Abstract
Trichoderma spp., a cosmopolitan fungal genus, has remarkable economic value in industry and agriculture. The resources of Trichoderma spp. in the grassland and forest ecosystems of northern Xinjiang were explored in this study. A total of 634 soil samples was collected, and 312 strains assigned to 23 species of Trichoderma spp. were identified. T. harzianum was the dominant species with 28.2% from all isolates. The principal components analysis indicated that ecosystem was the most dominant impact factor among longitude, latitude, altitude and ecosystems for the species diversities of Trichoderma spp. with the decreasing trend from the north to the south of northern Xinjiang (e.g., from Altay, followed by Yili, Changji, Bayingolin and finally Urumqi). Overall, Trichoderma spp. were more frequently encountered in forest ecosystems (coniferous forest and coniferous and broadleaf mixed forest) than in grassland ecosystems (desert steppe and temperate steppe). Frequency of Trichoderma spp. was significantly decreased along with increased altitude and only a few strains were isolated from altitudes above 3000 m. The results provided essential information on Trichoderma occurrence and distribution, which should benefit the application of Trichoderma in agriculture.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As a cosmopolitan fungal genus, Trichoderma (Ascomycetes, Hypocreales) spp. are found in different environments, such as soil, above-ground plants, fungal material, decaying wood, sediment, and various other substances (Cummings et al. 2016; Harman 2000; Jaklitsch 2009; Jaklitsch and Voglmayr 2015; Kubicek et al. 2008). Many studies have been published about promising applications of Trichoderma spp. in industry and agriculture due to their ability to produce enzymes and antibiotics, promote plant growth, induce plant resistance (Gajera et al. 2015; Mukherjee et al. 2013; Ortega-García et al. 2015; Vitti et al. 2016), and enhance the efficiency of nutrient use (Kashyap et al. 2017). In particular, Trichoderma strains are significant biocontrol agents against plant fungal pathogens(Bae et al. 2016; El-Hassan et al. 2013; Harman et al. 2004; Zhang et al. 2015a, b).
Recently, biodiversity of Trichoderma has been extensively investigated in different countries and regions, including Russia, Siberia, the Himalayas (Kullnig et al. 2000), a mid-European area (Wuczkowski et al. 2003), Egypt (Gherbawy et al. 2004), Tenerife (Zachow et al. 2009a, b), Colombia (Hoyos-Carvajal et al. 2009), Iran (Naeimi et al. 2011), South-East Asian (Kubicek et al. 2003), Manipur (Kamala et al. 2015), Southern Europe and Macaronesia (Jaklitsch and Voglmayr 2015), Malaysian Borneo (Cummings et al. 2016), and New Zealand (Braithwaite et al. 2017). These studies focused on the species diversity, phylogeny, geographic distribution, and habitat preference of Trichoderma as well as the effect of collection season and crop type on the distribution of Trichoderma spp. (Chaverri and Samuels 2013; Jiang et al. 2016; Kubicek et al. 2008). In China, species diversity and the distribution of Trichoderma have been investigated nationwide since the 1990s (Wen et al. 1993). Zhang et al. identified northern China as a potential center of origin of a unique haplotype of T. harzianum (Zhang et al. 2005a, b). Sun et al. further identified 23 Trichoderma spp. with significant ecological, biochemical, and genetic diversity in north, southwest, southeast, and middle China,and found that T. harzianum was the most widely distributed species in China, and the highest biodiversity of Trichoderma populations occurred in southwest China (Sun et al. 2012). In 2016, Jiang et al. surveyed Trichoderma biodiversity of 17 species in agricultural fields in four provinces of eastern China (Jiang et al. 2016).
The Xinjiang Uygur Autonomous Region in Northwestern China (73°E–96°E, 34°N–48°N) covers an area of 1.66 million square kilometers, and has rich natural resources because of its unique geographical and ecological environment, complex and diverse landforms and soils. Tianshan and Altai Mountains in northern Xinjiang are also covered with luxuriantly green primary forests and vast grasslands. The complex terrain and diverse ecological environment in this region provide capacity for Trichoderma spp. biodiversity. However, no system research have been conducted on the diversity of Trichoderma spp. In this study, we focused on the recovery of Trichoderma diversities in grassland (desert steppe and temperate steppe) and forest (coniferous forest and coniferous and broadleaf mixed forest) ecosystems of northern Xinjiang based on altitudes, longitude, latitude and ecosystems.
