Abstract
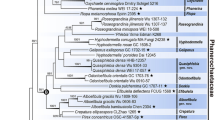
Myxomycetes, or plasmodial slime molds, are a monophyletic group of amoeboid protists whose classification is based mainly on morphological features of fruiting bodies. Although published phylogenies based on one or two genetic markers have clarified the boundaries of the main order-level systematic groups, the position and composition of some families and genera of myxomycetes are still a topic for discussion. In this study, we reconstructed the phylogeny of the family Didymiaceae based on three independent genetic markers: the 18S rDNA gene, the translation elongation factor 1-alpha, and the cytochrome c oxidase subunit 1 gene. Maximum likelihood and Bayesian inference phylogenetic analyses produced congruent topologies and showed that of the five major genera of the family, only species of the genus Diachea form a monophyletic clade, while the other four genera are clearly para- or polyphyletic. Species of the genus Didymium form a monophyletic clade with the only species of the genus Mucilago. The polymorphic species Lepidoderma tigrinum is clearly placed among 13 species of Diderma, including the type species of the genus. All other studied species of Lepidoderma form a separate clade together with Diderma fallax. We thus extend the latest nomenclatural revisions by disbanding the genera Mucilago and Lepidoderma, whereby the single species of Mucilago is transferred to the genus Didymium and L. tigrinum to Diderma. Extended taxon sampling allows the transfer of more nivicolous species of the former genus Lepidoderma to Polyschismium.












Similar content being viewed by others
Data availability
The DNA sequences generated during the current study are available in NCBI Genbank. The list of material used in the phylogenetic reconstruction, the concatenated alignment, partition file, and phylogenetic tree in the Newick format have been submitted as supplementary material and can also be found in FigShare (https://doi.org/10.6084/m9.figshare.21342834).
References
Adanson M (1763) Familles des plantes. Vincent, Paris
Adl SM, Simpson AGB, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, Heiss A, Hoppenrath M, Lara E, Gall LL, Lynn DH, McManus H, Mitchell EAD, Mozley-Stanridge SE, Parfrey LW et al (2012) The revised classification of eukaryotes. J Eukaryot Microbiol 59:429–514. https://doi.org/10.1111/j.1550-7408.2012.00644.x
Adl SM, Bass D, Lane CE, Lukeš J, Schoch CL, Smirnov AV, Agatha S, Berney C, Brown MW, Burki F (2019) Revisions to the classification, nomenclature, and diversity of eukaryotes. J Eukaryot Microbiol 66(1):4–119. https://doi.org/10.1111/jeu.12691
Ahmed S, Ibrahim M, Nantasenamat C, Nisar MF, Malik AA, Waheed R, Ahmed MZ, Ojha SC, Alam MK (2022) Pragmatic applications and universality of DNA barcoding for substantial organisms at species level: a review to explore a way forward. Biomed Res Int 2022:1846485. https://doi.org/10.1155/2022/1846485
Borg Dahl M, Shchepin ON, Schunk C, Menzel A, Novozhilov YK, Schnittler M (2018) A four year survey reveals a coherent pattern between occurrence of fruit bodies and soil amoebae populations for nivicolous myxomycetes. Sci Rep 8:11662. https://doi.org/10.1038/s41598-018-30131-3
Bortnikov FM, Shchepin ON, Gmoshinskiy VI, Prikhodko IS, Novozhilov YK (2018) Diderma velutinum, a new species of Diderma (Myxomycetes) with large columella and triple peridium from Russia. Botanica. Pacifica 7(2):1–5. https://doi.org/10.17581/bp.2018.07207
