Abstract
Heat resistance is the ability to survive short, extreme temperature stresses, exceeding the own growth temperature by far. Despite their occurrence in natural substrates and their relevance for the food and healthcare industry, the diversity of fungi with heat resistance abilities is poorly studied. Sampling of boreal forest soils in Canada in combination with a heat-shock treatment (75 °C, 30 min) yielded, among others, four heat resistant, mesophilic fungal isolates. Based on rDNA barcode sequences, the novel isolates were assigned to Basidiomycota. In this study, we use macromorphological and micromorphological observations, cultivation assays and comparative genomics for physiological characterization, interspecific differentiation, and phylogenetic placement of these isolates. A phylogeny of 38 single-copy orthologous genes, an orthology analysis, and septal pore type analysis revealed the isolates as representatives of two new basidiomycetous species of the novel class Peribolosporomycetes, a sister lineage to all other members of Ustilaginomycotina. Further genomic and phenotypic data support two different species (Peribolospora kevripleyi, Peribolospora baueri), which are heat resistant and osmotolerant.
Similar content being viewed by others
Introduction
Several ecosystems and habitats are characterized by spontaneous or reoccurring high temperatures. The increasing frequency and intensity of heat waves due to climate change, common phenomena like forest or bush fires in natural habitats, and the application of slash-and-burn methods in agriculture and forestry, flare pits in the oil and gas industry, or even the thermal changes during the process of composting cause heat stress to organisms in the respective ecosystems. Thus, substrates like soil (Piecková et al. 2020), plant debris, or compost (Witfeld et al. 2021), which are exposed to such fluctuating temperatures are often inhabited by fungi with high temperature adaptations, having a potential ecological advantage towards less heat resistant counterparts.
Most fungi are adapted to moderate temperatures up to maximum 40 °C, while some heat resistant species show remarkable resistance to temporary, extremely high temperatures. The physiological trait of heat resistance can be described as a function of stress intensity and time. Usually, a fungus can survive less intense heat-stresses for longer time periods, while extremely high temperatures can only be survived for shorter durations. For example, Aspergillus niger is reported to survive exposure to 60 °C for 60 min but shows no survival when exposed to 70 °C for 10 min (Jesenská et al. 1993). Due to this interaction of the two factors temperature and time, Samson et al. (2004) defined heat resistant fungi as such, which survive 75 °C for at least 30 min. And although survival of even higher temperatures is known for some fungi, e.g., Talaromyces avellaneus survives 90 °C for 10 min (Jesenská et al. 1993), this definition has been widely accepted and applied in the description and isolation of heat resistant fungi (Houbraken and Samson 2006; Yaguchi et al. 2012; Nguyen et al. 2013; Day et al. 2020; Ulusoy et al. 2022). Fungal heat resistance can be derived by various physiological and morphological adaptations, including especially heat-stable cell types and structures. The most heat resistant fungal structures known so far are thick-walled ascospores (Conner and Beuchat 1987; Dijksterhuis and Teunissen 2004; Dijksterhuis 2007; Rachon et al. 2021). Other examples are heat resistant sclerotia (Hind-Lanoiselet et al. 2005) and chlamydospores with thickened cell walls (Seifert et al. 2004). Moreover, heat and stress resistance of these fungal structures can be increased by the production and accumulation of compatible solutes with thermo-protective function, like glycerol, mannitol, trehalose, or trehalose-based oligosaccharides (Dijksterhuis 2007; Wyatt et al. 2013). Besides the synthesis of cell-stabilizing compounds, heat resistant fungi can produce toxic secondary metabolites (Tournas 1994; Puel et al. 2005), also called mycotoxins. Producers of such are, e.g., Byssochlamys species, which are among the most common fungal contaminants of heat processed food products (e.g., tomato paste, apple juice) (Tournas 1994; Kotzekidou 1997; Salomão et al. 2014; Santos et al. 2018). Therefore, heat resistant fungi represent a great concern for the food and healthcare industry. The natural occurrence of this ubiquitous and saprophytic soil fungi in agricultural substrates often leads to contamination of fruits and crops. Due to their increased stress and heat resistance, high-temperature treatments during food processing are insufficient to remove the contaminating fungi, or even trigger an activation of resting propagules (Dijksterhuis and Teunissen 2004). Subsequently, the occurrence and growth in food products and the production of toxins lead to economic losses (Piecková et al. 2020) and exposure of customers to potentially opportunistic pathogens and respective toxins (Yang et al. 2014).
While the known diversity of heat resistant fungi comprises predominantly soil inhabiting and food spoiling ascomycetes, descriptions of stress- and heat resistant basidiomycetes are rare. Despite the great diversity of the Agaricomycotina and Ustilaginomycotina lineages (Hibbett 2006; Begerow et al. 2014), most records of stress-resistant basidiomycetes account to the small subphylum of Wallemiomycotina. This group contains the two orders Geminibasidiales and Wallemiales, and its phylogenetic position is recently discussed, e.g., to retain the lineage of Wallemiomycetes within the Agaricomycotina or to represent a fourth subphylum of the Basidiomycota (Mishra et al. 2018, Naranjo-Ortiz and Gabaldón 2019). The species described for the Wallemiales were isolated from hypersaline environments (e.g., salted cod, solar salterns, sea water), sweets, soil, and air (Zalar et al. 2005), and include some of the most halophilic, osmophilic, and xerotolerant fungal species worldwide (Zajc and Gunde-Cimerman 2018). The few described species for the Geminibasidiales are exclusively soil-borne fungi and are moderately heat resistant and xerotolerant (Nguyen et al. 2013; Nasr et al. 2014). As seen in those taxa, physiological resistance often includes the resilience to multiple abiotic factors simultaneously like heat, drought, salinity, or osmolarity (Nguyen et al. 2013; Zajc and Gunde-Cimerman 2018). However, research on the diversity of stress- and heat resistant basidiomycetes, their evolution, and the ecological roles and physiological mechanisms is still underrepresented.
In this study, we describe and characterize four novel basidiomycetous mesophilic isolates with heat resistance abilities. The isolates were obtained from forest soils in Canada (Nova Scotia) by following a commonly recommended procedure to isolate heat resistant fungi from natural substrates (Seifert et al. 2004). BLAST searches using the ITS sequences of the isolates against the GlobalFungi database (Větrovský et al. 2020) and NCBI led to few hits of uncultured fungi from environmental samples, taken from forest soils in Australia and Taiwan (Tedersoo et al. 2014) and a forest fire region in Portugal (Buscardo et al. 2015). We provide, based on 38 orthologous single-copy genes using 63 fungal genomes, the phylogenetic hypothesis that these isolates represent a novel lineage named Peribolosporomycetes class. nov. within the Ustilaginomycotina, as well as two new species: Peribolospora kevripleyi sp. nov. and Peribolospora baueri sp. nov. Our taxonomic conclusions are supported by additional data from ultrastructure of the septal pore type of P. kevripleyi. We performed microscopic and colony observations, as well as scanning electron microscopy to assess the macromorphology and micromorphology and obtain insights into the putative lifestyle. Additionally, the relationship of the four isolates was studied, by analysis of barcode sequences, microsatellite fingerprinting, and the comparison of orthologous gene clusters with other fungal genomes.
Material and methods
Sampling
Forest soils associated with stands of Pinaceae trees were sampled at four locations in Canada (Shelburne, Nova Scotia) in 2001 and 2002 (Online Resource 1). For isolation of heat resistant, mesophilic fungi, a flotation approach with heat-treatment was used (Seifert et al. 2004). In brief, the soil substrate was put in a 0.1% (w/v) proteose peptone solution, heated to 75 °C in a water bath and then incubated at 75 °C for 30 min under constant shaking. Aliquots of the substrate solutions were mixed with melted and cooled (50 °C) potato dextrose agar (PDA, Difco) containing chloramphenicol (40 mg/l) and poured into Petri dishes. The plates were incubated at room temperature (RT) and monitored for growth. Fungal colonies were subcultured and purified from the environmental plates.
Morphological characterization
Macromorphology
For morphological description of cultures, fungal isolates were cultivated on potato dextrose agar (PDA) (39 g/l potato dextrose agar (Carl Roth); pH 6.2), malt extract agar (MEA) (30 g/l malt extract, 3 g peptone, 20 g/l agar; pH 6.2), corn meal agar (CMA) (30 g/l malt extract, 20 g/l agar, cornmeal water (25 g/l cornmeal in tap water at 60 °C overnight, filtered with cloth tissue); pH 6.2) and malt yeast 40% sucrose agar (M40Y) (20 g/l malt extract, 5 g/l yeast extract, 400 g/l sucrose, 15 g/l agar; pH 6.2) for 7 days at 25 °C. Inoculation of plates was performed with spore solutions of defined concentration. To obtain spore solutions, fungal isolates were grown on malt extract agar for 21 days. The cultures were covered with 1% (v/v) Tween 20 and 0.85% (w/v) NaCl solution and spores were carefully detached by scratching with an inoculation loop. The spore solutions were centrifuged (4000 g, 10 min, RT), and the sedimented spores were washed with 0.85% (w/v) NaCl solution. After a second centrifugation, the washed spores were diluted in 0.85% (w/v) NaCl solution and the spore titer was determined and standardized using a counting chamber (Neubauer improved) following the manufacturer’s recommendations. Cultures were pictured, and colony sizes were measured using an optical stereo microscope (Zeiss Axio Zoom.V16; Carl Zeiss Microscopy GmbH, Göttingen, Germany), a PlanApo Z 0.5 × objective, and Zen software (Zeiss, Oberkochen, Germany). Diameters of colonies on each growth medium were measured in triplicates. To test further growth characteristics in simulated natural conditions, the two species were grown in pots with twice autoclaved (first: 1 h, 121 °C; second: 25 min, 121 °C) pine forest soil. The substrate was taken from the O- and A-horizon containing both litter (e.g., needles, twigs) and soil. The samples were inoculated with spore solutions, incubated at RT, and growth was observed for three months.
