Abstract
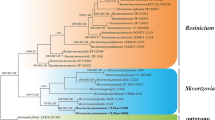
An endophytic fungus designated as EIT4T (MCC 9756T) was isolated from the asymptomatic stem tissue of Ephedra gerardiana collected from the Kargil district of Ladakh Union territory, India. Phylogenetic analysis based on concatenated nuclear ribosomal ITS (internal transcribed spacer) and LSU (large ribosomal subunit) sequence datasets revealed its placement within the genus Astragalicola. However, it formed a separate clade exhibiting strong bootstrap support value (80%). The highest nrITS sequence similarity between EIT4T and species of Astragalicola was 95.19% (A. vasilyevae) and 94.26% (A. amorpha), while nrLSU sequence similarity was 99.27% (A. amorpha). Morphologically, EIT4T differs from the other species of Astragalicola in having larger sub-globose to pyriform conidiomata, smaller and mostly unbranched conidiophores, and polymorphic translucent conidia with two terminal guttules. Based on combined cultural, micromorphological, molecular, and phylogenetic analyses, EIT4T represents a novel species in the genus Astragalicola proposed here as Astragalicola ephedrae sp. nov. Detailed description and illustrations of the novel species are provided. The type strain is EIT4T (= MCC 9756 T = MN29T).



Similar content being viewed by others
Availability of data and material
The nrITS and nrLSU sequence data was saved as FASTQ file and deposited in GenBank, National Center for Biotechnology Information (NCBI) under accession number MW205783 and OP476683, respectively. Holotype/ex-type (MCC 9756) culture is deposited in the culture collection of the National Center for Microbial Resource/National Center for Cell Science, Pune (Maharashtra), India.
Code availability
Not Applicable.
Change history
13 March 2023
A Correction to this paper has been published: https://doi.org/10.1007/s12223-023-01045-z
References
Adeleke BS, Babalola OO (2021) Pharmacological potential of fungal endophytes associated with medicinal plants: a review. J Fungi 7(2):147. https://doi.org/10.3390/jof7020147
Bacon CW, White Jr JF (2000) Physiological adaptations in the evolution of endophytism in the Clavicipitaceae. In: Microbial endophytes. CRC Press, pp. 251–276
Barr ME (1987) Prodromus to class Loculoascomycetes. University of Massachusetts, Amherst, Massachusetts
Bilański P, Grad B, Kowalski T (2022) Pyrenochaeta fraxinina as colonizer of ash and sycamore petioles, its morphology, ecology, and phylogenetic connections. Mycol Prog 21(9):1–22. https://doi.org/10.1007/s11557-022-01827-8
Burragoni SG, Jeon J (2021) Applications of endophytic microbes in agriculture, biotechnology, medicine, and beyond. Microbiol Res 245:126691. https://doi.org/10.1016/j.micres.2020.126691
de Gruyter J, Aveskamp MM, Woudenberg JH, Verkley GJ, Groenewald JZ, Crous PW (2009) Molecular phylogeny of Phoma and allied anamorph genera: towards a reclassification of the Phoma complex. Mycol Res 113(4): 508–519. https://doi.org/10.1016/jmycres200901002
de Gruyter J, Woudenberg JH, Aveskamp MM, Verkley GJ, Groenewald JZ, Crous PW (2010) Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma. Mycologia 102(5):1066–108. https://doi.org/10.3852/09-240
Doilom M, Liu JK, Jaklitsch WM, Ariyawansa H, Wijayawardene NN, Chukeatirote E, Zhang M, McKenzie EH, Geml J, Voglmayr H, Hyde KD (2013) An outline of the family Cucurbitariaceae. Sydowia 65(1):167–192
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hereme R, Morales-Navarro S, Ballesteros G, Barrera A, Ramos P, Gundel PE, Molina-Montenegro MA (2020) Fungal endophytes exert positive effects on Colobanthus quitensis under water stress but neutral under a projected climate change scenario in Antarctica. Front Microbiol 11:264. https://doi.org/10.3389/fmicb.2020.00264
Jia M, Chen L, Xin HL, Zheng CJ, Rahman K, Han T, Qin LP (2016) A friendly relationship between endophytic fungi and medicinal plants: a systematic review. Front Microbiol 7:906. https://doi.org/10.3389/fmicb.2016.00906
Jaklitsch WM, Checa J, Blanco MN, Olariaga I, Tello S, Voglmayr H (2018) A preliminary account of the Cucurbitariaceae. Stud Mycol 90:71–118. https://doi.org/10.1016/jsimyco201711002
