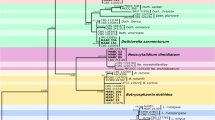
Abstract
Grapevine trunk diseases (GTDs) are destructive fungal diseases and economically important threat to grapevine industry worldwide. During a survey of vineyards in the Kurdistan Province, 233 fungal isolates were obtained that associated with various external and internal trunk diseases symptoms. Based on sequence data and morphological characteristics, 24 species belonging to 19 genera were characterized. Botryosphaeriaceae species were the most prevalent identified fungi followed by Alternaria, Sporocadaceae, Phaeoacremonium, and Didymellaceae taxa. At the species level, Botryosphaeria dothidea, Alternaria malorum, Phaeoacremonium aleophilum, Acremonium sclerotigenum, Alternaria chlamydosporigena, and Neoscytalidium dimidiatum were the most frequent identified species. Acremonium sclerotigenum, A. chlamydosporigena, A. malorum, Juxtiphoma eupyrena, and Paecilomyces formosus are new records associated with GTDs around the world. Among the four pestalotia-like species identified based on morphology and phylogenetic analyses of LSU, ITS, TEF1, and TUB2 sequence data, three species are newly described and introduced here as Seimatosporium marivanicum, Sporocadus kurdistanicus, and Xenoseimatosporium kurdistanicum. Furthermore, three new combinations are proposed, including Sporocadus corni, Spo. italicus, and Spo. pseudocorni. Pathogenicity of 15 species was studied on cultivars Rasha and Bidaneh Sefid in field conditions. The pathogenicity of Neoscytalidium novahollandiae, B. dothidea, Ph. aleophilum, and Phaeomoniella chlamydospora was confirmed in field conditions.
Graphical abstract








Similar content being viewed by others
Data availability
Requests for data and materials should be addressed to Jafar Abdollahzadeh.
References
Abdollahzadeh J, Goltapeh EM, Javadi A, Shams-Bakhsh M, Zare R, Phillips AJL (2009) Barriopsis iraniana and Phaeobotryon cupressi: two new species of the Botryosphaeriaceae from trees in Iran. Persoonia 23:1–8. https://doi.org/10.3767/003158509X467552
Abed Ashtiani F, Narmani A, Arzanlou M (2019) Analysis of Kalmusia variispora associated with grapevine decline in Iran. Eur J Plant Pathol 154:787–799. https://doi.org/10.1007/s10658-019-01702-5
Akgül DS, Savaş NG, Özarslandan M (2019) First report of wood canker caused by Lasiodiplodia exigua and Neoscytalidium novaehollandiae on grapevine in Turkey. Plant Dis 103:1036. https://doi.org/10.1094/PDIS-11-18-1938-PDN
Alves A, Crous PW, Correia A, Phillips AJL (2008) Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Divers 28:1–13
Amarloo OA, Mohammadi H, Mahdian SA, Ghanbary T (2020) Identification and pathogenicity of fungal species associated with grapevine trunk diseases in Khorasan-Razavi province, Iran. Mycol Iran 7:83–94. https://doi.org/10.22043/MI.2020.122800
Arabnezhad M, Mohammadi H, Massumi H, Farahmand F (2013) Fungal flora associated with internal symptoms of grapevine decline in Kerman province. Agric Biotechnol J 6:115–132. https://doi.org/10.22103/JAB.2014.1316
Armengol J, Vicent A, Torné L, García-Figueres F, García-Jiménez J (2001) Fungi associated with esca and grapevine declines in Spain: a three-year survey. Phytopathol Mediterr 40:S325–S329. https://doi.org/10.14601/Phytopathol_Mediterr-1621
