Abstract
Downy mildew is a serious threat to corn (maize) production in the tropics and subtropics. Corn is native to Central America, and was introduced into South-East Asia by the Spanish colonisers in the 1700s. Corn is evolutionarily naïve to downy mildews of the genus Peronosclerospora. Consequently, corn monocultures are particularly susceptible to a variety of Peronosclerospora species, which spread to the crop from local grasses. Globally, corn is one of the most important crops for both humans and livestock. Several downy mildews of corn have been identified as potential threats to global food security, and trade with corn seeds is strictly regulated to avoid spreading the pathogens. Despite their importance, little is known about the biodiversity of graminicolous downy mildews, because their identification has often relied on variable morphological features, such as conidial dimensions. DNA barcodes for most species have become available only recently. During surveys for downy mildews on corn in Indonesia, a previously unrecognised species of Peronosclerospora was found and investigated using a combination of morphological characters and molecular phylogenetic analyses. The new species, introduced here as Peronosclerospora neglecta, is widely distributed in South-East Asia from Thailand to eastern Indonesia. The impact of this downy mildew can be severe, with complete crop losses in heavily affected fields. Given the aggressiveness of the species, close surveillance is warranted to restrict its further spread.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Corn (maize, Zea mays) was first domesticated from the wild grass teosinte (Z. mays subsp. parviglumis) in central America about 9000 years ago (Ramos-Madrigal et al. 2016; Wang et al. 2017; Stitzer and Ross-Ibarra 2018). It has become one of the most important crops globally, for both humans and livestock, due to its fast growth and high yield (Nuss and Tanumihardjo 2010). Graminicolous mildews of the genus Peronosclerospora are naturally absent in Central America, the native range of maize and teosinte, and consequently, specific resistance to Peronosclerospora spp. is lacking in commercial cultivars and most current corn cultivars are susceptible to several Peronosclerospora spp. (Kenneth 1981; Thines 2014). Peronosclerospora spp. are characterised by the production of conidia that form germ tubes to infect host plants (Shaw 1978). The centre of origin of the genus is likely to be South-East Asia or parts of the Australasian realm (Spencer and Dick 2002; Thines 2014), where it parasitises a wide range of C4 grasses, such as sugarcane and sorghum species (Telle et al. 2011; Suharjo et al 2020; Ryley et al. 2022). The grasses parasitised by Peronosclerospora are largely unrelated, even though most of them are members of the tribe Andropogonoideae. It can be assumed that Peronosclerospora spp. have colonised this tribe by host jumps and subsequent radiation, as described for other genera of downy mildew (Thines 2019). The pathogenicity effectors that triggered the successful radiation within Andropogonoideae likely also enable parasitism of corn, which has been observed for most species described in Peronosclerospora (Kenneth 1981; Shivas et al. 2012).
Downy mildew of corn caused by Peronosclerospora spp. has been reported in many countries in South-East Asia and Australasia (Sharma et al. 1993; Spencer and Dick 2002; Suharjo et al. 2020; Crouch et al. 2022). The native grass species that serve as the primary host for most of the Peronosclerospora species that infect corn are known. Some examples are Eriochloa pseudoacrotricha parasitised by P. eriochloae (Telle et al. 2011); Heteropogon contortus parasitised by P. heteropogonis (Siradhana et al. 1980); Sorghum spontaneum parasitised by P. spontanea (Weston 1921); and Sorghum timorense parasitised P. maydis (Suharjo et al. 2020), which was first described from corn (Raciborski 1897). The native host grass for corn downy mildew caused by P. philippinensis remains unknown (Weston 1920). Recently, Suharjo et al. (2020) reported a potentially undescribed downy mildew on corn in Indonesia. However, due to the absence of specimens, the species could not be morphologically compared to the known species of Peronosclerospora and was not formally introduced. More recently, ex-type sequences of additional Peronosclerospora species have been made available (Crouch et al. 2022), but none of them matched the new lineage found in Indonesia. It was the aim of the current study to clarify the identity of this Peronosclerospora species using the cox2 gene sequence barcode (Choi et al. 2015) and specimen morphology.