Jaklitsch WM, Voglmayr H (2020) Fenestelloid clades of the Cucurbitariaceae. Persoonia 44:1–40. https://doi.org/10.3767/persoonia20204401
Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform (4):1160–1166. https://doi.org/10.1093/bib/bbx108
Khare E, Mishra J, Arora NK (2018) Multifaceted interactions between endophytes and plant: developments and prospects. Front Microbiol 9:2732. https://doi.org/10.3389/fmicb.2018.02732
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120. https://doi.org/10.1007/BF01731581
Mattoo AJ, Nonzom S (2022a) Parathyridaria ephedrae sp. nov. (Thyridariacaeae, Pleosporales), endophytic to Ephedra gerardiana in India. Phytotaxa 556(2):149–159. https://doi.org/10.11646/PHYTOTAXA55624
Mattoo AJ, Nonzom S (2022b) Investigating diverse methods for inducing sporulation in endophytic fungi. Studies in Fungi 7(16):1–10. https://doi.org/10.48130/SIF-2022b-0016
Nissan S, Boelens J, Lagrou K, Roels D (2022) Pyrenocheata unguis-hominis: a new cause of fungal keratitis in a contact lens wearer. Am J Ophthalmol 28:101731. https://doi.org/10.1016/j.ajoc.2022.101731
Petrini O (1986) Taxonomy of endophytic fungi of aerial plant tissues. In: Van den Heuvel J (ed) Fokkema NJ. Microbiology of the Phyllosphere, Cambridge University Press, pp 175–187
Sadrati, N (2021) Isolement, identification et culture des champignons endophytes isolés à partir des plantes médicinales algériennes pour la production des métabolites secondaires biologiquement actifs (Doctoral dissertation).
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Sharma P, Kumar S (2021) Bioremediation of heavy metals from industrial effluents by endophytes and their metabolic activity: Recent advances. Bioresour Technol 339:125589. https://doi.org/10.1016/j.biortech.2021.125589
Su W, Xu R, Bhunjun CS, Tian S, Dai Y, Li Y, Phukhamsakda C (2022) Diversity of Ascomycota in Jilin: Introducing Novel Woody Litter Taxa in Cucurbitariaceae. J Fungi 8(9):905. https://doi.org/10.3390/jof8090905
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38(7):3022–7. https://doi.org/10.1093/molbev/msab120
Ustiatik R, Nuraini Y, Suharjono S, Jeyakumar P, Anderson CW, Handayanto E (2021) Mercury resistance and plant growth promoting traits of endophytic bacteria isolated from mercury-contaminated soil. Bioremediat J 26(3):208–227. https://doi.org/10.1080/10889868.2021.1973950
Valenzuela-Lopez N, Cano-Lira JF, Guarro J, Sutton DA, Wiederhold N, Crous PW, Stchigel AM (2018) Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Stud Mycol 90(1):1–69. https://doi.org/10.1016/jsimyco201711003
Verkley GJ, Dukik K, Renfurm R, Göker M, Stielow JB (2014) Novel genera and species of coniothyrium-like fungi in Montagnulaceae (Ascomycota). Persoonia 32(1):25–251. https://doi.org/10.3767/003158514X679191
Verma H, Kumar D, Kumar V, Kumari M, Singh SK, Sharma VK, Droby S, Santoyo G, White JF, Kumar A (2021) The potential application of endophytes in management of stress from drought and salinity in crop plants. Microorganisms 9(8):1729. https://doi.org/10.3390/microorganisms9081729
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol Res 172: 4238–4246. https://doi.org/10.1128/jb17284238-42461990
Vu D, Groenewald M, De Vries M, Gehrmann T, Stielow B, Eberhardt U, Al-Hatmi A, Groenewald JZ, Cardinali G, Houbraken J, Boekhout T (2018) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud Mycol 91(1):23–36. https://doi.org/10.1016/jsimyco201805001
Wanasinghe DN, Phookamsak R, Jeewon R, Li WJ, Hyde KD, Jones EB, Camporesi E, Promputtha I (2017) A family level rDNA based phylogeny of Cucurbitariaceae and Fenestellaceae with descriptions of new Fenestella species and Neocucurbitaria gen. nov. Mycosphere 8:397–414. https://doi.org/10.5943/mycosphere/8/4/2
Wanasinghe DN, Phukhamsakda C, Hyde KD, Jeewon R, Lee HB, Gareth Jones EB, Tibpromma S, Tennakoon DS, Dissanayake AJ, Jayasiri SC, Gafforov Y (2018) Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers 89(1):1–236. https://doi.org/10.1007/s13225-018-0395-7
White TJ, Bruns T, Lee SJ, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: MA, Gelfand DH, Sninsky JJ, J. White TJ (eds): PCR protocols: A Guide to Methods and Applications, Academic Press, pp. 315–322.