Arzanlou M, Narmani A, Moshari S, Khodaei S, Babai Ahari A (2013) Truncatella angustata associated with grapevine trunk disease in northern Iran. Arch Phytopathol Pflanzenschutz 46:1168–1181. https://doi.org/10.1080/03235408.2012.761417
Bahmani Z, Abdollahzadeh J, Amini J, Evidente A (2021) Biscogniauxia rosacearum the charcoal canker agent as a pathogen associated with grapevine trunk diseases in Zagros region of Iran. Sci Rep 11:14098. https://doi.org/10.1038/s41598-021-93630-w
Barber PA, Crous PW, Groenewald JZ, Pascoe IG, Keane P (2011) Reassessing Vermisporium (Amphisphaeriaceae), a genus of foliar pathogens of eucalypts. Persoonia 27:90–118. https://doi.org/10.3767/003158511X617381
Bertsch C, Ramírez-Suero M, Magnin-Robert M, Larignon P, Chong J, Abou-Mansour E, Spagnolo A, Clément C, Fontaine F (2013) Grapevine trunk diseases: complex and still poorly understood. Plant Pathol 62:243–265. https://doi.org/10.1111/j.1365-3059.2012.02674.x
Bonthond G, Sandoval-Denis M, Groenewald JZ, Crous PW (2018) Seiridium (Sporocadaceae): an important genus of plant pathogenic fungi. Persoonia 40:96–118. https://doi.org/10.3767/persoonia.2018.40.04
Carbone I, Kohn LM (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556. https://doi.org/10.2307/3761358
Casieri L, Hofstetter V, Viret O, Gindro K (2009) Fungal communities living in the wood of different cultivars of young Vitis vinifera. Phytopathol Mediterr 48:73–83. https://doi.org/10.14601/Phytopathol_Mediterr-2876
Cimmino A, Bahmani Z, Masi M, Di Lecce R, Amini J, Abdollahzadeh J, Tuzi A, Evidente A (2020) Massarilactones D and H, phytotoxins produced by Kalmusia variispora, associated with grapevine trunk diseases (GTDs) in Iran. Nat Prod Res. https://doi.org/10.1080/14786419.2020.1791116
Cimmino A, Bahmani Z, Masi M, Abdollahzadeh J, Amini J, Tuzi A, Evidente A (2021) Phytotoxins produced by Didymella glomerata and Truncatella angustata, associated with grapevine trunk diseases (GTDs) in Iran. Nat Prod Res. https://doi.org/10.1080/14786419.2021.1979544
Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G (2004) MycoBank: an online initiative to launch mycology into the 21st century. Stud Mycol 50:19–22
Crous PW, Carris LM, Giraldo A, Groenewald JZ, Hawksworth DL, Hemández-Restrepo M, Jaklitsch WM, Lebrun MH, Schumacher RK, Stielow JB, Van der Linde EJ, Vilcāne J, Voglmayr H, Wood AR (2015) The genera of fungi-fixing the application of the type species of generic names–G 2: Allantophomopsis, Latorua, Macrodiplodiopsis, Macrohilum, Milospium, Protostegia, Pyricularia, Robillarda, Rotula, Septoriella, Torula, and Wojnowicia. IMA Fungus 6:163–198. https://doi.org/10.5598/imafungus.2015.06.01.11
Crous PW, Liu F, Cai L, Barber PA, Thangavel R, Summerell BA, Wingfield MJ, Edwards J, Carnegie AJ, Groenewald JZ (2018) Allelochaeta (Sporocadaceae): pigmentation lost and gained. Fungal Sys Evol 2:273–309. https://doi.org/10.3114/fuse.2018.02.08
De Hoog GS, Guarru J, Gene J, Figueras MJ (2000) Atlas of clinical fungi. Centralbureau voor Schimmel Cultures.
Dubos B, Larignon P (1988) Esca and black measles. In: Pearson RC, Goheen AC (eds) Compendium of grape diseases. American Phytopathological Society, St. Paul, pp 34–35
Farr DF, Rossman AY (2021) Fungal Databases, U.S. National Fungus Collections, ARS, USDA. https://nt.ars-grin.gov/fungaldatabases/. Accessed May 2021.