Materials and Methods
Plant material and microscopy
Downy mildew specimens were collected from corn fields in Indonesia during 2018–2019, and deposited in the internationally recognised herbaria at the Research Centre for Biology, Cibinong, Indonesia (BO) and the Queensland Plant Pathology Herbarium (BRIP), Dutton Park, Australia. Detailed information on the specimens is given in the Taxonomy section. Microscopy was done as described in Ryley et al. (2022), using specimen pieces rehydrated in hot lactic acid. Due to the evanescent nature of the asexual morph of the newly discovered corn downy mildew, only the maximum and minimum dimensions of 25 conidia were recorded from the type specimen.
DNA extraction PCR, sequencing, and phylogenetic reconstruction
DNA extraction, PCR, and sequencing methods followed Ryley et al. (2022), using the AnalyticJena plant DNA extraction kit (AnalyticJena, Germany) and the cox2 amplification procedure described in Telle et al. (2011). The sequences obtained were added to the dataset of Suharjo et al. (2020), to which additional sequences with high similarity to the new lineage identified by Suharjo et al. (2020) were added, as identified by BLAST (Altschul et al. 1990) searches against sequences deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/). Alignments were done in MUSCLE (Edgar 2004) as implemented in MEGA7 (Kumar et al. 2016), which resulted in an alignment without internal gaps. Leading and trailing gaps were removed before phylogenetic analyses, and the final alignment is available as Supplemental File 1. Phylogenetic reconstruction using the Minimum Evolution algorithm was done in MEGA7, with 500 bootstrap replicates and default settings, except for choosing the Tamura-Nei substitution model (Tamura and Nei 1993), which is the most complex standard model offered by the programme. The phylogenetic reconstructions using Bayesian Inference and Maximum Likelihood were done using MrBayes (Ronquist et al. 2012) and RAxML (Stamatakis 2014), respectively, as implemented on the TrEase webserver (http://thines-lab.senckenberg.de/trease), using default settings.
Results
Morphology
Plants infected by downy mildew exhibited chlorotic streaks and were stunted (Fig. 1). Evanescent conidiophores protruded from stomata, predominantly on the abaxial leaf surface. The morphology of conidiophores and conidia was typical for Peronosclerospora. The conidiophores were often intertwined on the abaxial surface of leaf blades, up to 180 µm long, swollen at the base, and dichotomously branched 2–4 times. The ultimate ramification gave rise to 2–3 (mostly 2) ultimate branchlets which were 5–11 µm long and 2–4 µm broad. Conidia were hyaline, sub-globose to ovoid, 23–30 µm long and 13–19 µm broad and thin-walled. Conidia germinated by producing germ tubes. Oospores were not found.
Symptoms and morphological features of Peronosclerospora neglecta on maize. A stunted maize plant with chlorotic streaks. B close-up of symptoms, with whitish conidiophores emerging from stomata on the abaxial leaf surface. C conidia and ultimate branchlets. Scale bars equal to 10 mm in B and 10 µm in C
Phylogeny
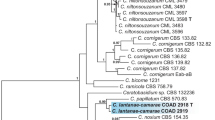
In the phylogenetic reconstruction based on partial cox2 sequences (Fig. 2), all five genera of graminicolous downy mildews were resolved as monophyletic with high to maximum support. Within the genus Peronosclerospora, the monophyly of all species represented by more than one sequence received moderate to strong support. None of the infrageneric splits received significant support. Both P. maydis and the corn-parasitic lineage reported by Suharjo et al. (2020) were represented by samples from several different regions. Within P. maydis, some variation was present, but no clear-cut association of the variants within a specific region was found. Samples of P. maydis were obtained from both Z. mays and Sorghum timorense, but no strict correlation between genotype and host was found. The lineage reported by Suharjo et al. (2020) was genetically uniform, despite being sampled throughout a wide geographic range. It included samples from Thailand and Indonesia, where the pathogen was found on three different islands. All samples of this lineage originated from Z. mays.
