification and direct sequencing of ribosomal RNA genes. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
Wijayawardene NN, Hyde KD, Al-Ani LKT, Tedersoo L, Haelewaters D, Rajeshkumar KC, Zhao RL, Aptroot A, Leontyev DV, Saxena RK, Tokarev YS, Dai DQ, Letcher PM, Stephenson SL, Ertz D, Lumbsch HT, Kukwa M, Issi IV, Madrid H, Phillips AJL, Selbmann L, Pfliegler WP, Horváth E, Bensch K, Kirk PM, Kolaříková K, Raja HA, Radek R, Papp V, Dima B, Ma J, Malosso E, Takamatsu S, Rambold G, Gannibal PB, Triebel D, Gautam AK, Avasthi S, Suetrong S, Timdal E, Fryar SC, Delgado G, Réblová M, Doilom M, Dolatabadi S, Pawłowska J, Humber RA, Kodsueb R, Sánchez-Castro I, Goto BT, Silva DKA, de Souza FA, Oehl F, da Silva GA, Silva IR, Błaszkowski J, Jobim K, Maia LC, Barbosa FR, Fiuza PO, Divakar PK, Shenoy BD, Castañeda-Ruiz RF, Somrithipol S, Lateef AA, Karunarathna SC, Tibpromma S, Mortimer PE, Wanasinghe DN, Phookamsak R, Xu J, Wang Y, Tian F, Alvarado P, Li DW, Kušan I, Matočec N, Maharachchikumbura SSN, Papizadeh M, Heredia G, Wartchow F, Bakhshi M, Boehm E, Youssef N, Hustad VP, Lawrey JD, Santiago ALCMA, Bezerra JDP, Souza-Motta CM, Firmino AL, Tian Q, Houbraken J, Hongsanan S, Tanaka K, Dissanayake AJ, Monteiro JS, Grossart HP, Suija A, Weerakoon G, Etayo J, Tsurykau A, Vázquez V, Mungai P, Damm U, Li QR, Zhang H, Boonmee S, Lu YZ, Becerra AG, Kendrick B, Brearley FQ, Motiejūnaitė J, Sharma B, Khare R, Gaikwad S, Wijesundara DSA, Tang LZ, He MQ, Flakus A, Rodriguez-Flakus P, Zhurbenko MP, McKenzie EHC, Stadler M, Bhat DJ, Liu JK, Raza M, Jeewon R, Nassonova ES, Prieto M, Jayalal RGU, Erdoğdu M, Yurkov A, Schnittler M, Shchepin ON, Novozhilov YK, Silva-Filho AGS, Liu P, Cavender JC, Kang Y, Mohammad S, Zhang LF, Xu RF, Li YM, Dayarathne MC, Ekanayaka AH, Wen TC, Deng CY, Pereira OL, Navathe S, Hawksworth DL, Fan XL, Dissanayake LS, Kuhnert E, Grossart HP, Thines M (2020) Outline of fungi and fungus-like taxa. Mycosphere 11:1060–1456. https://doi.org/10.5943/mycosphere/11/1/8
Zoller S, Scheidegger C, Sperisen C (1999) PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. Lichenologist 31:511–516. https://doi.org/10.1006/lich.1999.0220
Funding
This work was supported in part by the U.S. National Science Foundation (DEB-2018098 to D.H.). P.R.J. and D.P. were supported through the Manaaki Whenua–Landcare Research Biota Portfolio with funding from the Science and Innovation Group of the New Zealand Ministry of Business, Innovation and Employment.
Author information
Authors and Affiliations
Contributions
D.P. and P.R.J. generated sequence data. D.H. performed phylogenetic analyses. D.H. and P.R.J. wrote the first draft of the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Section editor: Roland Kirschner
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Haelewaters, D., Park, D. & Johnston, P.R. Multilocus phylogenetic analysis reveals that Cyttariales is a synonym of Helotiales. Mycol Progress 20, 1323–1330 (2021). https://doi.org/10.1007/s11557-021-01736-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-021-01736-2