Abstract
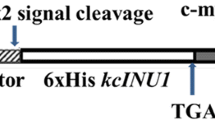
Chaetomium globosum can inhibit the growth of fusarium by means of their extracellular proteins. Two novel β-glucanases, designated Cgglu17A and Cgglu16B, were separated from the supernatant of C. globosum W7 and verified to have the ability to hydrolyze cell walls of Fusarium sporotrichioides MLS-19. Cgglu17A (397 amino acids) was classified as glycoside hydrolase family 17 while Cgglu16B belongs to the family16 (284 amino acids). Recombinant protein Cgglu17A was successfully expressed in Escherichia coli, and the enzymes were purified by affinity chromatography. Maximum activity of Cgglu17A appeared at the pH 5.5 and temperature 50 °C, but Cgglu16B shows the maximum activity at the pH 5.0 and temperature 50 °C. Most of heavy metal ions had inhibition effect on the two enzymes, but Cgglu17A and Cgglu16B were respectively activated by Ba2+ and Mn2+. Cgglu17A exhibited high substrate specificity, almost only catalyzing the cleavage of β-1,3-glycosidic bond, in various polysaccharose, to liberate glucose. However, Cgglu16B showed high catalytic activities to both β-1,3-glycosidic and β-1,3-1,4-glycosidic bonds. Cgglu17A was an exo-glucanase, but Cgglu16B was an endo-glucanase based on hydrolytic properties assay. Both of two enzymes showed potential antifungal activity, and the synergistic effect was observed in the germination experiment of pathogenic fungus. In conclusion, Cgglu17A (exo-1,3-β-glucanase) and Cgglu16B (endo-1,3(4)-β-glucanase) were confirmed to play a key role in the process of C. globosum controlling fusarium and have potential application value on industry and agriculture for the first time.






Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this article (and its supplementary information files).
References
Apiraksakorn, J., Nitisinprasert, S., & Levin, R. E. (2008). Grass degrading beta-1,3-1,4-D-glucanases from Bacillus subtilis GN156: purification and characterization of glucanase J1 and pJ2 possessing extremely acidic pI. Applied Biochemistry and Biotechnology, 149, 53–66.
Bailey, M. J., Biely, P., & Poutanen, K. (1992). Interlaboratory testing of methods for assay of xylanase activity. Journal of Biotechnology, 23, 257–270.
Bara, M. T., Lima, A. L., & Ulhoa, C. J. (2003). Purification and characterization of an exo-beta-1,3-glucanase produced by Trichoderma asperellum. FEMS Microbiology Letters, 219, 81–85.
Barran, L. R., Schneider, E. F., Wood, P. J., Madhosingh, C., & Miller, R. W. (1975). Cell wall of Fusarium sulphureum. Biochimica et Biophysica Acta, 392, 148–158.
Chen, H., Li, X. L., & Ljungdahl, L. G. (1997). Sequencing of a 1,3-1,4-beta-D-glucanase (lichenase) from the anaerobic fungus Orpinomyces strain PC-2: properties of the enzyme expressed in Escherichia coli and evidence that the gene has a bacterial origin. Journal of Bacteriology, 179, 6028–6034.
Chen, X., Meng, K., Shi, P., Bai, Y., Luo, H., Huang, H., Yuan, T., Yang, P., & Yao, B. (2012). High-level expression of a novel Penicillium endo-1,3(4)-beta-D-glucanase with high specific activity in Pichia pastoris. Journal of Industrial Microbiology & Biotechnology, 39, 869–876.
Genta, F. A., Bragatto, I., Terra, W. R., & Ferreira, C. (2009). Purification, characterization and sequencing of the major β-1,3-glucanase from the midgut of Tenebrio molitor larvae. Insect Biochemistry and Molecular Biology, 39, 861–874.
Guo, Y., Yan, Q., Yang, Y., Yang, S., Liu, Y., & Jiang, Z. (2015). Expression and characterization of a novel beta-glucosidase, with transglycosylation and exo-beta-1,3-glucanase activities, from Rhizomucor miehei. Food Chemistry, 175, 431–438.