Phylogenetic reconstruction in Minimum Evolution, based on partial cox2 sequences. Numbers on branches denote support from Minimum Evolution bootstrapping, Maximum Likelihood bootstrapping, and Bayesian posterior probabilities, in the respective order. A minus sign denotes a lack of support for the topology (bootstrap support < 50%, posterior probabilities < 0.8)
Taxonomy
Peronosclerospora neglecta Muis, Ryley, Suharjo, Y.P. Tan, Thines and R.G. Shivas, sp. nov., MycoBank MB 846809.
Etymology: from Latin neglecta meaning neglected, overlooked.
Classification: Peronosporaceae, Peronsporales, Oomycota.
Description: Asexual sporulation on abaxial surfaces of leaf blades. Conidiophores emerging from stomata, evanescent, up to 180 µm long, swollen base, dichotomously branched, ultimate branchlets 5–11 × 2–4 µm. Conidia hyaline, sub-globose or ovoid, 23–30 × 13–19 µm, thin-walled, germination by a germ tube. Oospores not seen.
Typus: Indonesia, East Java, Grogol, Kediri, 07° 49′ 01″ S, 111° 58′ 12″ E, on leaves of Zea mays, 18 Jan. 2019, A. Muis, N. Nonci, S.H. Kalqutny, Aminah, M.J. Ryley, M.D.E. Shivas and R.G. Shivas (holotype BO 24212, isotype BRIP 71135; cox2 sequence GenBank OK336429).
Additional specimens examined: Indonesia, East Java, Kediri, 07° 45′ 13″ S, 111° 58′ 20″ E, on leaves of Z. mays, 18 Jan. 2018, A. Muis, N. Nonci, S.H. Kalqutny, Aminah (BRIP 71133, BO 24210, cox2 sequence GenBank OK336427); East Nusa Tenggara, Timor, 10° 08′ 01″ S, 123° 41′ 37″ E, on leaves of Z. mays, 09 May 2018, A. Muis, N. Nonci (BRIP 71115, BO 24192, cox2 sequence GenBank OK336428); South Kalimantan, Banjabaru, 03° 26′ 11″ S, 114° 48′ 00″ E on leaves of Z. mays, 16 Oct. 2018, Suriani (BRIP 71118, BO 24195, cox2 sequence GenBank OK336430); South Sulawesi, Indonesian Cereals Research Institute, Bajeng Experimental Station, 05° 18′ 34″ S, 119° 30′ 26″ E, on leaves of Z. mays, 23 Aug 2018, A. Muis (BRIP 71112, BO 24189, cox2 sequence GenBank OK336425); Bone, 04° 32′ 08″ S, 119° 57′ 18″ E, on leaves of Z. mays, 29 Dec. 2018, Suriani (BRIP 71123, BO 24200, cox2 sequence GenBank OK336426); Maros, 04° 58′ 55″ S, 119° 34′ 32″ E, on leaves of Z. mays, 14 Jan. 2018, S.H. Kalqutny (BRIP 71124, BO 24201, cox2 sequence GenBank OK336431).
Known distribution: Indonesia and Thailand.
Notes: Based on a BLASTn search, P. neglecta differed from P. maydis (ex-type specimen KRAM O-5859(J)) in cox2 (GenBank MW025835; Identities 580/594 (98%), 0 gaps); from P. miscanthi sensu Telle et al. (2011) in cox2 (GenBank HQ261811; Identities 532/541 (98%), 0 gaps); from P. philippinensis (specimen Pp04, Telle et al. (2011)) in cox2 (GenBank KX702314 Identities 518/528 (98%), 0 gaps); and from P. sorghi sensu Telle et al. (2011) (specimen HUH 897) in cox2 (GenBank EU116055; Identities 251/257 (98%), 0 gaps). A summary of the morphological features of the species known to affect maize is given in Table 1.