Methods
Study regions and sample collection
A total of 634 soil samples were collected in July 2014, 2015, and 2016 from grassland (desert steppe and temperate steppe) and forest (coniferous forest and coniferous and broadleaf mixed forest) ecosystems in Xinjiang Uygur Autonomous Region. This region consists of Urumqi Nanshan (hereinafter referred to as Urumqi), Changji Tianchi (Changji), Ili Kazak Autonomous Prefecture (Yili), Altay Prefecture (Altay), and Mongolian Autonomous Prefecture of Bayingolin (Bayingolin). Each sample contained about 200 g of mixed soil from all five locations covering about 400 m2, a depth of approximately 5−20 cm. The longitude, latitude, and altitude of the collection locations, vegetation families, and geographical coordinates were recorded and are shown in Table S1. Samples were placed into sterile polyethylene bags, transported to the laboratory, and stored at 4 °C.
Isolation and storage of Trichoderma strains
PDAm(Vargas Gil et al. 2009) and Rose Bengal Agar were used as selective media and Trichoderma strains were then isolated using the soil dilution plating method; colonial morphology was observed after 5–10 days. Trichoderma and non-Trichoderma colonies were observed. Putative Trichoderma colonies were purified by two rounds of subculture on potato-dextrose agar (PDA). All isolates described in this study were stored in liquid storage medium containing glycerol (final concentration 20%).
Identification of Trichoderma spp.
Species were identified with a combination of morphological analysis and molecular methods. The morphological characteristics were based on the key by Gams and Bissett(Gams 1998). Colony characteristics were examined in cultures grown on PDA, CMD, and SNA, after 10 days of incubation at 25 °C. Microscopic observations were performed in cultures grown on PDA. As recommended by Gazis (Gazis et al. 2011), molecular identification was first done based on sequences of internal transcribed spacer regions 1 and 2 (ITS1 and ITS2) of the rRNA gene cluster. In case of failure in unambiguous species identification with ITS1 and ITS2, we also sequenced a fragment of the translation elongation factor 1-alpha (tef1-α) gene and the RNA polymerase II subunit B (rpb2) gene for further identification. Mycelia for DNA extraction were obtained on PDA through 5–7 days of incubation at 28 °C in a mildew incubator. Total DNA was extracted using the Fungal DNA Kit (Aidlab Biotechnologies Co., Ltd).
The ITS region of the rDNA was amplified using primers ITS4 and ITS5(White et al. 1990). A fragment of the tef1 gene was amplified using the primers TEF1-728F(Druzhinina et al. 2004) and TEF1LLErev(Jaklitsch et al. 2005). A fragment of rpb2 was amplified using the primer pair RPB2-250 (forward) and RPB2-1150 (reverse). Amplifications were performed in either a PTC-200 or PTC-100 thermocycler (MJ Research, USA) under the following conditions: initial denaturation 3 min at 94 °C, 35 cycles of 1 min at 94 °C, 1 min at 49 °C (for the ITS region), or 55 °C (for the tef1-α and rpb2 fragment), 1 min at 72 °C, with a final extension of 10 min at 72 °C.
Sequence and phylogenetic analysis
ITS rDNA sequences of all isolates were submitted to the oligonucleotide barcode program TrichOKEY(Druzhinina et al. 2005), tef1 and rpb2 sequences were submitted to TrichoBLAST(Kopchinskiy et al. 2005) and blasted in NCBI (https://www.ncbi.nlm.nih.gov/). ITS rDNA sequences of all Trichoderma strains were used to obtain haplotypes in Dna.sp ver. 5.1(Librado and Rozas 2009). Sequences of known species including type strains and outgroup were downloaded from the NCBI database. Type haplotypes and known species were aligned using Clustal W(Larkin et al. 2007), and then rechecked and adjusted manually as necessary using MEGA6.0 software(Tamura et al. 2013). Phylogenetic relationships were reconstructed with MEGA6.0 using the maximum likelihood approach (Kimura 2-parameter model and gamma distributed, with complete deletion in gaps/missing data treatment). All reconstructions were tested with 1000 bootstrap replicates. All sequences were deposited in GenBank with the accession numbers given in Supplemental Table 1.
Diversity analysis
The degree of dominance index (Y) was used to quantitatively describe the adaptation of Trichoderma to all fungi in soil. The dominance values were calculated using the following formula:
where ‘N’ is the total count of fungal strains, ‘ni’ is the count of genus (species) i, and ‘fi’ is the frequency with which genus (species) i appears in the samples. The genus or species i is dominant when Y > 0.02.