Fischer M, Kassemeyer HH (2003) Fungi associated with esca disease of grapevine in Germany. Vitis 42:109–116. https://doi.org/10.5073/vitis.2003.42.109-116
Fischer M, Peighami Ashnaei S (2019) Grapevine, esca complex, and environment: the disease triangle. Phytopathol Mediterr 58:17–37. https://doi.org/10.14601/Phytopathol_Mediterr-25086
Fontaine F, Pinto C, Vallet J, Clement C, Gomes AC, Spagnolo A (2016) The effects of grapevine trunk diseases (GTDs) on vine physiology. Eur J Plant Pathol 144:707–721. https://doi.org/10.1007/s10658-015-0770-0
Glass NL, Donaldson GC (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61:1323–1330. https://doi.org/10.1128/aem.61.4.1323-1330.1995
Gramaje D, Muñoz RM, Lerma ML, Garcia-Jimenez J, Armengol J (2009) Fungal grapevine trunk pathogens associated with Syrah decline in Spain. Phytopathol Mediterr 48:396–402. https://doi.org/10.14601/Phytopathol_Mediterr-2934
Gramaje D, Mostert L, Groenewald JZ, Crous PW (2015) Phaeoacremonium: from esca disease to phaeohyphomycosis. Fungal Biol 119:759–783. https://doi.org/10.1016/j.funbio.2015.06.004
Graniti A, Surico G, Mugnai L (2000) Esca of grapevine: a disease complex or a complex of diseases? Phytopathol Mediterr 39:16–20. https://doi.org/10.14601/Phytopathol_Mediterr-1539
Hall T (2004) BioEdit version 7.0.0. In: Department of Microbiology. North Carolina State University, North Carolina
Hatakeyama S, Harada Y (2004) A new species of Discostroma and its anamorph Seimatosporium with two morphological types of conidia, isolated from the stems of Paeonia suffruticosa. Mycoscience 45:106–111. https://doi.org/10.1007/s10267-003-0158-4
Hergholi N, Ghosta Y, Javan-Nikkhah M, Campisano A, Pancher M (2015) New species of endophytic fungi from grapevine (Vitis vinifera) in Iran. Rostaniha 16:17–35. https://doi.org/10.22092/botany.2015.101977
Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. https://doi.org/10.1093/bioinformatics/17.8.754
Jaklitsch WM, Gardiennet A, Voglmayr H (2016) Resolution of morphology based taxonomic delusions: Acrocordiella, Basiseptospora, Blogiascospora, Clypeosphaeria, Hymenopleella, Lepteutypa, Pseudapiospora, Requienella, Seiridium and Strickeria. Persoonia 37:82–105. https://doi.org/10.3767/003158516X690475
Jayawardene RS, Liu M, Maharachchikumbura SSN, Zhang W, Xing Q, Hyde KD, Nilthong S, Li X, Yan J (2016) Neopestalotiopsis vitis sp. nov. causing grapevine leaf spot in China. Phytotaxa 258(1):63–74. https://doi.org/10.1007/s00284-015-0955-y
Jeewon R, Liew EC, Hyde KD (2002) Phylogenetic relationships of Pestalotiopsis and allied genera inferred from ribosomal DNA sequences and morphological characters. Mol Phylogenet Evol 25:378–392. https://doi.org/10.1016/s1055-7903(02)00422-0
Jeewon R, Edward CY, Hyde KD (2003) Molecular systematics of the Amphisphaeriaceae based on cladistic analyses of partial LSU rDNA gene sequences. Mycol Res 107:1392–1402. https://doi.org/10.1017/s095375620300875x
Larignon P, Dubos B (1997) Fungi associated with esca disease in grapevine. Eur J Plant Pathol 103:147–157. https://doi.org/10.1023/A:1008638409410
Lawrence DP, Travadon R, Baumgartner K (2018) Novel Seimatosporium species from grapevine in northern California and their interactions with fungal pathogens involved in the trunk-disease complex. Plant Dis 102:1081–1092. https://doi.org/10.1094/PDIS-08-17-1247-RE
Lee S, Crous PW, Wingfield MJ (2006) Pestalotioid fungi from Restionaceae in the Cape Floral Kingdom. Stud Mycol 55:175–187. https://doi.org/10.3114/sim.55.1.175
Liu YJ, Whelen S, Hall BD (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol 16:1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
Liu F, Bonthond G, Groenewald JZ, Cai L, Crous PW (2019) Sporocadaceae, a family of coelomycetous fungi with appendage-bearing conidia. Stud Mycol 92:287–415. https://doi.org/10.1016/j.simyco.2018.11.001
Lo Piccolo S, Alfonzo A, Giambra S, Conigliaro G, Lopez-Llorca LV, Burruano S (2015) Identification of Acremonium isolates from grapevines and evaluation of their antagonism towards Plasmopara viticola. Ann Microbiol 65:2393–2403. https://doi.org/10.1007/s13213-015-1082-5
Maharachchikumbura SSN, Larignon P, Hyde KD, Al-Sadi AM, Liu ZY (2016) Characterization of Neopestalotiopsis, Pestalotiopsis and Truncatella species associated with grapevine trunk diseases in France. Phytopathol Mediterr 55:380–390. https://doi.org/10.14601/Phytopathol_Mediterr-18298
Miller MA, Pfeiffer W, Schwartz T (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. In: Proceedings of the 1st conference of the extreme science and engineering discovery environment: Bridging from the extreme to the campus and beyond. Association for Computing Machinery, USA, pp 1–8
Mirabolfathy M, Hosseinian L, Peighami Ashnaei S (2021) Fungal communities of grapevine decline in the main grape-growing regions of Iran. Indian Phytopathol. https://doi.org/10.1007/s42360-020-00314-y
Mohammadi H, Banihashemi Z, Gramaje D, Armengol J (2013) Fungal pathogens associated with grapevine trunk diseases in Iran. J Agric Sci Technol 15:137–150. https://doi.org/10.22043/MI.2020.122800
Mostert L, Halleen F, Fourie P, Crous PW (2006) A review of Phaeoacremonium species involved in petri disease and esca of grapevines. Phytopathol Mediterr 45:S12–S29. https://doi.org/10.3852/08-222
Mugnai L, Graniti A, Surico G (1999) Esca (black measles) and brown wood-streaking: two old and elusive diseases of grapevines. Plant Dis 83:404–418. https://doi.org/10.1094/PDIS.1999.83.5.404
Nag Raj TR (1993) Coelomycetous anamorphs with appendage-bearing conidia. Mycologue publications, Canada
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Sweden.