Discussion
Peronosclerospora neglecta causes a serious disease of corn (maize) in Indonesia, especially in Java and South Sulawesi. The symptoms of infection are chlorotic leaf lesions, leaf distortion, and stunted plants, which can lead to complete crop loss. The conidia of P. neglecta differ in shape and size from those recorded for P. maydis, P. philippinensis, P. sorghi, and P. spontanea, which have been previously recorded on maize in South-East Asia (Weston 1921; Suharjo et al. 2020). The subglobose to ovoid conidia of P. neglecta are larger than those of P. maydis (Raciborski 1897) and smaller than the conidia of P. sorghi (Weston and Uppal 1932), P. spontanea (Weston 1921), and P. philippinensis (Weston 1920). Sequence data must be obtained for unambiguous identification of P. neglecta and other Peronosclerospora spp., as it is known that morphology can be unreliable for identification, because the dimensions of downy mildew conidia may be influenced by environmental conditions (Dudka et al. 2007) and host (Runge and Thines 2011).
In contrast to P. maydis, no variation in the cox2 sequences was observed in P. neglecta, which hints at a rather uniform metapopulation. An explanation for this uniformity might be that the downy mildew only recently spread throughout South-East Asia with infested corn seeds. The potential of seed infection and spread by other Peronosclerospora species is well known (Advincula and Exconde 1975; Sommartaya et al. 1975; Adenle and Cardwell 2000). Peronosclerospora neglecta appears to be the dominant downy mildew of corn in Java (Suharjo et al. 2020, this study), which raises the possibility that Lukman et al. (2016) and Sommartaya et al. (1975) detected P. neglecta in corn seeds rather than P. maydis or P. sorghi. Sequences available on GenBank showed that P. neglecta occurs in Thailand, reported as P. maydis (Janruang and Unartngam 2018).
Corn is alien to Asia and cannot be the primary host for Peronosclerospora species, which likely evolved in South-East Asia (Spencer and Dick 2002). Peronosclerospora spp. are absent from the native range of maize and its wild progenitor, teosinte. In addition, the high virulence potential of P. sacchari is noteworthy. A Taiwanese isolate of P. sacchari was able to infect eight genera of Andropogonoideae (Andropogon, Bothriochloa, Eulalia, Saccharum, Schizachyrium, Sorghum, Tripsacum, and Zea) at different levels of susceptibility under laboratory conditions (Bonde 1981). The susceptibility of corn is likely due to the long-standing evolutionary interaction of Peronosclerospora with grasses of the tribe Andropogonoideae that leads to a high virulence potential, coupled with the absence of corn varieties that have high resistance to downy mildew (Thines 2014). Apart from P. neglecta and P. philippinensis, the primary hosts of all graminicolous downy mildews infecting corn are known (Kenneth 1981; Spencer and Dick 2002; Telle et al. 2011; Suharjo et al. 2020). Oospores on corn are rarely formed or not at all (Kenneth 1981). It should follow that infections of corn result from the spread of the pathogen into the crop population from native grass host(s). For some species, e.g. P. eriochloae, this might be the case as only sporadic outbreaks have been reported (Telle et al. 2011). However, vertical transmission by seeds might explain a wider distribution and occurrence than expected through the spread from primary to secondary hosts. For example, the downy mildew Bremia lactucae on cultivated lettuce has been known to often spread from infected populations of wild lettuce (Runge et al. 2021), which has resulted in the pathogen becoming widely distributed in cultivated lettuce (e.g. Trimboli and Nieuwenhuis 2011; Marin et al. 2020). Fungicide-containing seed cover is frequently applied for disease management, but its long-term usage might also lead to the emergence of fungicide-tolerant strains (Pakki and Jainuddin 2019). Apart from fungicide treatments, the removal of weedy and proximal primary hosts could be an important measure to prevent disease spread into crops. Knowledge of the primary host(s) and the frequency of the spread of downy mildew to cultivated corn crops is a prerequisite for understanding the epidemiology of these diseases. Considering the potential damage P. neglecta can inflict, it is advisable to maintain vigilance by monitoring the distribution of the species and to further develop diagnostic tools (Lukman et al. 2016) for standardised detection in seed lots and soil.
Data availability
Sequence data have been deposited in GenBank.