Simpson biodiversity index (Dr)(Simpson 1949), Shannon–Weiner biodiversity index (H)(Shannon 1948), Pielou species evenness index (J)(Pielou 1966) and Margalef’s abundance index (E)(Margalef 1958), were used to quantitatively describe the diversity of Trichoderma species in different environments and regions. Margalef’s abundance index was used to represent richness, Simpson index was applied as a measure of the probability of diversity, the Shannon–Wiener index was followed to measure community diversity, and the Pielou index was used to measure evenness of the community. The calculation formulas of the biological diversity indexes are as follows(Gomes et al. 2018; Geml et al. 2014; Shi et al. 2014):
where ‘S’ is the number of Trichoderma species, ‘N’ is the sum of all Trichoderma species strains, ‘Pi’ is relative quantity of Trichoderma species ‘i’, and ‘ni’ is the number of strains of Trichoderma species ‘i’. All statistical analyses were performed using Microsoft Excel 2010.
Principal components analysis
To determine the dominant factor among altitudes, longitudes, latitudes and ecosystems for the distribution of Trichoderma spp., a principal components analysis (PCA) was conducted in R package vegan (Garrido-Benavent et al. 2020).
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Results
Trichoderma isolation and species identification
We obtained 2,859 Trichoderma isolates from samples collected from five regions of northern Xinjiang in three consecutive years from 2014 to 2016. Due to the large number of isolates, the fungi were initially grouped according to morphological characteristics (type of mycelium, colony color, presence of spores) and then representative isolates with the same morphological characteristics from the soil sample were selected for subsequent analysis, resulting in 312 Trichoderma isolates. With the combination of morphological analysis and molecular methods based on the ITS, tef1-α and rpb2 genes, we identified 23 species: T. harzianum (88 strains), T. paraviridescens (46), T. longibrachiatum (26), T. polysporum (24), T. asperellum (20), T. afroharzianum (20), T. oblongisporum (17), T. citrinoviride (17), T. rossicum (14), T. viridescens (7), T. saturnisporum (6), T. gamsii (5), T. semiorbis (4), T. pleurotum (3), T. koningii (3), T. atroviride (3), T. ghanense (2), T. brevicompactum (2), T. piluliferum (1), T. hamatum (1), T. pararogersonii (1), T. fertile (1), and T. caerulescens (1) (Supplemental Table 1). Of these species, T. pararogersonii was identified as a new species record in China.
Phylogenetic analysis
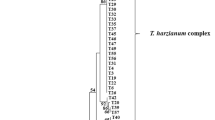
To infer a phylogenetic tree, we first calculated haplotypes from ITS1 and ITS2 sequences of the 312 strains. Finally, 51 haplotypes (Supplemental Table 1) were subjected to maximum likelihood analysis; Nectria eustromatica and N. berolinensis were used as outgroup taxa to root the tree. The phylogenetic tree is shown in Fig. 1. The 51 haplotypes belonging to the 23 Trichoderma species were placed in seven groups. The phylogenetic structure was consistent with previously established sections and clades in most cases(Druzhinina et al. 2006; Druzhinina et al. 2012).
The first group included the Viride clade with T. asperellum, T. hamatum, T. caerulescens, T. atroviride, T. gamsii, T. koningii, T. viridescens, and T. paraviridescens. Within this clade, T. asperellum and T. hamatum (except AP5) formed a separate branch named the Hamatum Clade as described previously(Druzhinina et al. 2006). However, many of the species on the first branch, eventually identified from analysis of tef1-α sequences and phenetic data, which were not differentiated by the phylogenetic analysis of their ITS sequences: (a) T. gamsii and T. koningii; (b) T. paraviridescens and T. viridescens; (c) T. caerulescens; and (d) T. pararogersonii. The second group, lacking bootstrap support, comprises two species in clade Polysporum- T. polysporum and T. piluliferum. The six haplotypes of T. rossicum constituted the third group with a bootstrap value of 89. The fourth group is clade Brevicompactum, which only includes one species (T. brevicompactum) with strong bootstrap support (99%). The fifth group is the Green spored clade, including T. harzianum, T. afroharzianum and T. pleurotum; this clade had 99% bootstrap support. Within this clade, the six haplotypes of T. harzianum formed a moderately well-supported (89%) clade. The sixth group (Semiorbis), which lacked significant bootstrap support, includes T. semiorbis, T. fertile, and T. oblongisporum. Within Semiorbis, three haplotypes of T. oblongisporum formed a separate clade with 79% bootstrap support. The last group (clade Longibrachiatum) includes T. longibrachiatum, T. ghanense, T. saturnisporum, and T. citrinoviride, and had low support (73%).