Micromorphology
For assessment of micromorphological structures, fungal isolates were grown on MEA and CMA medium for 2–3 weeks at RT. Scanning electron microscopy (SEM) was performed with a Sigma VP electron microscope (Carl Zeiss Microscopy GmbH, Göttingen, Germany) on colony sections. After freezing in liquid nitrogen, samples were sublimated at − 90 °C for 8 min, platinum sputtered for 90 s at 30 mA, and recorded with the Zeiss-smartSEM-User-Interface. Measurements were performed with SmartTiffV3 software (v.02.01) from Zeiss. For septal pore type analysis, transmission electron microscopy (TEM) was performed. High-pressure freezing and freeze substitution were conducted as described by Bauer et al. (1995).
To check for indicators of meiosis, the nuclear behavior of the four fungal isolates was studied by using DAPI staining. The isolates were grown in liquid PD medium for up to 1 week on RT. Mycelium was taken with a sterile inoculation loop, mixed with 10 µl of ddH2O on a glass slide, and the cells were quickly heat fixed by using a Bunsen burner. The slides were cooled down, and 5 µl of 1 × DAPI solution was added to the cells, which were afterwards covered by a coverslip. The cells were examined by fluorescence microscopy using an Axioplan microscope (Carl Zeiss Microscopy GmbH, Göttingen, Germany).
Physiological characterization
Cardinal growth temperatures of the isolates were tested on PDA and yeast peptone dextrose agar with additional 10% glucose (YPD + 10) (10 g/l yeast extract, 20 g/l peptone, 120 g/l dextrose, 20 g/l agar; pH 6.2) in five replicates with one point inoculation. Plates were inoculated with spore solutions and incubated at 10, 16, 20, 25, 30, and 36 °C (PD) and 25, 27, 30, and 33 °C (YPD + 10). Colonies were photographed after 14 days with a Samsung J5 smartphone, and growth was measured using Image J (Abramoff et al. 2004). Heat resistance of the fungal isolates was investigated with different approaches. First, maximum 3-week-old culture sections with mycelium and spores from CMA medium were transferred into 1.5 ml reaction tubes and heat-treated at 60 °C and 75 °C for 30 min in a heating shaker. Heated material was spread on PDA, incubated at RT, and examined periodically for growth. Second, soil, inoculated with spore solutions and incubated (RT) for 3 months, was mixed with 0.1% (w/v) protease peptone solution and heated at 75 °C (30 min) in a water bath as described previously. Plating of the solution on PDA was followed by incubation (RT) for 21 days and periodical examination for fungal growth. To test the fermentation abilities of the strains, phenol red assays were performed using liquid media, which contained protease peptone (10 g/l), sodium chloride (5 g/l), phenol red (0.018 g/l), and a single carbon source trehalose, sucrose, or xylose (each 10 g/l). For sterilization, 25 ml of the phenol red carbohydrate broth were filled in test tubes containing an inverted Durham tube and autoclaved (121 °C, 3 min). Each test tube was inoculated with three inoculation loops of freshly grown mycelium, and incubated for three days at RT. The tests were performed in triplicates. The osmotolerance, xerotolerance, and halotolerance of the four isolates was tested on MEA plates with increasing sucrose, sodium chloride (NaCl) and magnesium chloride (MgCl2) gradients (Nguyen et al. 2013), and glycerol medium (5 g/l peptone, 1 g/l KH2PO4, 200 g/l glycerol, 10 g/l glucose, 0.5 g MgSO4, 16 g/l agar; pH 5.8), similar to DG18 agar (Hocking and Pitt 1980).
Molecular studies
DNA extraction, barcode amplification, and fingerprinting
For genome sequencing, gDNA was extracted from 150 mg mycelium per sample using an adapted cetyl-trimethylammonium-bromide (CTAB) protocol, provided in scope of the 1000 fungal genomes project (Grigoriev et al. 2014) by Kohler et al. (2011), modified from Fulton et al. (1995). Mycelium (150 mg), grown on MEA (2 weeks, RT), was cooled in liquid nitrogen and homogenized with a ball mill (Retsch). Homogenized material was roughly mixed with 5.25 ml CTAB (2%) lysis buffer and incubated at 65 °C for 30 min, under frequent inverting of the tubes. Then, 0.33 volumes of potassium acetate (5 M) were added and samples were incubated on ice (30 min) and centrifuged (40 min, 3.220 g, 4 °C). The supernatant was transferred in a fresh tube and mixed with one volume of chloroform:isoamylalcohol (24:1) and centrifuged again (30 min, 3.220 g, 4 °C). Then, the aqueous phase was mixed with 20 µl RNAse A (20 mg/ml) (NEB) and incubated (2 h, 35 °C). For precipitation, the samples were mixed with 1/10 volume of sodium acetate (3 M) and 1 volume isopropanol and incubated at RT (5 min). After centrifugation (30 min, 10.000 g, 4 °C), the supernatant got discarded, and the pellet washed with 0.6 ml EtOH (70%) and centrifuged (10 min, 10.000 g, 4 °C). After air drying (5 min, RT), the DNA pellet was dissolved in 100 µl EDTA TE buffer at 65 °C and stored at − 80 °C for further processing. For barcode analysis and microsatellite fingerprinting, gDNA was extracted by a phenol–chloroform-based approach, as in Witfeld et al. (2021). Amplification of the ITS-rDNA region (Schoch et al. 2012; Stielow et al. 2015) was performed with the primer combination ITS1F (Gardes and Bruns 1993) and ITS4 (White et al. 1990), as described in Witfeld et al. (2021). The LSU was amplified using the primers LROR (Cubeta 1991) and LR6 (Vilgalys and Hester 1990). Partial amplification of RPB1 was performed with the primers RPB1-af and RPB1-cr (Stiller and Hall 1997; Matheny et al. 2002), the RPB2 gene was partially amplified with the primers RPB2-5f and RPB2-7cr (Liu et al. 1999), and partial MCM7 gene region was amplified using the primers MCM7-709for and MCM7-1348rev (Schmitt et al. 2009). Amplicons were purified using Exonuclease I and Shrimp alkaline phosphatase (NEB) with 1:5 diluted enzyme concentration and sequenced with a capillary sequencer (3130xl Genetic Analyzer, Applied Biosystems) by the sequencing facility of the Ruhr-University Bochum. Sequences were manually checked and corrected with BioEdit (v.7.0.5.3 28.10.2005) (Hall 1999). For alignments, MEGA X (v. 10.1.8, Build 10200331) was used to perform a multiple sequence alignment by MUSCLE on standard settings. Additionally, sequences were checked using BLASTn against the GenBank database (https://blast.ncbi.nlm.nih.gov).
For assessment of the global distribution, ITS barcode sequences of the four isolates were used in a BLAST search in the GlobalFungi Database (Větrovský et al. 2020). For genetic fingerprinting of the isolates, microsatellite-primed PCRs (MSP-PCR) were conducted using the primers (ATG)5, (GTG)5 (Lieckfeldt et al. 1993; Vuyst et al. 2008), and (GAC)5 (Baleiras Couto et al. 1996). The amplicon fingerprints of the different samples were compared by agarose gel electrophoresis with standardized conditions. Barcode sequences of the four isolates were deposited at the European Nucleotide Archive (ENA) database and received accession numbers, which are associated with the study PRJEB50042 (Online Resource 1). Genome assemblies and genome raw reads are publicly available at NCBI (Online Resource 1). The annotated genomes will be provided by request. Cultures of the four isolates were deposited at the German Collection of Microorganisms and Cell Cultures (DSMZ) (DSM 113856, DSM 113857, DSM 113858, DSM 113859) and Canadian Collection of Fungal Cultures (DAOMC) (DAOMC 252470, DAOMC 252471, DAOMC 252472, DAOMC 252473). Moreover, dried specimens were deposited in the Canadian National Mycological Herbarium (DAOM) and received respective accession numbers (DAOM 984983, DAOM 984984, DAOM 984985, DAOM 984986) (Online Resource 1).