Cainelli R, de Haan M, Meyer M, Bonkowski M, Fiore-Donno AM (2020) Phylogeny of Physarida (Amoebozoa, Myxogastria) based on the small-subunit ribosomal RNA gene, redefinition of Physarum pusillum s. str. and reinstatement of P. gravidum Morgan. J Eukaryot Microbiol 67:327–336. https://doi.org/10.1111/jeu.12783
Che J, Chen H, Yang JX, Jin JQ, Jiang K, Yuan ZY, Murphy R, Zhang YP (2011) Universal COI primers for DNA barcoding amphibians. Mol Ecol Resour 12:247–258. https://doi.org/10.1111/j.1755-0998.2011.03090.x
Chen C, Frankhouser D, Bundschuh R (2012) Comparison of insertional RNA editing in Myxomycetes. PLoS Comput Biol 8(2):e1002400. https://doi.org/10.1371/journal.pcbi.1002400
Clark J, Haskins EF (2014) Sporophore morphology and development in the myxomycetes: a review. Mycosphere 5(1):153–170. https://doi.org/10.5943/mycosphere/5/1/7
Dagamac NHA, Rojas C, Novozhilov YK, Moreno GH, Schlueter R, Schnittler M (2017) Speciation in progress? A phylogeographic study among populations of Hemitrichia serpula (Myxomycetes). PLoS One 12(4):e0174825. https://doi.org/10.1371/journal.pone.0174825
Feng Y, Schnittler M (2015) Sex or no sex? Group I introns and independent marker genes reveal the existence of three sexual but reproductively isolated biospecies in Trichia varia (Myxomycetes). Org Divers Evol 15:631–650. https://doi.org/10.1007/s13127-015-0230-x
Fiore-Donno AM, Berney C, Pawlowski J, Baldauf SL (2005) Higher-order phylogeny of plasmodial slime molds (Myxogastria) based on elongation factor 1-A and small subunit rRNA gene sequences. J Eukaryot Microbiol 52(3):201–210. https://doi.org/10.1111/j.1550-7408.2005.00032.x
Fiore-Donno AM, Meyer M, Baldauf SL, Pawlowski J (2008) Evolution of dark-spored Myxomycetes (slime-molds): molecules versus morphology. Mol Phylogenet Evol 46:878–889. https://doi.org/10.1111/j.1550-7408.2005.00032.x
Fiore-Donno AM, Kamono A, Chao EE, Fukui M, Cavalier-Smith T (2010a) Invalidation of Hyperamoeba by transferring its species to other genera of Myxogastria. J Eukaryot Microbiol 57:189–196. https://doi.org/10.1111/j.1550-7408.2009.00466.x
Fiore-Donno AM, Nikolaev SI, Nelson M, Pawlowski J, Cavalier-Smith T, Baldauf SL (2010b) Deep phylogeny and evolution of slime moulds (Mycetozoa). Protist 161:55–70. https://doi.org/10.1016/j.protis.2009.05.002
Fiore-Donno AM, Novozhilov YK, Meyer M, Schnittler M (2011) Genetic structure of two protist species (Myxogastria, Amoebozoa) suggests asexual reproduction in sexual amoebae. PLoS One 6(8):e22872. https://doi.org/10.1371/journal.pone.0022872
Fiore-Donno AM, Kamono A, Meyer M, Schnittler M, Fukui M, Cavalier-Smith T (2012) 18S rDNA phylogeny of Lamproderma and allied genera (Stemonitales, Myxomycetes, Amoebozoa). PLoS One 7:e35359. https://doi.org/10.1371/journal.pone.0035359
Fiore-Donno AM, Clissmann F, Meyer M, Schnittler M, Cavalier-Smith T (2013) Two-gene phylogeny of bright-spored Myxomycetes (Slime Moulds, Superorder Lucisporidia). PLoS One 8:e62586. https://doi.org/10.1371/journal.pone.0062586
Fiore-Donno AM, Tice AK, Brown MW (2019) A non-flagellated member of the Myxogastria and expansion of the Echinosteliida. J Eukaryot Microbiol 66:538–544. https://doi.org/10.1111/jeu.12694
Flatau L (1984) Myxomyceten aus Nord-Hessen – III. Ein neuer Myxomycet aus der Umgebung von Kassel Beiträge zur Kenntnis der Pilze Mitteleuropas 1:193–196
Flatau L, Massner W, Schirmer P (1987) Myxomyceten aus Nordhessen – IV. Ein neuer Myxomycet aus der Umgebung von Kassel Zeitschrift für Mykologie 53:145–149