Species diversity, evenness, and abundance
The analysis was based on the 312 Trichoderma strains isolated from soil of selected regions in northern Xinjiang. A total of 212 soil samples (isolation rate = 33.28%) produced Trichoderma isolates from 634 varied soil samples, and 2,859 Trichoderma isolates (relative rate = 3%) were identified from 19,848 fungal colonies. The total biodiversity of Trichoderma spp. from grassland and forest in northern Xinjiang is shown in Fig. 2. A total of 23 species, including one new species record in China (T. pararogersonii), were recorded. The dominance value (Y) was 0.038 (> 0.02), indicating that the genus Trichoderma was dominant in soil samples. T. harzianum was the most abundant species (28.21%) followed by T. paraviridescens (14.74%).
Biodiversity of Trichoderma spp. with 23 species from grassland and forest in Northern Xinjiang. 23 species: T. harzianum (88 strains), T. paraviridescens (46), T. longibrachiatum (26), T. polysporum (24), T. asperellum (20), T. afroharzianum (20), T. oblongisporum (17), T. citrinoviride (17), T. rossicum (14), T. viridescens (7), T. saturnisporum (6), T. gamsii (5), T. semiorbis (4), T. pleurotum (3), T. koningii (3), T. atroviride (3), T. ghanense (2), T. brevicompactum (2), T. piluliferum (1), T. hamatum (1), T. pararogersonii (1), T. fertile (1), and T. caerulescens (1)
The obtained data were used to calculate the Simpson’s biodiversity index (Dr), the Shannon-Weiner biodiversity index (H), the Pielou species evenness index (J), and Margalef’s abundance index (E) for each region (Supplemental Table 3). The highest species diversity and evenness were in the following order: Yili > Changji > Altay > Urumqi > Bayingolin. For the J index, Urumqi and Altay were characterized by lower homogeneity of species, while Yili, Bayingolin, and Changji were more homogenous. These results revealed that the grassland and forest ecosystem of northern Xinjiang had high diversity of Trichoderma spp. with optimum variation.
Distribution of Trichoderma spp. in different regions
The distribution of Trichoderma strains in different regions is presented in supplemental Fig. 1. We found that the numbers of Trichoderma strains had a decreasing trend from north to south. The proportion and composition of Trichoderma species varied between different regions: Altay Prefecture had the largest number of Trichoderma species (17), with a slightly lower number (16) in Yili; Changji (ten) was in the middle, while Bayingolin and Urumqi had fewer species (seven and six, respectively). However, the proportion of strains obtained from Altay was the highest (182 strains, 58%), followed by Yili (48 strains, 15%), Changji (36 strains, 12%), Urumqi (31 strains, 10%), and Bayingolin (15 strains, 5%).
A total of 17 species were identified from Altay: T. harzianum, T. paraviridescens, T. asperellum, T. citrinoviride, T. polysporum, T. oblongisporum, T. rossicum, T. saturnisporum, T. gamsii, T. longibrachiatum, T. viridescens, T. brevicompactum, T. afroharzianum, T. caerulescens, T. ghanense, T. pararogersonii, and T. piluliferum. Trichoderma from Yili comprised 16 species: T. harzianum, T. longibrachiatum, T. pleurotum, T. viridescens, T. asperellum, T. atroviride, T. citrinoviride, T. paraviridescens, T. polysporum, T. rossicum, T. semiorbis, T. fertile, T. gamsii, T. hamatum, T. koningii, and T. oblongisporum. Ten species were found in Changji: T. afroharzianum, T. harzianum, T. longibrachiatum, T. asperellum, T. atroviride, T. citrinoviride, T. paraviridescens, T. polysporum, T. rossicum, and T. semiorbis. Seven species were found in Bayingolin: T. harzianum, T. saturnisporum, T. asperellum, T. paraviridescens, T. polysporum, and T. koningii. Six species were found in Urumqi: T. afroharzianum, T. harzianum, T. longibrachiatum, T. pleurotum, T. saturnisporum, and T. viridescens.(Supplemental Table 1).