O'Donnell K, Cigelnik E (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol 7:103–116. https://doi.org/10.1006/mpev.1996.0376
O'Donnell K, Kistler HC, Cigelnik E, Ploetz RC (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci U S A 95:2044–2049. https://doi.org/10.1073/pnas.95.5.2044
Pintos C, Redondo V, Costas D, Aguin O, Mansilla P (2018) Fungi associated with grapevine trunk diseases in nursery-produced Vitis vinifera plants. Phytopathol Mediterr 57:407–424. https://doi.org/10.14601/Phytopathol_Mediterr-22964
Pirozynski KA, Shoemaker RA (1970) Seimatosporium leaf spot of Ledum and Rhododendron. Can J Bot 48:2199–2203
Raeder U, Broda P (1985) Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol 1:17–20. https://doi.org/10.1111/j.1472-765X.1985.tb01479.x
Rayner RW (1970) A mycological colour chart. CMI and British Mycological Society 34 pp.
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Senanayake IC, Maharachchikumbura SSN, Hyde KD, Bhat JD, Gareth Jones EB, McKenzie EHC, Dai DQ, Daranagama DA, Dayarathne MC, Goonasekara ID, Konta S, Li WJ, Shang QJ, Stadler M, Wijayawardene NN, Xiao YP, Norphanphoun C, Li Q, Liu XZ et al (2015) Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers 73:73–144. https://doi.org/10.1007/s13225-015-0340-y
Shoemaker RA (1964) Seimatosporium (= Cryptostictis) parasites of Rosa, Vitis, and Cornus. Can J Bot 42:411–417
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Steel CC, Greer LA, Savocchia S (2007) Studies on Colletotrichum acutatum and Greeneria uvicola: two fungi associated with bunch rot of grapes in sub-tropical Australia. Aust J Grape Wine Res 13:23–29. https://doi.org/10.1111/j.1755-0238.2007.tb00068.x
Surico G (2009) Towards a redefinition of the diseases within the esca complex of grapevine. Phytopathol Mediterr 48:5–10. https://doi.org/10.14601/Phytopathol_Mediterr-2870
Surico G, Mugnai L, Marchi G (2006) Older and more recent observations on esca: a critical review. Phytopathol Mediterr 45:68–86. https://doi.org/10.14601/Phytopathol_Mediterr-1847
Sutton BC (1975) Coelomycetes V. Coryneum. Mycol Pap 138:1–224
Tanaka K, Endo M, Hirayama K, Okane I, Hosoya T, Sato T (2011) Phylogeny of Discosia and Seimatosporium, and introduction of Adisciso and Immersidiscosia genera nova. Persoonia 26:85–98. https://doi.org/10.3767/003158511X576666
Trouillas FP, Úrbez-Torres JR, Gubler WD (2010) Diversity of diatrypaceous fungi associated with grapevine canker diseases in California. Mycologia 102:319–336. https://doi.org/10.3852/08-185
Úrbez-Torres JR, Adams P, Kamas J, Gubler WD (2009) Identification, incidence, and pathogenicity of fungal species associated with grapevine dieback in Texas. Am J Enol Vitic 60:497–507
Úrbez-Torres JR, Peduto F, Striegler RK, Urrea-Romero KE, Rupe JC, Cartwright RD, Gubler WD (2012) Characterization of fungal pathogens associated with grapevine trunk diseases in Arkansas and Missouri. Fungal Divers 52:169–189. https://doi.org/10.1007/s13225-011-0110-4
Úrbez-Torres JR, Peduto F, Smith RJ, Gubler WD (2013) Phomopsis dieback: a grapevine trunk disease caused by Phomopsis viticola in California. Plant Dis 97:1571–1579. https://doi.org/10.1094/PDIS-11-12-1072-RE
Úrbez-Torres JR, Haag P, Bowen P, O’Gorman DT (2014) Grapevine trunk diseases in British Columbia: Incidence and characterization of the fungal pathogens associated with esca and Petri diseases of grapevine. Plant Dis 98:469–482. https://doi.org/10.1094/PDIS-05-13-0523-RE
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
Wanasinghe DN, Phukhamsakda C, Hyde KD, Jeewon R, Lee HB, Gareth Jones EB, Tibpromma S, Tennakoon DS, Dissanayake AJ, Jayasiri SC, Gafforov Y, Camporesi E, Bulgakov TS, Ekanayake AH, Perera RH, Samarakoon MC, Goonasekara ID, Mapook A, Li W-J et al (2018) Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers 89:1–236. https://doi.org/10.1007/s13225-018-0395-7