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3(5):294–299
Fries EM (1825) Systema Orbis Vegetabilis. Pars I. Plantae Homonemeae, Typographia Academica, Lund
Furulund BMN, Karlsen BO, Babiak I, Johansen SD (2021) A phylogenetic approach to structural variation in organization of nuclear group I introns and their ribozymes. Non-Coding RNA 7:43. https://doi.org/10.3390/ncrna7030043
García-Cunchillos I, Carlos Zamora J, Ryberg M, Lado C (2022) Phylogeny and evolution of morphological structures in a highly diverse lineage of fruiting-body-forming amoebae, order Trichiales (Myxomycetes, Amoebozoa). Mol Phylogenet Evol 177:107609. https://doi.org/10.1016/j.ympev.2022.107609
Gott JM, Visomirski LM, Hunter JL (1993) Substitutional and insertional RNA editing of the cytochrome c oxidase subunit 1 mRNA of Physarum polycephalum. J Biol Chem 268:25483–25486. https://doi.org/10.1016/S0021-9258(19)74417-2
Hagelstein R (1944) The Mycetozoa of North America. Published by the author, Mineola, N. Y
Hebert PD, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc Lond B 270:313–321. https://doi.org/10.1098/rspb.2002.2218
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS (2018) UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol Biol Evol 35(2):518–522. https://doi.org/10.1093/molbev/msx281
Horton TL, Landweber LF (2000) Evolution of four types of RNA editing in myxomycetes. RNA 6(10):1339–1346. https://doi.org/10.1017/S135583820000087X
Horton TL, Landweber LF (2002) Rewriting the information in DNA: RNA editing in kinetoplastids and myxomycetes. Curr Opin Microbiol 5(6):620–626. https://doi.org/10.1016/s1369-5274(02)00379-x
Huang D, Meier R, Todd PA, Chou LM (2008) Slow mitochondrial COI sequence evolution at the base of the metazoan tree and its implications for DNA barcoding. J Mol Evol 66(2):167–174. https://doi.org/10.1007/s00239-008-9069-5
Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogeny. Bioinformatics 17:754–755. https://doi.org/10.1093/bioinformatics/17.8.754
Ing B (1999) The Myxomycetes of Britain and Ireland. The Richmond Publishing Co. Ltd, Slough
Kang S, Tice AK, Spiegel FW, Silberman JD, Pánek T, Čepička I, Kostka M, Kosakyan A, Alcântara DMC, Roger AJ, Shadwick LL, Smirnov A, Kudryavtsev A, Lahr DJG, Brown MW (2017) Between a pod and a hard test: the deep evolution of amoebae. Mol Biol Evol 34(9):2258–2270. https://doi.org/10.1093/molbev/msx162
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. https://doi.org/10.1038/nmeth.4285
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20(4):1160–1166. https://doi.org/10.1093/bib/bbx108
Kosakyan A, Heger TJ, Leander BS, Todorov M, Mitchell EAD, Lara E (2012) COI barcoding of nebelid testate amoebae (Amoebozoa: Arcellinida): extensive cryptic diversity and redefinition of the Hyalospheniidae Schultze. Protist 163(3):415–434. https://doi.org/10.1016/j.protis.2011.10.003
Kowalski DT (1971) The genus Lepidoderma. Mycologia 63:490–516. https://doi.org/10.1080/00275514.1971.12019131
Lado C (2005–2022) An on line nomenclatural information system of Eumycetozoa. Real Jardín Botánico, CSIC. Madrid. Available from: http://www.eumycetozoa.com. Accessed 28 Oct 2022
Lado C, Treviño-Zevallos I, García-Martín JM, Wrigley de Basanta D (2022) Diachea mitchellii: a new myxomycete species from high elevation forests in the tropical Andes of Peru. Mycologia 4:798–811. https://doi.org/10.1080/00275514.2022.2072140