References
Adenle VO, Cardwell KF (2000) Seed transmission of maize downy mildew (Peronosclerospora sorghi) in Nigeria. Plant Pathol 49:628–634
Advincula BA, Exconde OR (1975) Seed transmission of Sclerospora philippinensis Weston in maize. Philipp Agric 59:244–255
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bonde MR (1981) Host range of Taiwanese isolates of Peronoschlerospora sacchari. Plant Dis 65:739–740
Choi YJ, Beakes G, Glockling S, Kruse J, Nam B, Nigrelli L, Ploch S, Shin HD, Shivas RG, Telle S, Voglmayr H, Thines M (2015) Towards a universal barcode of oomycetes – a comparison of the cox1 and cox2 loci. Mol Ecol Resour 15:1275–1288
Crouch JA, Davis WJ, Shishkoff N, Castroagudín VL, Martin F, Michelmore RW, Thines M (2022) Peronosporaceae species causing downy mildew diseases of Poaceae, including nomenclature revisions and diagnostic resources. Fungal Syst Evol 9:43–86
Dudka IO, Anishchenko IM, Terent’eva NG (2007) The variability of Peronospora alta Fuckel conidia in dependence on the ecological conditions. In: Lebeda A, Spencer-Phillips PTN (eds) Advances in Downy Mildew Research, vol 3. Palacký University in Olomouc and JOLA, Kostelec na Hané, pp 39–46
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 32:1792–1797
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Ito S (1913) Kleine Notizen über parasitische Pilze Japans. Bot Mag 27:217–223
Janruang P, Unartngam J (2018) Morphological and molecular based identification of corn downy mildew distributed in Thailand. Int J Agr Tech 14:845–860
Kenneth RG (1981) Downy mildews of graminaceous crops. In: Spencer DM (ed) The Downy Mildews. Academic, London, pp 367–394
Lukman R, Afifuddin A, Lübberstedt T (2016) Tracing the signature of Peronosclerospora maydis in maize seeds. Australas Plant Pathol 45(1):73–82
Marin MV, Franco CA, Smilde D, Panizzi RC, Braz LT (2020) Distribution of races and virulence factors of Bremia lactucae in the main lettuce production area in Brazil. J Plant Pathol 102(2):395–407
Nuss ET, Tanumihardjo SA (2010) Maize: a paramount staple crop in the context of global nutrition. Compr Rev Food Sci Food Saf 9(4):417–436
Pakki S, Jainuddin N (2019) The effectiveness combination of resistant varieties and metalaxyl fungicide in controlling downy mildew disease (Peronosclerospora maydis) in maize plant. J HPT Tropika 19:42–51
Raciborski M (1897) Lijer, eine gefährliche Maiskrankheit. Berichte der Deutschen Botanischen Gesellschaft XV: 475–478
Ramos-Madrigal J, Smith BD, Moreno-Mayar JV, Gopalakrishnan S, Ross-Ibarra J, Gilbert MTP, Wales N (2016) Genome sequence of a 5,310-year-old maize cob provides insights into the early stages of maize domestication. Curr Biol 26(23):3195–3201
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Runge F, Thines M (2011) Host matrix has major impact on the morphology of Pseudoperonospora cubensis. Eur J Plant Pathol 129:147–156
Runge F, Gärber U, Lebeda A, Thines M (2021) Bremia lactucae populations on cultivated lettuce originate from prickly lettuce and are interconnected with the wild pathosystem. Eur J Plant Pathol 161:411–426
Ryley MJ, Langdon RFN (2001) Peronosclerospora eriochloae sp. nov. and other downy mildews on native grasses in Queensland Australia. Mycotaxon 79:87–99
Ryley MJ, Tan YP, Kruse J, Thines M, Shivas RG (2022) More than meets the eye—unexpected diversity in downy mildews (Oomycetes) on grasses in Australia. Mycol Prog 21:297–310
Sharma RC, De Leon C, Payak MM (1993) Diseases of maize in South and South-East Asia: problems and progress. Crop Prot 12:414–422
Shaw CG (1978) Peronosclerospora species and other downy mildews of the Gramineae. Mycologia 70:594–604
Shivas RG, Ryley MJ, Telle S, Liberato JR, Thines M (2012) Peronosclerospora australiensis sp. nov. and Peronosclerospora sargae sp. nov., two newly recognised downy mildews in northern Australia, and their biosecurity implications. Australas Plant Pathol 41:125–130