Some of the species were unique to one region—T. caerulescens, T. pararogersonii, T. brevicompactum, and T. piluliferum were only collected from Altay; T. fertile and T. hamatum only from Yili; and T. ghanense only from Bayingolin. T. harzianum, however, was found in all regions and was the most widely distributed species. Another widely distributed species was T. longibrachiatum, which was distributed in all regions except Bayingolin. Among the 23 species described in this study, the T. harzianum complex (T. harzianum and T. afroharzianum) was the most abundant species (35% of all strains) with dominancy in Altay and Bayingolin. Other regions had a different dominant species: Urumqi and Yili’s dominant species were T. longibrachiatum and T. polysporum, respectively. Another very common species complex (17% of all strains) occurring in the five regions was T. viridescens (T. viridescens and T. paraviridescens). T. longibrachiatum, T. polysporum, and T. asperellum were distributed in four regions (Altay,Yili, Changji and Urumqi) with relatively lower proportions (8%, 8%, and 6%, respectively). The other species were represented by several strains and accounted for the minority of isolates (26% of all strains).
Distribution of Trichoderma spp. in different ecosystems
The distribution of Trichoderma varied with ecosystems. According to the different ecological environments, the sampling sites can be divided into grassland and forest ecosystems. The grassland also included two sub-ecosystems—desert steppe and temperate steppe—while the forest ecosystem consisted of coniferous forest as well as coniferous and broadleaf mixed forest sub-ecosystems. There were 446 samples from the grassland ecosystem with 163 strains (52.24% of total strains) and 20 species of Trichoderma. A total of 188 samples were collected from the forest ecosystem with 149 strains (47.76% of total strains) and 16 species. The number of samples collected in the forest ecosystem was lower and the number of Trichoderma strains was lower than in the grassland ecosystem. However, the isolation frequency of Trichoderma in the forest ecosystem was 79.26%, which was significantly higher than that of grassland at 36.55%. Therefore, the forest ecosystem is the dominant ecosystem of Trichoderma, and it is more suitable for the survival and colonization of Trichoderma spp.
In total, 312 Trichoderma strains were collected from the four ecosystems; the differences among them are shown in Fig. 3a. The highest species’ numbers were obtained from temperate grassland soil (18 species) isolated from 127 strains, followed by coniferous forest (14 species isolated from 64 strains), coniferous broadleaf forest (11 species isolated from 85 strains), and the temperate-desert grassland had the fewest species (ten species isolated from 36 strains). In addition, the order of isolation frequency of Trichoderma strains from soil samples from high to low is: coniferous broadleaf forest (116.44%), coniferous forest (55.65%), temperate grassland (40.97%), and temperate-desert grassland (26.47%).
Distribution of Trichoderma species in different ecosystems. a The column curve picture for the positive detection of Trichoderma strains and species in different ecosystems. The numbers written on the bars correspond to the number of isolates in every ecosystem. The numbers written on the yellow line correspond to the frequency of Trichoderma in every ecosystem. b Distribution of Trichoderma species in different ecosystems. The inlaid box indicates the distributions of T. viridescens, T. saturnisporum, T. gamsii, T. semiorbis, T. koningii, T. atroviride, T. pleurotum, T. ghanense, T. brevicompactum, T. piluliferum, T. pararogersonii, T. caerulescens, T. hamatum, and T. fertile according to the different ecosystem. The numbers written on the bars correspond to the number of isolates in every ecosystem
The distribution of Trichoderma species is also related to the ecosystem (Fig. 3b). Some species like T. harzianum, T. paraviridescens, T. asperellum, T. rossicum, T. oblongisporum and T. saturnisporum were distributed in all four ecosystems; and some species were distributed in only one ecosystem, for example, T. semiorbis, T. koningii, T. caerulescens, and T. hamatum were only found in temperate grassland; T. piluferum and T. paraogersonii were only found in coniferous broadleaf forest; and T. fertile was only found in coniferous forest.
Distribution of Trichoderma spp. at different altitudes
According to the altitude, all collection sites can be divided as follows: below 1000 m, 1000–2000 m, 2000 to 3000 m, and above 3000 m. There were significant differences in communities of Trichoderma species from soils collected at different altitudes (Fig. 4a). Twenty-one species of Trichoderma were isolated from 1000 to 2000 m, which covered most of the species described in this study, and the number of strains was the highest (212) at this altitude, accounting for 67.95% of all strains. At altitudes of 2000–3000 m, there were 15 species and 73 strains, the distribution of Trichoderma was dense, but the richness of Trichoderma spp. was reduced. At altitudes below 1000 m, only 66 soil samples were collected, but nine species and 19 strains of Trichoderma were isolated. However, at altitudes above 3000 m, there were 42 samples collected, fewer than below 1000 m, and only two species and three strains were isolated. According to the isolation frequency, the four altitude groups can be ranked from high to low as follows: 1000–2000 m, 2000–3000 m, below 1000 m, and above 3000 m. Based on these results, 1000–2000 m is a suitable living environment for Trichoderma, but above 3000 m might not be suitable for Trichoderma to survive in our selected sites.