Genomic analysis
Genome sequencing, assembly, and annotation
The quality and quantity of extracted gDNA were measured with Qbit (Life Technologies, Burlington, Canada). For library preparation, the NxSeq AmpFREE Low DNA Library Kit (Lucigen) was used according to manufacturer conditions. Whole-genome sequencing (2 × 300 bp paired end) was done at the Agriculture and Agri-Food Canada (AAFC) on an Illumina MiSeq. Read quality was assessed with FastQC (v.0.11.8), and Trimmomatic (v.0.39) was applied to remove Illumina adaptors, low quality bases, and reads shorter than 36 bp. Genomes were assembled de novo using SPAdes (v.3.12.0), and assembly statistics were obtained with QUAST (v.5.0.2) (Gurevich et al. 2013). Genome completeness was calculated using BUSCO (v.5.2.2) against the fungal database (v.odb10) (Manni et al. 2021). Average nucleotide identities were calculated by pairwise whole-genome alignment with NUCmer (v3.23) (MUMmer; Kurtz et al. 2004) with the “-mum” parameter. A delta filtering step with “ − 1” and “ − l100” was applied to select 1-to-1 alignments to allow rearrangements and to select a minimal alignment length of 100 bp, respectively. Gene predictions of the genomes and other species were performed with AUGUSTUS (v.3.3.3), trained with Cryptococcus neoformans (San Felice) Vuillemin or Ustilago maydis (DeCandolle) Corda and for ascomycetes Schizosaccharomyces pombe Lindner, Saccharomyces cerevisiae (Desmazières) Meyen, Chaetomium globosum Kunze, or Aspergillus fumigatus Fresenius, as appropriate. Secreted proteins were calculated using SignalP (v.6.0) (Teufel et al. 2022), and tRNAs and CAZymes were predicted using tRNAscan-SE (v.2.0.9) (Chan et al. 2021) and dbCAN2 (v.10) (Zhang et al. 2018), respectively.
Phylogenomics
Genome assemblies of fungal species were obtained from GenBank (https://www.ncbi.nlm.nih.gov/assembly) and Mycocosm (https://mycocosm.jgi.doe.gov/mycocosm/home). Accession numbers are provided (Online Resource 2). Orthologous protein sequences among the Peribolosporomycetes genomes and 59 fungal species from the Basidiomycota and Ascomycota were calculated using OrthoFinder (v.2.5.2) (Emms and Kelly 2019). A set of 38 single-copy orthologous genes was found to be present in all species, and these were individually aligned with MAFFT (v.7.273) (Katoh and Standley 2013). These alignments were subsequently concatenated, and a maximum likelihood tree was calculated with RAxML (v.8.2.12) (Stamatakis 2014), using 500 bootstrap replicates, the PROTGAMMAWAG model, 123 as seed number for the parsimony inferences and a random seed of 321. Shared orthologous genes were calculated with ComplexUpset (v.1.3.1) package (Krassowski et al. 2021) and the “exclusive intersection” option in R (v.4.1.0; R Core Team 2021).
Results
Molecular studies
Illumina (MiSeq) sequencing yielded 4.48–6.29 million paired end reads for each isolate. After quality control and trimming, the reads were used for de novo assemblies. Between 750 and 1288 contigs were obtained for the four genomes (Table 1), and genome coverages were between 89,4X and 112,6X. The genome size of P. baueri is 22.7 Mb, while P. kevripleyi has a genome size of 23.2 Mb. The GC content varies slightly between the two species (P. baueri 58.7%; P. kevripleyi 59.1%) but shows a distinctly higher GC content in coding regions than in non-coding regions (Table 1). BUSCO calculated a genome completeness of 92.9% (P. baueri) and 92.6% (P. kevripleyi). Gene predictions lead to 7054 and 7024 protein-coding genes for P. baueri isolates and 7283 and 7224 for P. kevripleyi.
Calculations of the similarity of the genomic sequences of the Peribolosporomycetes revealed that the similarity between the P. baueri isolates (99.98%) and P. kevripleyi isolates (99.69%) is distinctly higher than between the species (95.37–95.40%) (Table 2). Maximum likelihood phylogenetic analyses were performed using 38 single-copy orthologous genes of 63 species to resolve the phylogenetic relationship of the Peribolosporomycetes isolates within Basidiomycota (Fig. 1). The phylogeny indicates that the four isolates represent a new monophyletic lineage, which we named Peribolosporomycetes class. nov., and this group is a sister lineage to all classes in Ustilaginomycotina. Moreover, the four isolates seem to be members of two closely related sister species, for which we propose the names Peribolospora baueri sp. nov. (DAOMC 252473, DAOMC 252471) and Peribolospora kevripleyi sp. nov. (DAOMC 252472, DAOMC 252470).
Phylogenetic position of the Peribolosporomycetes and Peribolospora species within Basidiomycota. The phylogenetic position of the isolates was calculated based on 38 single-copy orthologous protein coding genes. The number of species included in collapsed groups are indicated by white numbers. Lineages are color-coded and correspond to the subphyla of Agaricomycotina (red), Wallemiomycotina (purple), Ustilaginomycotina (yellow), and Pucciniomycotina (green). Ascomycota (white) was used as the outgroup. Nodes with a bootstrap of ≥ 99% support are indicated by a black dot. Scale bar represents the number of substituted aminoacids per site. A phylogeny without collapsed groups (Online Resource 6), strain IDs of used species (Online Resource 2), and the original alignment file are provided (Online Resource 7)
We compared the gene contents of species representing distinct classes within the Basidiomycota and the Ascomycota with the Peribolospora species. We identified orthologous groups (OGs) based on the predicted proteomes of the 63 species used for the phylogeny (Fig. 2). Orthology analyses identified 18,913 OGs for all species used in our data set. From these, 956 OGs were shared between all analyzed classes of both Ascomycota and Basidiomycota, while in total, 965 genes were shared between all basidiomycetous classes and the Peribolosporomycetes. A total of 2,072 OGs were shared among all Ascomycota. The number of class-unique OGs varied according to the taxonomic groups. For instance, 2,734 OGs were solely found in the Agaricomycetes (represented by 8 species), 565 OGs only in the Exobasidiomycetes (8), and the Wallemiomycetes (2) had 233 unique OGs, not shared with other fungal classes. The four Peribolosporomycetes shared 530 OGs, which were not present in other taxonomic groups. The counts of OGs shared between the Peribolosporomycetes and the closest related taxa or taxa of putatively similar lifestyle are low. For instance, 41 OGs are shared exclusively with the Exobasidiomycetes, two OGs with the Ustilaginomycetes, and 9 OGs with the Geminibasidiomycetes. Comparison of OGs within the Peribolospora species revealed almost no strain specific OGs. However, comparisons between the Peribolospora species showed substantial amounts of shared OGs within the species P. baueri and P. kevripleyi, indicated by the phylogeny (Fig. 1). Within P. baueri (DAOMC 252473, DAOMC 252470) 362 unique OGs are shared, while within P. kevripleyi (DAOMC 252472, DAOMC 252470) 309 OGs are shared.
Orthology analysis of the Peribolospora species, representing the Peribolosporomycetes, and related fungal lineages. Total number of orthogroups shared among datasets are indicated by bars. Bars representing orthogroups exclusively found in the Peribolospora isolates are highlighted in gray. Background colors correspond to Agaricomycotina (red), Wallemiomycotina (purple), Ustilaginomycotina (yellow), Pucciniomycotina (green), and Ascomycota (white). Taxonomic groups sharing a specific set of orthologous genes are highlighted by black dots, while not included taxa are grayed out. The number of species included in each group is indicated after the lineage name
The higher genetic similarity within the two isolates of P. baueri (DAOMC 252473 and DAOMC 252470) and P. kevripleyi (DAOMC 252472, DAOMC 252471) was also observed in barcode sequence alignments. While the ITS, MCM7, RPB1, and RPB2 sequences were highly similar or identical among the two isolates of each species, the similarity between the species was between 97.78% (RPB1) and 99.67% (ITS). The LSU sequences were identical for all isolates. Microsatellite-primed PCR-fingerprints revealed no differences between the isolates within one species but showed distinct patterns between the two species P. baueri and P. kevripleyi (Online Resource 3). Additionally, BLAST searches for mating-type genes and comparative analyses led to the detection of the A2 allele of the pheromone-receptor gene in all four Peribolospora strains.
Morphological and physiological studies
Colony growth
The Peribolospora species show relatively slow growth and a high phenotypic variability (Fig. 3). Young colonies are white or light beige. Older cultures are often gray-brown with purplish tints and tough, sometimes coriaceous, hyphal growth. After 1 week on PDA, flatly grown colonies of P. baueri consist of light beige, agglutinated hyphae, while P. kevripleyi colonies show more centrally convex growth. On MEA and CMA, P. baueri appears gray and P. kevripleyi gray-brownish. Characteristically, both species depict convex and grooved rosettes, and especially for P. kevripleyi, the sulcate growth continues peripherally. On M40Y, both species appear as flat-growing white hyphae, with P. kevripleyi showing centrally denser growth.