Siradhana BS, Dange SRS, Rathore RS, Singh SD (1980) A new downy mildew on maize in Rajasthan, India. Curr Sci 49:316–317
Sommartaya T, Pupipat U, Intrama S, Renfro BL (1975) Seed transmission of Sclerospora sorghi (Weston & Uppal) the downy mildew of corn in Thailand. Kasetsart J 9:12–25
Spencer MA, Dick MW (2002) Aspects of graminicolous downy mildew biology: perspectives for tropical plant pathology and Peronosporomycetes phylogeny. In: Watling R, Frankland JC, Ainsworth AM, Isaac S, Robinson CH (eds) Tropical Mycology, vol 2, Micromycetes. CABI, Wallingford, pp 63–81
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Stitzer MC, Ross-Ibarra J (2018) Maize domestication and gene interaction. New Phytol 220:395–408
Suharjo R, Swibawa G, Prasetyo J, Fitriana Y, Danaatmadja Y, Budiawan A, Roberts S, Noorhajati N, Amad M, Thines M (2020) Peronosclerospora australiensis is a synonym of P. maydis, which is widespread on Sumatra, and distinct from the most prevalent Java maize downy mildew pathogen. Mycol Prog 19:1309–1315
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evolution 10:512–526
Telle S, Shivas RG, Ryley MJ, Thines M (2011) Molecular phylogenetic analysis of Peronosclerospora (Oomycetes) reveals cryptic species and genetically distinct species parasitic to maize. Eur J Plant Pathol 130:521–528
Thines M (2014) Phylogeny and evolution of plant pathogenic oomycetes—a global overview. Eur J Plant Pathol 138:431–447
Thines M (2019) An evolutionary framework for host shifts–jumping ships for survival. New Phytol 224:605–617
Trimboli DS, Nieuwenhuis J (2011) New races of Bremia lactucae on lettuce in Australia. Australas Plant Dis Notes 6:62–63
Wang L, Beissinger TM, Lorant A, Ross-Ibarra C, Ross-Ibarra J, Hufford MB (2017) The interplay of demography and selection during maize domestication and expansion. Genome Biol 18(215):1–13
Waterhouse GM (1966) The genus Sclerospora, diagnoses (or descriptions) from the original papers and a key. CMI, Kew.
Weston WH (1920) Philippine downy mildew of maize. J Agr Res 19:97–122
Weston WH (1921) Another conidial Sclerospora of Philippine maize. J Agric Res 20:669–684
Weston WH, Uppal BN (1932) The basis for S. sorghi as a species. Phytopathol 22:578–586
Acknowledgements
MR and RS thank Susie Collins, Wendy Lee, Craig Marston, Ian Naumann, and James Walker (Department of Agriculture, Water and the Environment, Canberra, Australia) for making much of the field and laboratory work possible.
Funding
Open Access funding enabled and organized by Projekt DEAL. This study was supported by the grant AGRIFUTURE of the German Ministry BLE provided to MT, and by funds from the Department of Agriculture, Water and the Environment, Canberra, Australia, as well as the Centre for Crop Health, University of Southern Queensland, Toowoomba, Queensland, Australia to YT, MR, and RS.
Author information
Authors and Affiliations
Contributions
AM, MR, RS, and MT conceived the study; AM and NN provided plant material; AM, MR, and RS did the morphological examination; YPT did the molecular biology work; MT conducted the phylogenetic reconstructions; MT wrote the manuscript with major contributions from AM, MR, and RS, as well as contributions from the other authors; all authors discussed the findings and contributed to the final draft.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Section Editor: Tanay Bose
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Muis, A., Ryley, M.J., Tan, Y.P. et al. Peronosclerospora neglecta sp. nov.—a widespread and overlooked threat to corn (maize) production in the tropics. Mycol Progress 22, 12 (2023). https://doi.org/10.1007/s11557-022-01862-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11557-022-01862-5