Leontyev DV, Schnittler M, Stephenson SL, Novozhilov YK, Shchepin ON (2019) Towards a phylogenetic classification of the Myxomycetes. Phytotaxa 399:209–238. https://doi.org/10.11646/phytotaxa.399.3.5
Lister A (1911) A monograph of the Mycetozoa. A descriptive catalogue of the species in the herbarium of the British Museum, Ed. 2nd (revised by G. Lister). Witherby & Co, London
Lister A (1925) A monograph of the Mycetozoa, a descriptive catalogue of the species in the herbarium of the British Museum. Ed. 3rd (revised by G. Lister). Oxford Univ. Press, London
Liu QS, Yan SZ, Chen SL (2015) Further resolving the phylogeny of Myxogastria (slime molds) based on COI and SSU rRNA genes. Genetika 51:46–53. https://doi.org/10.7868/s0016675814110071
Martin GW, Alexopoulos CJ (1969) The Myxomycetes. University of Iowa Press, Iowa
Massee G (1892) A monograph of the Myxogastres. Methuen & Co., London
Meylan C (1910) Myxomycetes du Jura (Suite). Bulletin de la Société botanique de Genève II 2:263
Meylan C (1931) Contribution a la connaissance des Myxomycetes du Jura et des Alpes. Bulletin de la Société vaudoise des sciences naturelles 57:304
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gatew Comp Environ Work (GCE):1–8. https://doi.org/10.1109/GCE.2010.5676129
Moreno G, Singer H, Illana C (2004) A taxonomic review on the nivicolous myxomycetes species described by Kowalski. II. Physarales and Trichiales. Österr Z Pilzk 13:61–74
Moreno G, López-Villalba Á, Stephenson SL, Castillo A (2018a) Lepidoderma cristatosporum, a new species of myxomycete from Australia. Mycoscience 59:386–391
Moreno G, Sánchez A, Meyer M, López-Villalba Á, Castillo A (2018b) Revision of the nivicolous species of the genus Lepidoderma. Boletín de la Sociedad Micológica de Madrid 42:39–77
Nandipati SCR, Haugli K, Coucheron DH, Haskins EF, Johansen SD (2012) Polyphyletic origin of the genus Physarum (Physarales, Myxomycetes) revealed by nuclear rDNA mini-chromosome analysis and group I intron synapomorphy. BMC Evol Biol 12:166. https://doi.org/10.1186/1471-2148-12-166
Nassonova E, Smirnov A, Fahrni J, Pawlowski JK (2010) Barcoding amoebae: comparison of SSU, ITS and COI genes as tools for molecular identification of naked lobose amoebae. Protist 161(1):102–115. https://doi.org/10.1016/j.protis.2009.07.003
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32(1):268–274. https://doi.org/10.1093/molbev/msu300
Novozhilov YK, Okun MV, Erastova DA, Shchepin ON, Zemlyanskaya IV, García-Carvajal E, Schnittler M (2013a) Description, culture and phylogenetic position of a new xerotolerant species of Physarum. Mycologia 105:1535–1546. https://doi.org/10.3852/12-284
Novozhilov YK, Schnittler M, Erastova DA, Okun MV, Schepin ON, Heinrich E (2013b) Diversity of nivicolous myxomycetes of the Teberda State Biosphere Reserve (Northwestern Caucasus, Russia). Fungal Divers 59:109–130. https://doi.org/10.1007/s13225-012-0199-0
Novozhilov YK, Prikhodko IS, Shchepin ON (2019) A new species of Diderma from Bidoup Nui Ba National Park (southern Vietnam). Protistology 13:126–132. https://doi.org/10.21685/1680-0826-2019-13-3-2
Novozhilov YK, Shchepin ON, Prikhodko IS, Schnittler M (2022a) A new nivicolous species of Diderma (Myxomycetes) from Kamchatka, Russia. Nova Hedwigia 1-2:181–196. https://doi.org/10.1127/nova_hedwigia/2022/0670
Novozhilov YK, Prikhodko IS, Fedorova NA, Shchepin ON, Gmoshinskiy VI, Schnittler M (2022b) Lamproderma vietnamense: a new species of myxomycetes with reticulate spores from Phia Oắc – Phia Đén National Park (northern Vietnam) supported by molecular phylogeny and morphological analysis. Mycoscience 63:149–155. https://doi.org/10.47371/mycosci.2022.05.003