Colonies of Peribolospora baueri and Peribolospora kevripleyi. a–b Colonies of P. baueri strains (DAOMC 252471, DAOMC 252473) on PDA, MEA, CMA, and M40Y after 7 days at 25 °C. c–d Colonies of P. kevripleyi strains (DAOMC 252472, DAOMC 252470) on PDA, MEA, CMA, and M40Y after 7 days at 25 °C. Scale bars = 5 mm
Abiotic stress resistance
Growth on M40Y indicated osmotolerance for both Pelibolospora species, but growth on DG18-like glycerol medium was strongly reduced and no growth was observed for P. kevripleyi strain DAOMC 252472. Addition of different compounds, which lower the water activity (aw) of a medium, was used to assess the degree of xerotolerance, osmotolerance and halotolerance of the Peribolospora species (Fig. 4). Increasing amounts of sodium chloride (NaCl) and glycerol strongly inhibited growth of P. baueri and P. kevripleyi. In comparison, better growth on sodium chloride was observed up to an aw of 0.95 (7.8% NaCl, 1.3 M), while on glycerol, growth was slower and observed up to an aw of 0.98 (16.5% glycerol, 1.8 M). In contrast to that, reduction of the aw by addition of sucrose had a positive effect on the growth of both Peribolospora species, up to an aw of 0.95 (26% sucrose, 0.7 M), and growth on sucrose was observed up to a concentration of 60.8% (1.8 M), respectively, an aw of 0.87. Both species showed growth on media with tested pH values between 3.5 and 9. Thereby, growth was only marginally affected by pH values between 4.5 and 7.5, and growth at the rather extreme pH values 3.5 and 9 was slower, but not weakened. Notably, only a small growth reduction of P. kevripleyi strain DAOMC 252470 was observed at pH 9, while the other strains showed clearly inhibited growth.
Growth of Peribolospora baueri and Peribolospora kevripleyi at different water activities and pH. Water activities between 1.00 and 0.83 were reached by adding defined amounts of a sucrose, b sodium chloride, and c glycerol to the media. d Growth on pH 3.5–9. Cultures were incubated at room temperature for 14 days
Temperature adaptation
Growth of P. baueri and P. kevripleyi was observed from 10 to 35 °C. Growth of Peribolospora kevripleyi strain DAOMC 252472 was observed up to 36 °C. On PDA, the optimum temperature of all isolates is between 25 and 30 °C. Therefore, all Peribolospora isolates are considered as mesophilic. Heat resistance tests (75 °C, 60 °C; 30 min) using spore solutions or stamped mycelia from 3-week-old solid-state cultivations failed. However, for both P. baueri and P. kevripleyi (DAOMC 252473, DAOMC 252470), cultures which grew up to 3 months on sterilized pine forest soil with a pH of 3.65 (Fig. 5) survived the heat-treatment at 75 °C (30 min), were re-isolated from the soil substrate, and regrew on PDA.
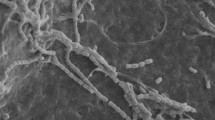
Growth of Peribolospora on sterilized coniferous forest soil. a P. kevripleyi DAOMC 252472 and b P. baueri DAOMC 252473 growing on soil samples, including coniferous litter. a (i–ii) Growth of P. kevripleyi on a piece of wood within the soil sample. (iii–iv) Chlamydospores and conidiophores of P. kevripleyi. (v–vi) SEM pictures of P. kevripleyi growing on soil substrate and spore formation. b (i–ii) Growth of P. baueri on soil samples, including pine needles and wood. (iii) Chlamydospores and conidiophores of P. baueri on soil samples. (iv–v) SEM pictures of P. baueri growing on soil samples and chlamydospores. Inoculated soil samples were incubated (RT) in moderate light for 90 days. Unlabeled black scale bars = 1 mm, white scale bars = 10 µm
Spore morphology
Chlamydospores and conidia were observed for cultures grown on corn meal agar (CMA), malt extract agar (MEA) with or without addition of sucrose for at least 2 weeks (Fig. 7), and on pine forest soils after 3 months (Fig. 5). Micromorphological features of Peribolospora baueri and Peribolospora kevripleyi are highly similar. In both species, chlamydospores are terminally produced and are carried by a coiled hypha, often wrapping around the fully developed spore. Characteristically, mature chlamydospores are hyaline, triangular, pear-like shaped with a narrow proximal side and broadening distally (Fig. 8). Chlamydospores of P. baueri had a length of 7.2–11.4 µm (mean ± SE = 9.0 ± 0.8) and a width of 7–11.4 µm (mean ± SE = 9.4 ± 0.4), while the measurements for P. kevripleyi were 7.6–11.7 µm (mean ± SE = 9.8 ± 0.9) and 7.1–10.2 µm (mean ± SE = 8.7 ± 0.7). Both species produce ovoid or cylindrical, petiolate conidia, sometimes with apical scars after bearing secondary conidia (Fig. 6; Fig. 8; Online Resource 4). Primary conidia of P. baueri are 4.3–9.7 µm (mean ± SE = 6.6 ± 1.3) in length and 2–3 µm (mean ± SE = 2.4 ± 0.2) in width, while P. kevripleyi primary conidia have a size of 4.3–13.7 µm (mean ± SE = 7 ± 2) and 1.8–3.8 µm (mean ± SE = 2.4 ± 0.4). Secondary conidia of P. baueri have a length of 2.6–5 µm (mean ± SE = 3.7 ± 0.5) and a width of 1.9–3.4 µm (mean ± SE = 2.5 ± 0.4), and secondary conidia of P. kevripleyi have a size of 2–5.1 µm (mean ± SE = 3.4 ± 0.6) length and 1.8–3.2 µm (mean ± SE = 2.2 ± 0.3) width. Nuclear staining and fluorescence microscopy of the micromorphological structures did not lead to the observation of any indicators of meiosis.
Chlamydospores and conidiogenous cells with conidia of Peribolospora baueri and Peribolospora kevripleyi. Scanning electron micrographs of P. baueri chlamydospores (a DAOMC 252473, b DAOMC 252471) and conidiophores (c DAOMC 252743, d DAOMC 252471), and P. kevripleyi chlamydospores (e DAOMC 252472, f DAOMC 252470) and conidiophores (g DAOMC 252472, h DAOMC 252470). Cultures were grown for at least 2 weeks on CMA or MEA at RT. Scale bars = 2 µm
Septal pore morphology
The septal pores of P. kevripleyi (DAOMC 252470) were observed with transmission electron microscopy, as a representative for all Peribolosporomycetes (Fig. 7). P. kevripleyi has relatively simple pore type characteristics. Septum membranes, adjacent to the pore openings, are not thickened, but unequally layered. Parenthesomes, forming distinct outer septal pore caps or tubular extensions, are absent. However, low contrasted elements margin the pore openings and could be related to the septal pore functions. Additionally, supposedly non-membranous areas of increased electron-density are visible in the pore openings and occasionally slightly extend towards the cytoplasm of connected cells (Fig. 7A, C). The imaged septal pores of P. kevripleyi had widths between 0.06 and 0.09 µm.
Septal pore morphology of Peribolospora. Transmission electron micrographs of P. keyripleyi (DAOMC 252470) septal pores (a–c). A tubular structure, visible in the pore, is indicated by white arrows. Low contrasted, non-membranous structures around the pore opening are highlighted by a black arrow. Cultures were grown for at least two weeks on CMA or MEA at RT. Scale bars = 100 nm
Taxonomy
Peribolosporomycetes Witfeld, M. A. Guerreiro, H.D.T. Nguyen, Begerow, class. nov.
Mycobank: 843630
Description: Class of mesophilic, heat resistant and osmotolerant basidiomycetes with slow hyphal growth and high phenotypic variability. Characteristically, triangular shaped chlamydospores are distally produced on coiled hyphae. Sympodial, ovoid conidia are produced. Relatively simple septal pores, without thickened septum membranes. Parenthesomes, forming distinct septal pore caps are absent, but low contrasted elements indicate non-membranous structures around the pore openings and within the pore. Based on the analysis of 38 protein coding genes, orthology analysis, and septal pore type analysis, this class is placed within the Ustilaginomycotina.
Type species: Peribolospora baueri
Peribolosporales Witfeld, M. A. Guerreiro, H.D.T. Nguyen, Begerow, ord. nov.
Mycobank: 843631
Description: According to the characters given above for the class Peribolosporomycetes.
Peribolosporaceae Witfeld, M. A. Guerreiro, H.D.T. Nguyen, Begerow, fam. nov.
Mycobank: 843632
Description: According to the characters given above for the class Peribolosporomycetes.
Peribolospora Witfeld, M. A. Guerreiro, H.D.T. Nguyen, Begerow, gen. nov.
Mycobank: 842991
Etymology: “Peribolos” refers to the characteristic enclosure or embracement of the chlamydospores (“spora”) by the coiled, spore-carrying hyphae.
Type species: Peribolospora baueri
Description: Class of mesophilic, heat resistant, and osmotolerant basidiomycetes with high phenotypic variability. Characteristically, the hyaline, triangular shaped chlamydospores are distally produced on coiled hyphae. Basidiomata or basidiospores are not observed. Sympodial, hyaline, and ovoid conidia are produced. Species have relatively simple septal pores, without thickened septum membranes. Parenthesomes, forming distinct septal pore caps, are absent, but low contrasted elements indicate non-membranous structures around the pore openings and within the pore. White–gray or beige mycelia are formed by slow growing, young colonies. A high phenotypic variability, depending strongly on cultivation medium and culture age, is observed. In older colonies brown and purplish pigmentation and coriaceous growth of agglutinated hyphae are often observed. Xylose, trehalose, or sucrose are not fermented. Based on the analysis of 38 protein coding genes, orthology analysis, and septal pore type analysis, this class is placed within the Ustilaginomycotina.
Notes: The two described species P. baueri and P. kevripleyi can be identified and distinguished using molecular barcodes (e.g., RPB1, RPB2, MCM7, ITS). Additionally, a reliable differentiation of the two species is possible based on MSP-PCR fingerprinting (e.g., ATG5, GAC5, GTG5) and orthology analysis.