Distribution of Trichoderma species on different altitudes. a The column curve picture for the positive detection of Trichoderma strains and species on different altitudes. The numbers written on the bars correspond to the number of isolates in every ecosystem. The numbers written on the yellow line correspond to the isolate frequency of Trichoderma in every ecosystem. b Distribution of Trichoderma species on different altitudes. The inlaid box indicates the distributions of T. viridescens, T. saturnisporum, T. gamsii, T. semiorbis, T. koningii, T. atroviride, T. pleurotum, T. ghanense, T. brevicompactum, T. piluliferum, T. pararogersonii, T. caerulescens, T. hamatum, and T. fertile according to the different altitudes. The numbers written on the bars correspond to the number of isolates in every altitude
In addition, the composition and distribution of Trichoderma at different altitudes were quite different (Fig. 4b). T. harzianum was distributed at all altitudes. The highest number of T. harzianum strains occurred between 1,000 and 2000 m, accounting for 80.46% of the total number of strains. The distribution of T. paraviridescens was similar to T. harzianum, with the largest distribution at altitudes of 1000–2000 m with 40 strains isolated, accounting for 86.96% of the total number of strains. T. asperellum, T. citrinoviride, and T. rossicum were distributed in the same way. T. polysporum, T. afroharzianum, T. oblongisporum, T. viridescens, T. semiorbis, T. koningii, T. pleurotum, and T. ghanense were not isolated at altitudes below 1000 m, indicating that higher altitudes may be more suitable for these species. The number of strains of T. longibrachiatum was the highest at altitudes between 2000 and 3000 m lower at 1000–2000 m, and lowest at altitudes below 1000 m. T. caerulescens, T. piluliferum, T. pararogersonii, and T. hamatum were isolated only at altitudes of 1000–2000 m, but the strains were fewer. At altitudes above 3000 m, only three strains were isolated: two of T. oblongisporum and one of T. polysporum.
Principal components analysis of the distribution of Trichoderma spp
All of the data for isolates of Trichoderma spp. were analyzed using principal components analysis (PCA) in R package vegan. The results showed us the ecosystem is the most dominant impact factor for the distribution of Trichoderma spp. among longitude, latitude, altitude and ecosystems (Fig. 5). The grassland and forest ecosystems were contained two different ecosystems, respectively, the grassland had two sub-ecosystems as desert steppe and temperate steppe, while forest had two sub-ecosystems with coniferous forest as well as coniferous and broadleaf mixed forest. In the same ecosystem or had two focus distribution areas, which were induced by altitude.
Principal Components Analysis (PCA) for all of the isolates of Trichoderma spp. from Northern Xinjiang by the R package vegan. Ecosystem groups: 1. desert steppe; 2. temperate steppe; 3. coniferous forest; 4. coniferous and broadleaf mixed forest. The colors of isolates from location regions: Altay (deep red); Yili (orange); 3. Changji and Urumuqi (bright green); 4. Bayingolin (blue)
Discussion
In this study, we examined the biodiversity of Trichoderma associated with collection region, ecosystem, and altitude from grassland and forest in northern Xinjiang. Several studies have investigated the biodiversity of Trichoderma in different provinces of China(Jiang et al. 2016; Saravanakumar et al. 2016; Sun et al. 2012; Zhang et al. 2005a, b); however, little attention was paid to Xinjiang. In 2014, 2015, and 2016, 634 soil samples were collected from grassland and forest ecosystems in five major regions of northern Xinjiang, a total of 2,859 isolates were obtained, 312 strains were classified, and 23 species were identified, including a new species record in China (T. pararogersonii). Compared to studies on the biodiversity in other countries and regions, like Poland(Błaszczyk et al. 2011), Central Europe(Błaszczyk et al. 2016), and the Indo-Burma Biodiversity Hot Spot(Kamala et al. 2015), there were almost twice the number of Trichoderma species (23) detected in our study. In addition, our results showed that Trichoderma distributions were remarkably diversified: the dominant species and proportion of each species were significantly different in different regions, and such a varied distribution might be associated with differences in ecological environments such as ecosystem type and altitude of the large land mass in Xinjiang.