Wijayawardene NN, Hyde KD, Dai DQ, Sánchez-García M, Goto BT, Saxena RK, Erdoðdu M, Selçuk F, Rajeshkumar KC, Aptroot A, Błaszkowski J, Boonyuen N, da Silva GA, de Souza FA, Dong W, Ertz D, Haelewaters D, Jones EBG, Karunarathna SC, Kirk PM, Kukwa M, Kumla J, Leontyev DV, Lumbsch HT, Maharachchikumbura SSN, Marguno F, Martínez-Rodríguez P, Mešić A, Monteiro JS, Oehl F, Pawłowska, J, Pem D, Pfliegler WP, Phillips AJL, Pošta A, He MQ, Li JX, Raza M, Sruthi OP, Suetrong S, Suwannarach N, Tedersoo L, Thiyagaraja V, Tibpromma S, Tkalčec Z, Tokarev YS, Wanasinghe DN, Wijesundara DSA, Wimalaseana SDMK, Madrid H, Zhang GQ, Gao Y, Sánchez-Castro I, Tang LZ, Stadler M, Yurkov A, Thines M (2022) Outline of fungi and fungus-like taxa–2021. Mycosphere 13:53–453. https://doi.org/10.5943/mycosphere/13/1/2
Winter HG (1885) Pilze II Abtheilung Ascomyceten: Gymnoasceen und Pyrenomyceten In: Rabenhorst GL (ed) Rabenhorst’s Kryptogamen-Flora von Deutschland, Oesterreich und der Schweiz, 2nd ed Kummer, Leipzig, pp. 65–528. https://doi.org/10.5962/bhltitle1356
Xia Y, Liu J, Chen C, Mo X, Tan Q, He Y, Wang Z, Yin J, Zhou G (2022) The multifunctions and future prospects of endophytes and their metabolites in plant disease management. Microorganisms 10(5):1072. https://doi.org/10.3390/microorganisms10051072
Yan L, Zhu J, Zhao X, Shi J, Jiang C, Shao D (2019) Beneficial effects of endophytic fungi colonization on plants. Appl Microbiol Biotechnol 103(8):3327–3340. https://doi.org/10.1007/s00253-019-09713-2
Acknowledgements
The authors are highly thankful to the Head, Department of Botany and UGC SAP-DRS-II for providing lab facilities and the Council of Scientific and Industrial Research (CSIR) for granting a fellowship to the first author.
Funding
This work was supported by the Council of Scientific and Industrial Research (CSIR), Grant No. 09/100(0241)/2019-EMR-I.
Author information
Authors and Affiliations
Contributions
Study conception was equally conceived. Aroosa Jan Mattoo carried out data analysis (morphological and phylogenetic analysis) and wrote the manuscript. Skarma Nonzom analyzed the data and critically revised the work. Both the authors reviewed and approved the final version of the paper.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mattoo, A.J., Nonzom, S. Astragalicola ephedrae sp. nov., isolated from the stem of Ephedra gerardiana in Ladakh, India. Folia Microbiol 68, 607–615 (2023). https://doi.org/10.1007/s12223-023-01041-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-023-01041-3