Okonechnikov K, Golosova O, Fursov M, UGENE team (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. https://doi.org/10.1093/bioinformatics/bts091
Poulain M, Meyer M, Bozonnet J (2011) Les Myxomycètes. Pressor, Delémont
Persoon CH (1794) Neuer Versuch einer systematischen Eintheilung der Schwämme. Neues Magazin für die Botanik 1:89
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67:901–904. https://doi.org/10.1093/sysbio/syy032
Ronikier A, García-Cunchillos I, Janik P, Lado C (2020) Nivicolous Trichiales from the austral Andes: unexpected diversity including two new species. Mycologia 112:753–780. https://doi.org/10.1080/00275514.2020.1759978
Ronikier A, Janik P, de Haan M, Kuhnt A, Zankowicz M (2022) Importance of type specimen study for understanding genus boundaries—taxonomic clarifications in Lepidoderma based on integrative taxonomy approach leading to resurrection of the old genus Polyschismium. Mycologia. https://doi.org/10.1080/00275514.2022.2109914
Rostafiński J (1873) Versuch eines Systems der Mycetozoen. Inagural dissertation. Druck von Friedrich Wolff, Strassburg
Rostafiński J (1874) Śluzowce (Mycetozoa). Kórnik Library, Paris
Schnittler M, Novozhilov Y (1996) The myxomycetes of boreal woodlands in Russian northern Karelia: a preliminary report. Karstenia 36(1):19–40
Schnittler M, Novozhilov YK (1999) Lepidoderma crustaceum, a nivicolous myxomycete, found on the island of Crete. Mycotaxon 71:387–392
Schnittler M, Unterseher M, Pfeiffer T, Novozhilov Y, Fiore-Donno AM (2010) Ecology of sandstone ravine myxomycetes from Saxonian Switzerland (Germany). Nova Hedwigia 90:277–302. https://doi.org/10.1127/0029-5035/2010/0090-0277
Schnittler M, Shchepin O, Dagamac NH, Borg Dahl M, Novozhilov Y (2017) Barcoding myxomycetes with molecular markers: challenges and opportunities. Nova Hedwigia 104:323–341. https://doi.org/10.1127/nova_hedwigia/2017/0397
Shchepin ON, Novozhilov YK, Schnittler M (2016) Disentangling the taxonomic structure of the Lepidoderma chailletii-carestianum species complex (Myxogastria, Amoebozoa): genetic and morphological aspects. Protistology 10:117–129
Shchepin O, Novozhilov Y, Woyzichovski J, Bog M, Prikhodko I, Fedorova N, Gmoshinskiy V, Borg Dahl M, Dagamac NHA, Yajima Y, Schnittler M (2021) Genetic structure of the protist Physarum albescens (Amoebozoa) revealed by multiple markers and genotyping by sequencing. Mol Ecol 31:372–390. https://doi.org/10.1111/mec.16239
Shrader HA (1797) Nova genera plantarum. Apvd S. L. Crvsivm, Lipsiae
Stephenson SL, Novozhilov YK, Prikhodko IS (2020) A new species of Physarum (Myxomycetes) from Christmas Island (Australia). Novosti Sistematiki Nizshikh Rastenii 54(2):397–404. https://doi.org/10.31111/nsnr/2020.54.2.397
Tekle YI (2014) DNA barcoding in amoebozoa and challenges: the example of Cochliopodium. Protist 165(4):473–484. https://doi.org/10.1016/j.protis.2014.05.002
Traphagen SJ, Dimarco MJ, Silliker ME (2010) RNA editing of 10 Didymium iridis mitochondrial genes and comparison with the homologous genes in Physarum polycephalum. RNA 16:828–838. https://doi.org/10.1261/rna.1989310
Vaidya G, Lohman DJ, Meier R (2011) SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27:171–180. https://doi.org/10.1111/j.1096-0031.2010.00329.x