Peribolospora baueri Witfeld, M. A. Guerreiro, H.D.T. Nguyen, Begerow, sp. nov.
Mycobank: 843634
Etymology: Named after the mycologist Robert Bauer for his extensive research in mycology and simple pored fungi.
Type: Canada: Nova Scotia, Shelbourne County, Colquist Road, isolated from soil associated with a stand of Pinus resinosa Aiton by a 75 °C heat-shock (30 min) approach, isolation date: Oct 1, 2002. Holotype: P. baueri DAOM 984984, deposition as dried culture of DAOMC 252473; Ex-type: P. baueri DAOMC 252473. DSMZ: DSM 113857. ENA database: Study: PRJEB50042, ITS: OW455115, LSU: OW455119, Genome assembly: JAMDHS000000000. Strains examined: DAOMC 252473 (ex-type), DAOMC 252471.
Description: Colony growth to a diameter of 1.73 cm ± 0.03 cm on PDA, 1.76 cm ± 0.04 cm on MEA, 1.76 cm ± 0.02 cm on CMA, and 0.99 cm ± 0.02 cm on M40Y after 1 week at RT, was observed. Cardinal temperatures are 10–35 °C, and optimum temperature is between 25 and 30 °C with colony sizes of 3.68 ± 0.21 cm at 30 °C. Increasing pigmentation from white and beige colored young colonies to gray, gray-brown, or purplish colored mycelia in older colonies. Growth is often coriaceous or shows agglutinated hyphae forming sulcate growing colonies with sharp margins. Chlamydospores and conidia are produced on MEA and CMA after 7–14 days. Chlamydospore development is observed to start with a distally swelling hyphae tip. During chlamydospore formation, spore carrying hyphae increase coiling. Mature, triangular shaped chlamydospores have a length of 7.2–11.4 µm (mean ± SE = 9 ± 0.8) and a width of 7–11.4 µm (mean ± SE = 9.3 ± 0.4). Ovoid or cylindrical conidia are petiolate with apical scars when secondary conidia detached. Primary conidia are 4.3–9.7 µm (mean ± SE = 6.6 ± 1.3) × 2–3 µm (mean ± SE = 2.4 ± 0.2) and secondary conidia are 2.6–5 µm (mean ± SE = 3.6 ± 0.5) × 1.9–3.4 µm (mean ± SE = 2.5 ± 0.4).
Peribolospora kevripleyi Witfeld, M. A. Guerreiro, H.D.T. Nguyen, Begerow sp. nov.
MycoBank: 843633
Etymology: Named after Kevin Ripley, the partner of one of the co-authors (H.D.T. Nguyen) of the study.
Type: Canada: Nova Scotia, Shelbourne County, Colquist Road, isolated from soil associated with a stand of Pinus banksiana Lambert by a 75 °C heat-shock (30 min) approach, isolation date: 17 Oct 2001. Holotype: P. kevripleyi DAOM 984985, deposition as dried culture of DAOMC 252472; Ex-type: P. kevripleyi DAOMC 252472. DSMZ: DSM 113856. ENA database: Study: PRJEB50042 ITS: OW455118, LSU: OW455122, Genome assembly: JAMDHT000000000. Strains examined: DAOMC 252472 (ex-type), DAOMC 252470.
Description: Colony growth to a diameter of 2.10 cm ± 0.06 cm on PDA, 1.81 cm ± 0.04 cm on MEA, 2.06 cm ± 0.02 cm on CMA, and 1.23 cm ± 0.01 cm on M40Y after 1 week at RT was observed. Cardinal temperatures are 10–36 °C, and optimum temperature is between 25 and 30 °C, with colony sizes of 3.18 ± 0.43 cm at 30 °C. Chlamydospore and conidia production on MEA and CMA after 7–14 days. During ontogenesis, increasing colony pigmentation from white and beige young colonies to gray-brown or purplish older colonies. Agglutinated hyphae with the tendency to form coriaceous appearing colonies with sulcate growth and often centrally rosette growth with sharp margins. Formation of chlamydospores begins with swelling hyphae tips and coiling of the spore-carrying hyphae. Mature, triangular shaped chlamydospores have a length of 7.6–11.7 µm (mean ± SE = 9.8 ± 0.9) and a width of 7.1–10.2 µm (mean ± SE = 8.7 ± 0.7). For ovoid or cylindrical, petiolate conidia apical scars are observed when secondary conidia detached. From proximal to distal, primary conidia are 4.3–13.7 µm (mean ± SE = 7 ± 2) and the width varies between 1.8–3.8 µm (mean ± SE = 2.4 ± 0.4). Secondary conidia are 2–5.1 µm (mean ± SE = 3.4 ± 0.6) × 1.8–3.2 µm (mean ± SE = 2.2 ± 0.3).
Discussion
A heat treatment approach (Seifert et al. 2004) used on Canadian boreal forest soils led to the isolation of four heat resistant, undescribed fungal strains, which based on molecular barcodes were classified as basidiomycetes. Morphological, physiological, phylogenetic, and genomic analyses provide evidence for the description of a novel class, Peribolosporomycetes, and two new species, Peribolospora baueri and P. kevripleyi.
Heat- and stress-resistance of Peribolospora species
Due to their high morphological similarity and overall phenotypic variability (Figs. 3, 6, and 8), we aimed for additional features for species differentiation of P. baueri and P. kevripleyi. Physiological traits are often used to differentiate morphologically similar and cryptic species (Peterson and Jurjević 2013; Jančič et al. 2015; Peintner et al. 2019). However, the growth temperature ranges of both species are typical for mesophilic fungi and highly similar (Online Resource 5), and the stress resistance to differently lowered water activities is also comparable. Xerotolerance or osmotolerance are the ability to grow at a water activity of 0.85 or lower (Pitt 1975). Both Peribolospora species grow on low NaCl concentration up to 7.8% (aw 0.95) (Fig. 4), which is high for the majority of terrestrial basidiomycetes, while ascomycetes often tolerate higher salt concentrations (Tresner and Hayes 1971; Jaouani et al. 2014). Theoretically, that enables Peribolospora to colonize saline soils and share habitats with halotolerant plants, which flourish at sea water salinity of 3.5% NaCl. Nonetheless, the ability to withstand high sucrose concentrations is far more pronounced in both Peribolospora species, showing osmotolerant capabilities with growth on M40Y and even up to 60.8% sucrose (aw 0.87) (Fig. 4). Although several common mechanisms are involved in adaptation to high salinity and sugar concentrations, the lopsided tolerance to high osmotic pressure caused by one of these factors is a common phenomenon in fungi (Kunčič et al. 2010, 2013; Bubnová et al. 2014). The ability to grow at high sugar concentrations is typical for many food spoilage fungi (e.g., Zygosaccharomyces or Xeromyces) (Snyder et al. 2019). Other traits of spoilage fungi, isolated frequently from industrially processed food products (e.g., canned fruits, juices), are tolerance of low pH values and heat resistance (Scaramuzza and Berni 2014; Santos et al. 2018). Both, P. kevripleyi and P. baueri show growth at pH 3 (Fig. 4) and colonized coniferous soils of pH 3.65 in an in vitro experiment (Fig. 5).
Micromorphological traits of Peribolospora. a–b Hyphae and conidiophores with primary and secondary conidia. After conidia release, apical scars are visible. c–d Young, developing chlamydospores with an increase in hyphal coiling. e–g Fully developed chlamydospores at coiled hyphae. Scale bars = 10 µm
While heat treatments (60 °C, 75 °C), using maximum 3-week-old spore solutions and mycelium stamps obtained from Peribilospora species grown on PDA were not survived, exposure to a heat treatment of the colonized soil substrate, imitating natural growth conditions and the commonly applied procedure for the isolation of heat resistant fungi (Seifert et al. 2004; Yaguchi et al. 2012; Nguyen et al. 2013), verified the heat resistance of Peribolospora. It is well known that the heat resistance abilities of fungi are highly dependent on manifold intrinsic and extrinsic factors. For instance, culture age can positively correlate with the resilience of spores to high temperature stresses (Conner and Beuchat 1987; Dijksterhuis and Teunissen 2004). Conner et al. (1987) demonstrated the increased heat resistance of older spores and the accumulation of thermoprotective sugars, morphological changes of the cell wall, and extractable proteins in aging spores. Further, a dependency between the heat resistance abilities of fungi and the availability of certain solutes (e.g., salts, sugars), organic acids, or the pH value (Bayne and Michener 1979) in the cultivation substrate or the heated test substrate has been observed before (Conner and Beuchat 1987; Engel and Teuber 1991; Tournas 1994). This variation of the heat resistance of a fungus is a major problem in food industry, since different food products require different temperature treatments to successfully inactivate contaminants of the same fungal species (Dijksterhuis 2007).