Hong, M. R., Kim, Y. S., Joo, A. R., Lee, J. K., Kim, Y. S., & Oh, D. K. (2009). Purification and characterization of a thermostable beta-1,3-1,4-glucanase from Laetiporus sulphureus var. miniatus. Journal of Microbiology and Biotechnology, 19, 818–822.
Hua, C., Yi, H., & Jiao, L. (2011). Cloning and expression of the endo-1,3(4)-beta-glucanase gene from Paecilomyces sp. FLH30 and characterization of the recombinant enzyme. Bioscience, Biotechnology, and Biochemistry, 75, 1807–1812.
Jiang, C., Song, J., Cong, H., Zhang, J., & Yang, Q. (2017). Expression and characterization of a novel antifungal exo-beta-1,3-glucanase from Chaetomium cupreum. Applied Biochemistry and Biotechnology, 182, 261–275.
Li, J., Xu, X., Shi, P., Liu, B., Zhang, Y., & Zhang, W. (2017). Overexpression and characterization of a novel endo-beta-1,3(4)-glucanase from thermophilic fungus Humicola insolens Y1. Protein Expression and Purification, 138, 63–68.
Luo, H., Yang, J., Yang, P., Li, J., Huang, H., Shi, P., Bai, Y., Wang, Y., Fan, Y., & Yao, B. (2010). Gene cloning and expression of a new acidic family 7 endo-beta-1,3-1,4-glucanase from the acidophilic fungus Bispora sp. MEY-1. Applied Microbiology and Biotechnology, 85, 1015–1023.
Monteiro, V. N., & Ulhoa, C. J. (2006). Biochemical characterization of a beta-1,3-glucanase from Trichoderma koningii induced by cell wall of Rhizoctonia solani. Current Microbiology, 52, 92–96.
Murray, P. G., Grassick, A., Laffey, C. D., Cuffe, M. M., Higgins, T., Savage, A. V., Planas, A., & Tuohy, M. G. (2001). Isolation and characterization of a thermostable endo-beta-glucanase active on 1,3-1,4-beta-D-glucans from the aerobic fungus Talaromyces emersonii CBS 814.70. Enzyme and Microbial Technology, 29, 90–98.
Ogunyewo, O. A., Okereke, O. E., Kumar, S., & Yazdani, S. S. (2022). Characterization of a GH5 endoxylanase from Penicillium funiculosum and its synergism with GH16 endo-1,3(4)-glucanase in saccharification of sugarcane bagasse. Scientific Reports, 12, 17219.
Ooi, T., Sato, H., Matsumoto, K., & Taguchi, S. (2009). A unique post-translational processing of an exo-beta-1,3-glucanase of Penicillium sp. KH10 expressed in Aspergillus oryzae. Protein Expression and Purification, 67, 126–131.
Otsuka, Y., Sato, K., Yano, S., Kanno, H., Suyotha, W., Konno, H., Makabe, K., & Taira, T. (2022). GH-16 type beta-1,3-glucanase from Lysobacter sp. MK9-1 enhances antifungal activity of GH-19 type chitinase, and its glucan-binding domain binds to fungal cell-wall. Journal of Applied Glycoscience, 69, 49–56.
Peng, Y., Liu, G. L., Yu, X. J., Wang, X. H., Jing, L., & Chi, Z. M. (2011). Cloning of exo-beta-1,3-glucanase gene from a marine yeast Williopsis saturnus and its overexpression in Yarrowia lipolytica. Marine Biotechnology, 13, 193–204.
Ruel, K., & Joseleau, J. P. (1991). Involvement of an extracellular glucan sheath during degradation of populus wood by Phanerochaete chrysosporium. Applied and Environmental Microbiology, 57, 374–384.
Sakamoto, Y., Nakade, K., & Konno, N. (2011). Endo-beta-1,3-glucanase GLU1, from the fruiting body of Lentinula edodes, belongs to a new glycoside hydrolase family. Applied and Environmental Microbiology, 77, 8350–8354.
Sietsma, J. H., & Wessels, J. G. H. (1981). Solubility of (1→3)-β-d/(1→6)-β-d-glucan in fungal walls: importance of presumed linkage between glucan and chitin. Journal of General Microbiology, 125, 209–212.