The composition and proportion of Trichoderma species and strains were distinct among different regions. Geographic distance is the dominant factor driving variation in fungal diversity at a regional scale (1000–4000 km)(Wu et al. 2013), which is consistent with our research. In this study, the total number of Trichoderma strains in northern Xinjiang varied from north to south. Altay had 17 species and had the largest number of Trichoderma strains (182), followed by Yili (16 species, 48 strains), Changji (10 species, 36 strains), Bayingolin (7 species, 15 strains) and Urumqi (6 species, 31 strains). Fungal diversity is related to vegetation zones and environmental factors (Zachow et al. 2009a, b), which might account for this distinct distribution. The ecological environment of Altay is diverse, including four ecological types: temperate grassland, temperate-desert grassland, coniferous forest, and coniferous broadleaf forest. The vegetation cover is also dense, which may have led to the high isolation frequency of Trichoderma. The ecological environment of some collection sites in Bayingolin is very poor, with ground cover of only a small amount of desert vegetation and soil types that are mostly sandy soil, but other parts of the region have more favorable ecological environments and dense vegetation cover. However, the high altitude of Bayingolin, which can reach more than 3000 m, may also negatively affect the colonization of Trichoderma.
The distribution of Trichoderma varies with changes in the ecosystem. Researchers have reported that fungal communities are associated with distinct plant species(de Souza Sebastianes et al. 2013), vegetation cover density, edaphic factors(Birhane et al. 2018), soil age, and ecosystem development(Courty et al. 2018). In this study, the vegetation types of forest ecosystems in northern Xinjiang were diverse, the soil was more fertile, and Trichoderma was widely distributed. Although the overall sampling number (188) in the forest was less than that of the grassland (446), the isolation frequency of Trichoderma was higher (79.26% and 36.55%, respectively). Therefore, the forest ecosystem is dominant for Trichoderma and is more suitable for survival and colonization of Trichoderma. This indicated that Trichoderma has an environmental preference, consistent with previous studies(Błaszczyk et al. 2011; Friedl and Druzhinina, 2011; Yu et al. 2010). Similarly, forest ecosystems subjected to natural dynamics for a longer period support a more diverse fungal community(Pioli et al. 2018).
Limited diversity of Trichoderma was found in temperate-desert grassland, which may be attributed to the poor ecological environment. In these ecosystems, there is only desert vegetation, such as shuttle, caraway grass and red willow (Tamirix chinensis), and soil types are sandy and sandy loam, which may be detrimental to the colonization of Trichoderma. In the forest ecosystem, the number of Trichoderma strains isolated from coniferous broadleaf forest (85) was higher than that of coniferous forest (64), while the number of Trichoderma species (14) obtained from coniferous forest was higher than that of coniferous broadleaf forest (11). This may be because more soil samples were collected in the coniferous forest, with a wider range of collection, including parts of Urumqi, Changji, Yili and Altay, but the only coniferous broadleaf forest collection site was in Kanas Nature Reserve in Altay. In addition, the isolation frequency of coniferous broadleaf forest was higher, which might be attributed to the better ecological environment (mean temperature, maximum and minimum relative humidity, cumulative rainfall) and higher vegetation coverage, or may be associated with the soil moisture, pH, and oxygen content.
The numbers of species and strains isolated from different altitudes were highly variable. Yu et al. investigated the diversity of Trichoderma in Guangxi and found that altitude had a minimal effect on richness(Yu et al. 2012). Their investigation in a forest in Guangxi was conducted at altitudes between 100-2,100 m, which was lower compared to our altitude range of 325-3,547 m, and Guangxi is a rainy region. Other recent evidence has revealed that community composition and diversity patterns of some soil microbiota correlate strongly with altitude: we found that the distribution of Trichoderma was related to the altitude of the sampling site based on a decline of quantity and species in different kinds of fungi (Shi et al. 2014; Dong et al. 2004) with altitudes above 3000 m or below 1000 m. The order of the Trichoderma diversity at different altitudes is: 2000–3000 m, 1000–2000 m, below 1000 m, and above 3000 m. This trend was in accordance with reports from Yungas forests, in which the total fungal diversity did not decrease significantly with increasing elevation from 400 to 2160 m a.s.l (Geml et al. 2014), but contrary to Yu et al. (2012). who found that fungal species richness declined gradually with increasing elevation. Our results indicated that altitudes under 3000 m was a suitable living environment, but above 3000 m was not suitable for Trichoderma to survive, which might be attributed to low temperature. Gomes et al. demonstrated that fungal community structure was also driven by mean temperatures at 10 (for endophytes) or 20 (for epiphytes) days before the sampling date (Gomes et al. 2018). The dominant species was different in every altitude gradient, indicating that lower altitudes should be more suitable for T. harzianum, while higher altitudes are more suitable for T. longibrachiatum and T. polysporum.