Wikmark OG, Haugen P, Haugli K, Johansen SD (2007a) Obligatory group I introns with unusual features at positions 1949 and 2449 in nuclear LSU rDNA of Didymiaceae myxomycetes. Mol Biol Evol 43:596–604. https://doi.org/10.1016/j.ympev.2006.11.004
Wikmark OG, Haugen P, Lundblad EW, Haugli K, Johansen SD (2007b) The molecular evolution and structural organization of group I introns at position 1389 in nuclear small subunit rDNA of myxomycetes. J Eukaryot Microbiol 54:49–56. https://doi.org/10.1111/j.1550-7408.2006.00145.x
Wrigley de Basanta D, Lado C, García-Martín JM, Estrada-Torres A (2015) Didymium xerophilum, a new myxomycete from the tropical Andes. Mycologia 107:157–168. https://doi.org/10.3852/14-058
Wrigley de Basanta D, Estrada-Torres A, García-Cunchillos I, Cano Echevarría A, Lado C (2017) Didymium azorellae, a new myxomycete from cushion plants of cold arid areas of South America. Mycologia 109:993–1002. https://doi.org/10.1080/00275514.2018.1426925
Yang C, Zheng Y, Tan S, Meng G, Rao W, Yang C, Bourne DG, O'Brien PA, Xu J, Liao S, Chen A, Chen X, Jia X, Zhang AB, Liu S (2020) Efficient COI barcoding using high throughput single-end 400 bp sequencing. BMC Genomics 21(1):862. https://doi.org/10.1186/s12864-020-07255-w
Zhao FY, Liu SY, Stephenson SL, Hsiang T, Qi B, Li Z (2021) Morphological and molecular characterization of the new aethaloid species Didymium yulii. Mycologia 113(5):926–937. https://doi.org/10.1080/00275514.2021.1922224
Acknowledgements
We acknowledge the use of equipment of the Core Facility Center “Cell and Molecular Technologies in Plant Science” at the Komarov Botanical Institute of the Russian Academy of Sciences (BIN RAS, St. Petersburg) and send personal thanks to Lyudmila Kartzeva and Elena Krapivskaya (lead engineers of the Core Facility Center). The authors are grateful to Marianne Meyer, Marja Pennanen and the staff of the Botanic Garden Meise (BR) for providing specimens and to Dr. F. J. Rejos, curator of the University of Alcalá Herbarium (AH), for his assistance with some specimens examined in the current study.
Funding
The molecular lab work of Ilya S. Prikhodko, Oleg. N. Shchepin, Nadezhda A. Bortnikova and Yuri K. Novozhilov was supported by the Russian Science Foundation (project No. 22-24-00747).
Author information
Authors and Affiliations
Contributions
Ilya S. Prikhodko, Yuri K. Novozhilov, and Oleg N. Shchepin contributed to the study conception and design. All authors participated in the collection of herbarium specimens for morphological and molecular phylogenetic analyses. Light and scanning electron microscopy was performed by Yuri K. Novozhilov, Vladimir I. Gmoshinskiy, Gabriel Moreno, and Ángela López-Villalba. Ilya S. Prikhodko, Oleg N. Shchepin, and Nadezhda A. Bortnikova were responsible for DNA extraction, PCR analysis, and sequencing. Ilya S. Prikhodko and Oleg N. Shchepin performed phylogenetic analyses. Ilya S. Prikhodko and Vladimir I. Gmoshinskiy prepared illustrations on the phylogeny and morphology of the studied material, respectively. The first draft of the manuscript was written by Ilya S. Prikhodko, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Section Editor: Tanay Bose
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Prikhodko, I.S., Shchepin, O.N., Bortnikova, N.A. et al. A three-gene phylogeny supports taxonomic rearrangements in the family Didymiaceae (Myxomycetes). Mycol Progress 22, 11 (2023). https://doi.org/10.1007/s11557-022-01858-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11557-022-01858-1