SEM of the colonized soils revealed exceedingly numerous amounts of chlamydospores in the samples (Fig. 5). We speculate that either the prolonged cultivation time itself, the low pH value of the soil substrate, or compounds within the soil led to physiological changes during the maturation of the numerous spores, which allowed the survival of the heat treatment at 75 °C for 30 min. The possibility of soil accumulates, which protected the fungal structures from the heat treatment can be excluded, due to the high dilution of the substrate and the constant shaking during the heat exposure. Notably, the trait of heat resistance is more commonly known for ascomycetous fungi, producing extremely heat resistant asco- or chlamydospores (Seifert et al. 2004; Berni et al. 2017; Biango-Daniels et al. 2019), while descriptions of heat resistant basidiomycetes are rare, with only a few species described so far (Nguyen et al. 2013, 2014). Further investigations (e.g., cell wall composition, thermoprotective compound accumulation, heat-shock proteins) are necessary to elucidate the potential mechanisms behind the heat resistance, presumably derived by heat resistant chlamydospores of the novel Peribolospora species. Also, additional sampling campaigns aiming for mesophilic, heat resistant basidiomycetes might reveal further heat resistant species belonging to the Peribolosporomycetes.
Taxonomic position of the new class Peribolosporomycetes
The isolates described in this study share certain physiological traits (e.g., heat resistance, pH growth range) with the Geminibasidiales within the subphylum of Wallemiomycotina, which is a sister lineage to the Agaricomycotina (Naranjo-Ortiz and Gabaldón 2019). Our phylogenetic analysis is congruent with the current knowledge of fungal taxonomy (Spatafora et al. 2017; Naranjo-Ortiz and Gabaldón 2019) and supports the placement of the Peribolospora isolates in a sister lineage to all other existing Ustilaginomycotina (Fig. 1; Online Resource 6). Additionally, it revealed two different Peribolospora species in our study. This is supported by the similarity of analyzed barcode sequences and the identical fingerprints within the P. baueri and P. kevripleyi isolates (Online Resource 3). Microsatellite-primed PCR fingerprinting has been already successfully used by several studies to differentiate isolates both on species or strain level, leading to the unraveling of genetic diversity and species complexes within the Ascomycota (Suh et al. 2013; Ramírez-Castrillón et al. 2014), and differentiation of basidiomycetous genera (Caldeira et al. 2009). Given that lifestyle adaptations and relatedness are reflected in genomic differences and gene content (Buijs et al. 2021), the orthology analysis further strengthens the creation of new lineage within the Basidiomycota, represented by two different species (Figs. 1 and 2). Differentiation of P. baueri and P. kevripleyi based on their orthologous groups is possible, and each chosen fungal class, as well as the combined Peribolospora isolates, had a unique set of orthologs, not shared with species from other groups. Moreover, individual comparisons of the set of orthologs of Peribolospora to the other classes revealed no compelling overlap, with the biggest overlap observed for the Exobasiodiomycetes. Respectively, this indicates unique genetic traits and a lower degree of relatedness of the Peribolospora species towards already existing fungal taxa. To substantiate the idea of a novel lineage within the Basidiomycota and to answer the question of the rank of the taxonomic group, for which the Peribolospora isolates are representative species, we investigated the septal pore type of a Peribolospora isolate (P. kevripleyi DAOMC 252470) (Fig. 7). Since a long time, septal pore morphology is essential for the arrangement of the major basidiomycetous taxa (Bauer et al. 1997) and is still used as an important micromorphological marker to solve phylogenetic relationships and describe novel classes and species (Scheuer et al. 2008; Nguyen et al. 2015). The simple septal pore type of Peribolospora without membrane swelling differs from the characteristic dolipores found in the Agaricomycotina and Wallemiomycotina (Van Driel et al. 2009; Nguyen et al. 2015). Instead, Peribolospora septal pores resemble rather the morphology of Ustilaginomycotina or Pucciniomycotina species, which have simple septal pores. Especially the members of the Ustilaginomycotina are characterized by outer pore caps and often variably intense, electron-dense areas or bands associated to the pore openings, as frequently described for various taxa in the Ustilaginomycetes and Exobasidiomycetes (Bauer et al. 1997; de Beer et al. 2006; Lutz et al. 2012). Notably, the septal pores of these classes usually contain very distinct, membranous outer septal pore caps. However, we observed only extremely low contrasted, thin structures in association to the pore opening (Fig. 7), which could be homologous to the septal pore caps, similar to Lutz et al. (2012). In addition, a tubular structure in the pore seems highly similar to the tubular ring described for Exobasidiomycetes (Begerow et al. 2014). Therefore, and based on the genetic analyses, further morphological observations and the physiological properties, we conclude that the Peribolospora isolates represent at least a novel class within the Ustilaginomycotina, called Peribolosporomycetes, and are a sister lineage to the existing classes of Ustilaginomycotina. Further, we describe the four Peribolospora isolates of our study as two different species (P. baueri and P. kevripleyi) of the novel genus Peribolospora.
In further studies, detailed genome analysis assessing genome composition and structure, functional gene annotation, transcriptomic, and CAZyme analyses could provide insights into the physiological adaptations and the lifestyle of the Peribolospora species. With their heat resistance, growth at low pH values, and osmotolerant properties, the Peribolospora species represent a novel taxonomic lineage within the Basidiomycota. These capabilities could serve as an ecological advantage and be conducive for Peribolosporomycetes to successfully live in environments with pronounced physiological stresses. Several BLAST hits for its sequences against fungal databases putatively indicate a global distribution of species related to Peribolospora. Notably, all hits were from environmental samples taken from forest soils, partially in forest fire regions. This supports the idea of forest soils, exposed to severe temperature stresses, as potentially one of the natural habitats of Peribolosporomycetes and explains the heat resistance as an adaptation to an ecological niche.
Data availability
All data and material generated during this study are included in this manuscript, on its supplementary information, or publicly available on databases or microbial culture collections. The study, including sampling, research work as well as culture and data transfer was conducted in compliance with the Nagoya protocol.
References
Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics Int 11:36–42
Baleiras Couto CMM, Eijsma B, Hofstra H, Huis in't VJH, van der Vossen JM (1996) Evaluation of molecular typing techniques to assign genetic diversity among Saccharomyces cerevisiae strains. Appl Environ Microbiol 62(1). https://doi.org/10.1128/aem.62.1.41-46.1996
Bauer R, Mendgen K, Oberwinkler F (1995) Septal pore apparatus of the smut Ustacystis waldsteiniae. Mycologia 87(1):18–24
Bauer R, Oberwinkler F, Vánky K (1997) Ultrastructural markers and systematics in smut fungi and allied taxa. Can J Bot 75(8):1273–1314. https://doi.org/10.1139/b97-842
Bayne HG, Michener HD (1979) Heat resistance of Byssochlamys ascospores. Appl Environ Microbiol 3:449–453
Begerow D, Schäfer AM, Kellner R, Yurkov A, Kemler M, Oberwinkler F et al (2014) “11 Ustilaginomycotina” in systematics and evolution. The Mycota (a comprehensive treatise on fungi as experimental systems for basic and applied research), eds. McLaughlin D, Spatafora J (Springer Berlin Heidelberg), pp 295–329 https://doi.org/10.1007/978-3-642-55318-9_11
Berni E, Tranquillini R, Scaramuzza N, Brutti A, Bernini V (2017) Aspergilli with Neosartorya-type ascospores: heat resistance and effect of sugar concentration on growth and spoilage incidence in berry products. Int J Food Microbiol 258:81–88. https://doi.org/10.1016/j.ijfoodmicro.2017.07.008
Biango-Daniels MN, Snyder AB, Worobo RW, Hodge KT (2019) Fruit infected with Paecilomyces niveus: a source of spoilage inoculum and patulin in apple juice concentrate? Food Control 97:81–86. https://doi.org/10.1016/j.foodcont.2018.10.020
Bubnová M, Zemančíková J, Sychrová H (2014) Osmotolerant yeast species differ in basic physiological parameters and in tolerance of non-osmotic stresses. Yeast 31(8):309–321. https://doi.org/10.1002/yea.3024
Buijs VA, Groenewald JZ, Haridas S, LaButti KM, Lipzen A, Martin FM et al (2021) Enemy or ally: a genomic approach to elucidate the lifestyle of Phyllosticta citrichinaensis. bioRxiv. https://doi.org/10.1101/2021.11.27.470207