Takahashi, M., Konishi, T., & Takeda, T. (2011). Biochemical characterization of Magnaporthe oryzae β-glucosidases for efficient β-glucan hydrolysis. Applied Microbiology & Biotechnology, 91, 1073–1082.
Takeda, T., Nakano, Y., Takahashi, M., Konno, N., Sakamoto, Y., Arashida, R., Marukawa, Y., Yoshida, E., Ishikawa, T., & Suzuki, K. (2015). Identification and enzymatic characterization of an endo-1,3-beta-glucanase from Euglena gracilis. Phytochemistry, 116, 21–27.
Takeuchi, K. (1983). Purification and characterization of exo-beta-1,3-glucanase from a hatching supernatant of Strongylocentrotus intermedius. Canadian Journal of Biochemistry and Cell Biology = Revue Canadienne de Biochimie et Biologie Cellulaire, 61, 54–62.
Tang, Y., Yang, S., Yan, Q., Zhou, P., Cui, J., & Jiang, Z. (2012). Purification and characterization of a novel beta-1,3-1,4-glucanase (lichenase) from thermophilic Rhizomucor miehei with high specific activity and its gene sequence. Journal of Agricultural and Food Chemistry, 60, 2354–2361.
Vijayendra, S. V., & Kashiwagi, Y. (2009). Characterization of a new acid stable exo-beta-1,3-glucanase of Rhizoctonia solani and its action on microbial polysaccharides. International Journal of Biological Macromolecules, 44, 92–97.
Wang, J., Kang, L., Liu, Z., & Yuan, S. (2017). Gene cloning, heterologous expression and characterization of a Coprinopsis cinerea endo-beta-1,3(4)-glucanase. Fungal Biology, 121, 61–68.
Wang, Y. J., & Yang, Q. (2009). Cloning and expression of a novel chitinase chi58 from Chaetomium cupreum in Pichia pastoris. Biochemical Genetics, 47, 547–558.
Wessels, J., & Sietsma, J. H. (1981). Fungal Cell Walls: A Survey. Plant Carbohydrates, II, 352–394.
Xie, Y. R., Raruang, Y., Chen, Z. Y., Brown, R. L., & Cleveland, T. E. (2015). ZmGns, a maize class I beta-1,3-glucanase, is induced by biotic stresses and possesses strong antimicrobial activity. Journal of Integrative Plant Biology, 57, 271–283.
Yan, Q., Yang, H., Jiang, Z., Liu, E., & Yang, S. (2018). A novel thermostable beta-1,3-1,4-glucanase from Thermoascus aurantiacus and its application in oligosaccharide production from oat bran. Carbohydrate Research, 469, 31–37.
Yang, S., Xiong, H., Yan, Q., Yang, H., & Jiang, Z. (2014). Purification and characterization of a novel alkaline beta-1,3-1,4-glucanase (lichenase) from thermophilic fungus Malbranchea cinnamomea. Journal of Industrial Microbiology & Biotechnology, 41, 1487–1495.
Zhang, H., & Li, M. (2012). Transcriptional profiling of ESTs from the biocontrol fungus Chaetomium cupreum. Scientific World Journal, 2012, 1–7.
Acknowledgments
The authors are thankful to the authorities of Huainan Normal University to provide scientific research platform. The authors are thankful to the Microbial Genetic Engineering Lab of Harbin Institute of Technology to provide microorganism Chaetomium globosum strainW7 and Fusarium sporotrichioides MLS-19.
Funding
The authors wish to thank the Chinese government for the financial support of this study under “Doctoral Research Foundation of Huainan Normal University” (bsqdj2018-JiangCheng) and “Natural Science Foundation of Anhui Province” (2108085MC84).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by CJ, GM, JL, ZZ, JL, SZ, JZ, and XZ. The first draft of the manuscript was written by CJ, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
All the authors agreed to participate in the preparation and submission of the manuscript.
Consent for Publication
All the authors agree to publish the data.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 1380 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang, ., Miao, G., Li, J. et al. Identification and Characterization of Two Novel Extracellular β-Glucanases from Chaetomium globosum against Fusarium sporotrichioides. Appl Biochem Biotechnol (2023). https://doi.org/10.1007/s12010-023-04698-1
Accepted:
Published:
DOI: https://doi.org/10.1007/s12010-023-04698-1