Watanabe K, Motohashi K, Ono Y (2010) Description of Pestalotiopsis pallidotheae: a new species from Japan. Mycoscience 51:182–188. https://doi.org/10.1007/S10267-009-0025-Z
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innes MA, Gelfand DH, Sninsky JJ et al (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
White CL, Halleen F, Fischer M, Mostert L (2011) Characterization of the fungi associated with esca diseased grapevines in South Africa. Phytopathol Mediterr 50:S204–S223. https://doi.org/10.14601/Phytopathol_Mediterr-8983
Wijayawardene NN, Goonasekara ID, Camporesi E, Wang Y, An YL (2016a) Two new Seimatosporium species from Italy. Mycosphere 7:204–213. https://doi.org/10.5943/mycosphere/7/2/9
Wijayawardene NN, Hyde KD, Wanasinghe DN, Papizadeh M, Goonasekara ID, Camporesi E, Bhat DJ, McKenzie EH, Phillips AJL, Diederich P, Tanaka K, Li WJ, Tangthirasunun N, Phookamsak R, Dai DQ, Dissanayake AJ, Weerakoon G, Maharachchikumbura SSN, Hashimoto A et al (2016b) Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Divers 77:1–316. https://doi.org/10.1007/s13225-016-0360-2
Woudenberg JHC, Aveskamp MM, de Gruyter J, Spiers AG, Crous PW (2009) Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia 22:56–62. https://doi.org/10.3767/003158509X427808
Acknowledgements
We thank Mr. Alireza Javadi for his assistance in preparation of holotypes and recording growth rate of new species introduced here. Some sequences were amplified and generated in the context of a sabbatical leave at the Westerdijk Fungal Biodiversity Institute (Utrecht, the Netherlands) in 2018.
Funding
This study was supported by the University of Kurdistan. Jafar Abdollahzadeh was supported by the Kurdistan Provincial Office under project number 65/6/64197/2011.
Author information
Authors and Affiliations
Contributions
Project conception and design, photography, phylogeny, GenBank, TreeBASE and MycoBank submissions and writing the manuscript: Jafar Abdollahzadeh. Sampling, fungal isolation and purification, slide preparation and molecular experiments: Esmail Khaledi and Jamal Nahvi Moghadam. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Hans-Josef Schroers
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Fig. S1
Single gene tree inferred from a Bayesian analysis based on LSU sequence data. Bayesian posterior probabilities (PP) values are indicated at the nodes. The scale bar represents the expected number of changes per site. The tree was rooted with Distononappendiculata banksiae (CBS 143906) (PDF 293 kb)
Fig. S2
Single gene tree inferred from a Bayesian analysis based on ITS sequence data. Bayesian posterior probabilities (PP) values are indicated at the nodes. The scale bar represents the expected number of changes per site. The tree was rooted with Distononappendiculata banksiae (CBS 143906) (PDF 304 kb)
Fig. S3
Single gene tree inferred from a Bayesian analysis based on TEF1 sequence data. Bayesian posterior probabilities (PP) values are indicated at the nodes. The scale bar represents the expected number of changes per site. The tree was rooted with Distononappendiculata banksiae (CBS 143906) (PDF 277 kb)
Fig. S4
Single gene tree inferred from a Bayesian analysis based on TUB2 sequence data. Bayesian posterior probabilities (PP) values are indicated at the nodes. The scale bar represents the expected number of changes per site. The tree was rooted with Distononappendiculata banksiae (CBS 143906) (PDF 293 kb)
Rights and permissions
About this article
Cite this article
Moghadam, J.N., Khaledi, E., Abdollahzadeh, J. et al. Seimatosporium marivanicum, Sporocadus kurdistanicus, and Xenoseimatosporium kurdistanicum: three new pestalotioid species associated with grapevine trunk diseases from the Kurdistan Province, Iran. Mycol Progress 21, 427–446 (2022). https://doi.org/10.1007/s11557-021-01764-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-021-01764-y