Buscardo E, Rodríguez-Echeverría S, Freitas H, de Angelis P, Pereira JS, Muller LAH (2015) Contrasting soil fungal communities in Mediterranean pine forests subjected to different wildfire frequencies. Fungal Divers 70(1):85–99. https://doi.org/10.1007/s13225-014-0294-5
Caldeira AT, Salvador C, Pinto F, Arteiro JM, Martins MR (2009) MSP-PCR and RAPD molecular biomarkers to characterize Amanita ponderosa mushrooms. Ann Microbiol 59(3):629–634. https://doi.org/10.1007/BF03175156
Chan PP, Lin BY, Mak AJ, Lowe TM (2021) tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res 49(16):9077–9096. https://doi.org/10.1093/nar/gkab688
Conner DE, Beuchat LR (1987) Heat resistance of ascospores of Neosartorya fischeri as affected by sporulation and heating medium. Int J Food Microbiol 4(4):303–312. https://doi.org/10.1016/0168-1605(87)90005-5
Conner DE, Beuchat LR, Chang CJ (1987) Age-related changes in ultrastructure and chemical composition associated with changes in heat resistance of Neosartorya fischeri ascospores. Trans Br Mycol Soc 89(4):539–550. https://doi.org/10.1016/S0007-1536(87)80088-8
Cubeta MA (1991) Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified Ribosomal RNA gene. Phytopathology 81(11):1395–1400. https://doi.org/10.1094/Phyto-81-1395
Day NJ, Cumming SG, Dunfield KE, Johnstone JF, Mack MC, Reid KA, Turetsky MR, Walker XJ, Baltzer JL (2020) Identifying functional impacts of heat-resistant fungi on boreal forest recovery after wildfire. Front For Glob Change 9(3):68. https://doi.org/10.3389/ffgc.2020.00068
de Beer ZW, Begerow D, Bauer R, Pegg GS, Crous PW, Wingfield MJ (2006) Phylogeny of the Quambalariaceae fam. nov., including important Eucalyptus pathogens in South Africa and Australia. Stud Mycol 55:289–298. https://doi.org/10.3114/sim.55.1.289
Dijksterhuis J, Teunissen PG (2004) Dormant ascospores of Talaromyces macrosporus are activated to germinate after treatment with ultra high pressure. J Appl Microbiol 96(1):162–169. https://doi.org/10.1046/j.1365-2672.2003.02133.x
Dijksterhuis J (2007) Heat resistant ascospores. In: Mycology series. CRC Press, pp 115–132
Dos Santos JL, Samapundo S, Biyikli A, Van Impe J, Akkermans S, Höfte M, Abatih EN, Sant’Ana AS, Devlieghere F (2018) Occurrence, distribution and contamination levels of heat resistant moulds throughout the processing of pasteurized high-acid fruit products. Int J Food Microbiol 281:72–81. https://doi.org/10.1016/j.ijfoodmicro.2018.05.019
Emms DM, Kelly S (2019) OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol 20(1):1–14. https://doi.org/10.1186/s13059-019-1832-y
Engel G, Teuber M (1991) Heat resistance of ascospores of Byssochlamys nivea in milk and cream. Int J Food Microbiol 12(2–3):225–233. https://doi.org/10.1016/0168-1605(91)90073-X
Fulton TM, Chunwongse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Rep 13:207–209. https://doi.org/10.1007/BF02670897
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidioales — application to the identification of mycorrhizae and rusts. Mol Ecol 2(2):113–118. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x
Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T (2014) MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42(1):699–704
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinform 29(8):1072–1075. https://doi.org/10.1093/bioinformatics/btt086
Hall TA (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hibbett DS (2006) A phylogenetic overview of the Agaricomycotina. Mycologia 98(6):917–925. https://doi.org/10.3852/mycologia.98.6.917
Hind-Lanoiselet TL, Lanoiselet VM, Lewington FK, Ash GJ, Murray GM (2005) Survival of Sclerotinia sclerotia under fire. Austral Plant Pathol 34(3):311–317. https://doi.org/10.1071/AP05048
Hocking AD, Pitt JI (1980) Dichloran-glycerol medium for enumeration of xerophilic fungi from low-moisture foods. Appl Environ Microbiol 39(3):488–492. https://doi.org/10.1128/AEM.39.3.488-492.1980
Houbraken J, Samson RA (2006) Standardization of methods for detecting heat resistant fungi. In: Hocking AD, Pitt JI, Samson RA, Thrane U (Eds) Adv Food Mycol. Adv Exp Med Biol. Springer, pp 107–111. https://doi.org/10.1007/0-387-28391-9_5
Jančič S, Nguyen HDT, Frisvad JC, Zalar P, Schroers HJ, Seifert KA et al (2015) A taxonomic revision of the Wallemia sebi species complex. PLoS ONE 10(5):e0125933. https://doi.org/10.1371/journal.pone.0125933
Jaouani A, Neifar M, Prigione V, Ayari A, Sbissi I, Ben Amor S et al (2014) Diversity and enzymatic profiling of halotolerant micromycetes from Sebkha El Melah, a Saharan salt flat in southern Tunisia. BioMed Res Int 1–11. https://doi.org/10.1155/2014/439197
Jesenská Z, Piecková E, Bernát D (1993) Heat resistance of fungi from soil. Int J Food Microbiol 19(3):187–192. https://doi.org/10.1016/0168-1605(93)90076-S
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780. https://doi.org/10.1093/molbev/mst010
Kohler A, Murat C, Costa M (2011) High quality genomic DNA extraction using CTAB and Qiagen genomic-tip. Nat Inst Agric Res (INRA), France
Kotzekidou P (1997) Heat resistance of Byssochlamys nivea, Byssochlamys fulva and Neosartorya fischeri isolated from canned tomato paste. J Food Sci 62(2):410–412. https://doi.org/10.1111/j.1365-2621.1997.tb04014.x
Krassowski M, Arts M, Lagger C (2021) krassowski/complex-upset: V1.3.3. Zenodo. https://doi.org/10.5281/zenodo.3700590.
Kunčič MK, Kogej T, Drobne D, Gunde-Cimerman N (2010) Morphological response of the halophilic fungal genus Wallemia to high salinity. Appl Environ Microbiol 76(1):329–337. https://doi.org/10.1128/AEM.02318-09
Kunčič MK, Zajc J, Drobne D, Tkalec ŽP, Gunde-Cimerman N (2013) Morphological responses to high sugar concentrations differ from adaptation to high salt concentrations in the xerophilic fungi Wallemia spp. Fungal Biol 117(7–8):466–478. https://doi.org/10.1016/j.funbio.2013.04.003
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C et al (2004) Versatile and open software for comparing large genomes. Genome Biol 5(2):1–9. https://doi.org/10.1186/gb-2004-5-2-r12
Lieckfeldt E, Meyer W, Börner T (1993) Rapid identification and differentiation of yeasts by DNA and PCR fingerprinting. J Basic Microbiol 33(6):413–425. https://doi.org/10.1002/jobm.3620330609
Liu YJ, Whelen S, Hall BD (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol 16(12):1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
Lutz M, Vánky K, Bauer R (2012) Melanoxa, a new genus in the Urocystidales (Ustilaginomycotina). Mycol Prog 11(1):149–158. https://doi.org/10.1007/s11557-010-0737-7
Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM (2021) BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol 38(10):4647–4654. https://doi.org/10.1093/molbev/msab199
Matheny PB, Liu YJ, Ammirati JF, Hall BD (2002) Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am J Bot 89(4):688–698. https://doi.org/10.3732/ajb.89.4.688
Mishra B, Choi YJ, Thines M (2018) Phylogenomics of Bartheletia paradoxa reveals its basal position in Agaricomycotina and that the early evolutionary history of basidiomycetes was rapid and probably not strictly bifurcating. Mycol Prog 17:333–341. https://doi.org/10.1007/s11557-017-1349-2
Naranjo-Ortiz MA, Gabaldón T (2019) Fungal evolution: diversity, taxonomy and phylogeny of the fungi. Biol Rev Camb Philos Soc 94(6):2101–2137. https://doi.org/10.1111/brv.12550
Nasr S, Soudi MR, Nasrabadi SMZ, Nikou MM, Salmanian AH, Nguyen HDT (2014) Basidioascus persicus sp. nov., a yeast-like species of the order Geminibasidiales isolated from soil. Int J Syst Evol Microbiol 64(9):3046–3052. https://doi.org/10.1099/ijs.0.062380-0
Nguyen HDT, Nickerson NL, Seifert KA (2013) Basidioascus and Geminibasidium: a new lineage of heat resistant and xerotolerant basidiomycetes. Mycologia 105(5):1231–1250. https://doi.org/10.3852/12-351
Nguyen HDT, Tanney JB, Chabot D, Nickerson NL, Seifert KA (2014) Paratritirachium curvibasidium, a new heat resistant basidiomycete from flare pit soils in Alberta, Canada. Mycol Prog 13(3):575–587. https://doi.org/10.1007/s11557-013-0941-3
Nguyen HDT, Chabot D, Hirooka Y, Roberson RW, Seifert KA (2015) Basidioascus undulatus: genome, origins, and sexuality. IMA Fungus 6(1):215–231. https://doi.org/10.5598/imafungus.2015.06.01.14
Peintner U, Kuhnert-Finkernagel R, Wille V, Biasioli F, Shiryaev A, Perini C (2019) How to resolve cryptic species of polypores: an example in Fomes. IMA Fungus 10(1):1–21. https://doi.org/10.1186/s43008-019-0016-4
Peterson SW, Jurjević Ž (2013) Talaromyces columbinus sp. nov., and genealogical concordance analysis in Talaromyces clade 2a. PLoS ONE 8(10):e78084. https://doi.org/10.1371/journal.pone.0078084
Piecková E, Lehotská R, Globanová M (2020) Heat resistant fungi, toxicity and their management by nanotechnologies. In: Nanomycotoxicology. Academic Press, pp 217–237
Pitt J (1975) Xerophilic fungi and the spoilage of foods of plant origin. In: Duckworth RB (Ed) Water relations of foods. Academic Press, London, pp 273–308
Puel O, Tadrist S, Galtier P, Oswald IP, Delaforge M (2005) Byssochlamys nivea as a source of mycophenolic acid. Appl Environ Microbiol 71(1):550–553. https://doi.org/10.1128/aem.71.1.550-553.2005
R Core Team (2021) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
Rachon G, Raleigh CP, Pawlowsky K (2021) Heat resistance of yeast ascospores and their utilisation for the validation of pasteurisation processes for beers. J Inst Brew 127(2):149–159. https://doi.org/10.1002/jib.646
Ramírez-Castrillón M, Mendes SDC, Inostroza-Ponta M, Valente P (2014) (GTG)5 MSP-PCR fingerprinting as a technique for discrimination of wine associated yeasts? PLoS ONE 9(8):e.105870. https://doi.org/10.1371/journal.pone.0105870
Salomão BD, Muller C, Amparo HC, Aragão GM (2014) Survey of molds, yeast and Alicyclobacillus spp. from a concentrated apple juice productive process. Braz J Microbiol 45:49–58. https://doi.org/10.1590/S1517-83822014000100008
Samson RA, Hoekstra ES, Lund F, Filtenborg O, Frisvad JC (2004) Methods for the detection, isolation and characterisation of food-borne fungi. In: Samson RA, Hoekstra ES, Frisvad JC, Filtenborg O (Eds) Introduction to food- and airborne fungi, 7 edn. Centraalbureau voor Schimmelcultures, pp 283–297
Scaramuzza N, Berni E (2014) Heat resistance of Hamigera avellanea and Thermoascus crustaceus isolated from pasteurized acid products. Int J Food Microbiol 168:63–68. https://doi.org/10.1016/j.ijfoodmicro.2013.10.007
Scheuer C, Bauer R, Lutz M, Stabentheiner E, Grube M (2008) Bartheletia paradoxa is a living fossil on Ginkgo leaf litter with a unique septal structure in the Basidiomycota. Mycol Res 112(11):1265–1279. https://doi.org/10.1016/j.mycres.2008.06.008
Schmitt I, Crespo A, Divakar PK, Fankhauser JD, Herman-Sackett E, Kalb K et al (2009) New primers for promising single-copy genes in fungal phylogenetics and systematics. Pers: Mol Phylogeny Evol Fungi 23:35–40. https://doi.org/10.3767/003158509X470602
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA et al (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A 109(16):6241–6246. https://doi.org/10.1073/pnas.1117018109
Seifert KA, Nickerson NL, Corlett M, Jackson ED, Louis-Seize G, Davies RJ (2004) Devriesia, a new hyphomycete genus to accommodate heat resistant, cladosporium-like fungi. Can J Bot 82(7):914–926. https://doi.org/10.1139/b04-070
Snyder AB, Churey JJ, Worobo RW (2019) Association of fungal genera from spoiled processed foods with physicochemical food properties and processing conditions. Food Microbiol 83:211–218. https://doi.org/10.1016/j.fm.2019.05.012
Spatafora JW, Aime MC, Grigoriev IV, Martin F, Stajich JE, Blackwell M (2017) The fungal tree of life: from molecular systematics to genome-scale phylogenies. Microbiol Spectr, pp 1–34. https://doi.org/10.1128/microbiolspec.FUNK-0053-2016
Stamatakis A (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinform 30(9):1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Stielow JB, Lévesque CA, Seifert KA, Meyer W, Irinyi L, Smits D et al (2015) One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Pers: Mol Phylogeny Evol Fungi 35(1):242–263. https://doi.org/10.3767/003158515x689135
Stiller JW, Hall BD (1997) The origin of red algae: Implications for plastid evolution. Proc Natl Acad Sci U S A 94(9):4520–4525. https://doi.org/10.1073/pnas.94.9.4520
Suh SO, Gujjari P, Beres C, Beck B, Zhou J (2013) Proposal of Zygosaccharomyces parabailii sp. nov. and Zygosaccharomyces pseudobailii sp. nov., novel species closely related to Zygosaccharomyces bailii. Int J Syst Evol Microbiol 63(5):1922–1929. https://doi.org/10.1099/ijs.0.048058-0
Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R et al (2014) Fungal biogeography Global diversity and geography of soil fungi. Science 346(6213):1256688. https://doi.org/10.1126/science.1256688
Teufel F, Almagro Armenteros JJ, Johansen AR, Gíslason MH, Pihl SI, Tsirigos KD et al (2022) SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol 1–3. https://doi.org/10.1038/s41587-021-01156-3
Tournas V (1994) Heat resistant fungi of importance to the food and beverage industry. Crit Rev Microbiol 20(4):243–263. https://doi.org/10.3109/10408419409113558
Tresner HD, Hayes J (1971) Sodium chloride tolerance of terrestrial fungi. Appl Microbiol 22(2):210–213
Ulusoy BH, Hamed NS, Yıldırım FK (2022) Heat-resistant moulds: Assessment, prevention and their consequences for food safety and public health. Czech J Food Sci 40(4):273–280. https://doi.org/10.17221/26/2022-CJFS
Van Driel KG, Humbel BM, Verkleij AJ, Stalpers J, Müller WH, Boekhout T (2009) Septal pore complex morphology in the Agaricomycotina (Basidiomycota) with emphasis on the Cantharellales and Hymenochaetales. Mycol Res 113(5):559–576. https://doi.org/10.1016/j.mycres.2008.12.007
Větrovský T, Morais D, Kohout P, Lepinay C, Algora C, Awokunle Hollá S et al (2020) GlobalFungi, a global database of fungal occurrences from high-throughput-sequencing metabarcoding studies. Sci Data 7:1–14. https://doi.org/10.1038/s41597-020-0567-7
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172(8):4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
Vuyst L, Camu N, Winter T, Vandemeulebroecke K, van de Perre V, Vancanneyt M et al (2008) Validation of the (GTG)5-rep-PCR fingerprinting technique for rapid classification and identification of acetic acid bacteria, with a focus on isolates from Ghanaian fermented cocoa beans. Int J Food Microbiol 125:79–90. https://doi.org/10.1016/j.ijfoodmicro.2007.02.030
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direkt sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: A guide to methods and applications, vol 18(1). Academic Press, pp 315–322. https://doi.org/10.1016/b978-0-12-372180-8.50042-1
Witfeld F, Begerow D, Guerreiro MA (2021) Improved strategies to efficiently isolate thermophilic, thermotolerant, and heat resistant fungi from compost and soil. Mycol Prog 20(3):325–339. https://doi.org/10.1007/s11557-021-01674-z
Wyatt TT, Wösten HAB, Dijksterhuis J (2013) Fungal spores for dispersion in space and time. Adv Appl Microbiol 85:43–91. https://doi.org/10.1016/b978-0-12-407672-3.00002-2
Yaguchi T, Imanishi Y, Matsuzawa T, Hosoya K, Hitomi J, Nakayama M (2012) Method for identifying heat-resistant fungi of the genus Neosartorya. J Food Prot 75(10):1806–1813. https://doi.org/10.4315/0362-028X.JFP-12-060
Yang J, Li J, Jiang Y, Duan X, Qu H, Yang B et al (2014) Natural occurrence, analysis, and prevention of mycotoxins in fruits and their processed products. Crit Rev Food Sci Nutr 54(1):64–83. https://doi.org/10.1080/10408398.2011.569860
Zajc J, Gunde-Cimerman N (2018) The Genus Wallemia - From contamination of food to health threat. Microorganisms 6(2):46. https://doi.org/10.3390/microorganisms6020046
Zalar P, Sybren HG, Schroers HJ, Frank JM, Gunde-Cimerman N (2005) Taxonomy and phylogeny of the xerophilic genus Wallemia (Wallemiomycetes and Wallemiales, cl. et ord. nov.). Antonie van Leeuwenhoek 87(4):311–328. https://doi.org/10.1007/s10482-004-6783-x
Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z et al (2018) dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46(1):95–101. https://doi.org/10.1093/nar/gky418
Acknowledgements
We thank Nancy Nickerson for isolating the strains, Kasia Dadej for performing the genome sequencing of the four Peribolospora strains, and Ekaterina Ponomareva and Rafik Assabgui for assistance with culture growth for deposition into the Canadian Collection of Fungal Cultures (DAOMC). We are grateful to Jean Sindern and Frauke Küster for help with electron microscopy and cultivation experiments and thank Tanja Rollnik and Sabine Adler for support in electron microscopy. Many thanks to Dr. Christian Schulze for philologic consulting in regard to genus name creation. We acknowledge support by the Open Access Publication Funds of the Ruhr-Universität Bochum. We thank the two anonymous reviewers for the constructive comments, which helped us improve the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. FW was partially funded by Stiftung Mercator (MERCUR: Pr 2017-0020), DFG (BE 2201/16-1) and the Ruhr-University Bochum. MAG was partly funded by the German Research Foundation (DFG PE 1673/6-1; GU 2252/1-1). QMW was funded by the National Natural Science Foundation of China (NSFC, No. 31961133020).
Author information
Authors and Affiliations
Contributions
DB, HN, and FW designed the study. FW conducted the molecular work, light microscopy, cultivation experiments, and micromorphological drawings. FW conducted the scanning electron microscopy together with undergraduate students, and the transmission electron microscopy, and supported by technicians. MAG and FW analyzed the genomic data. FN prepared the samples for transmission electron microscopy. QMW provided the genome of Robbauera albescens. FW wrote the draft manuscript and edited it. All authors read, reviewed, and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Meike Piepenbring
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Witfeld, F., Guerreiro, M.A., Nitsche, F. et al. Peribolosporomycetes class. nov.: description of a new heat resistant and osmotolerant basidiomycete lineage, represented by Peribolospora gen. nov., P. kevripleyi sp. nov., and P. baueri sp. nov.. Mycol Progress 22, 30 (2023). https://doi.org/10.1007/s11557-023-01879-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11557-023-01879-4