Here and in a previous study, T. harzianum was the dominant taxon(Jaklitsch and Voglmayr 2015; Kubicek et al. 2008). T. harzianum is the most commonly reported species in the genus, occurring in diverse ecosystems and ecological niches. High genetic diversity may contribute to the higher abundance of T. harzianum: we found six haplotypes in this study and it has been reported that T. harzianum is a species complex (Chaverri et al. 2015). We only identified this species according to the ITS region, so there might be new species within the species complex. The second most dominant taxon in Xinjiang was T. paraviridescens, which belongs to the T. viridescens complex that contains 13 species (Jaklitsch et al. 2013). In contrast, Wu et al. found that T. viridescens had the smallest communities in the soils of northwestern China (Wu et al. 2017), indicating that T. viridescens may only be distributed in Xinjiang, especially in Altay.
T. pararogersonii was first reported from China in this paper. Previously, this species was only reported by Jaklitsch et al. to be distributed in Mediterranean Europe (Jaklitsch et al. 2013), here we expanded its distribution range. There is no report of its ITS sequence, so we identified it by analyzing the sequences of tef and rpb2, which had similarities of 100% with the reported sequence of T. pararogersonii from GenBank.
To our knowledge, this is the first report that analyzes Trichoderma biodiversity from grassland and forest ecosystems in Xinjiang Uygur Autonomous Region, China, and this study includes the identification of a large number of Trichoderma strains based on molecular biology. The data generated in this study reveals a great reservoir of Trichoderma genetic diversity in the Chinese forests and grasslands and highlights substantial differences between Trichoderma communities from distinct sampling regions, ecosystems, and altitudes. This work could be important in the future to detect possible valuable Trichoderma resources in local environments. In the meantime, new experiments should be performed to better understand the biocontrol ability of Trichoderma to select some strains that can be used as inoculants for plant growth and health promotion, such as fungicide applications and/or introduction of biological control agents.
Abbreviations
- PDA:
-
Potato dextrose agar
- SNA:
-
Synthetic low nutrient agar
- CMD:
-
Corn meal dextrose agar
- NCBI:
-
National Center for Biotechnology Information
- BLAST:
-
Basic local alignment search tool
- PCR:
-
Polymerase Chain Reaction
- Bp:
-
Base pair
- ddH2O:
-
Double Distilled H2O
- TEF1-α:
-
Transcription Elongation Factor 1 α
- RPB2:
-
RNA Polymerase II second largest subunit
- ITS:
-
Internal Transcribed Spacer
- F:
-
Fusarium oxysporum
- R:
-
Rhizoctonia solani
- B:
-
Botrytis cinerea
- MS:
-
Murashige and Skoog
References
Bae S et al (2016) Trichoderma metabolites as biological control agents against Phytophthora pathogens. Biol Control 92:128–138
Birhane E et al (2018) Vegetation cover density and disturbance affected arbuscular mycorrhiza fungi spore density and root colonization in a dry Afromontane forest, northern Ethiopia. J Forest Res 29:675–686
Błaszczyk L et al (2011) Species diversity of Trichoderma in Poland. J Appl Genet 52:233–243
Błaszczyk L et al (2016) Trichoderma species occurring on wood with decay symptoms in mountain forests in Central Europe: genetic and enzymatic characterization. J Appl Genet 57:397–407
Braithwaite M et al (2017) Trichoderma down under: species diversity and occurrence of Trichoderma in New Zealand. Australas Plant Pathol 46:11–30
Chaverri P, Samuels GJ (2013) Evolution of habitat preference and nutrition mode in a cosmopolitan fungal genus with evidence of interkingdom host jumps and major shifted in ecology. Evolution 67(10):2823–2837
Chaverri P et al (2015) Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia 107:558–590
Courty PE et al (2018) Impact of soil pedogenesis on the diversity and composition of fungal communities across the California soil chronosequence of Mendocino. Mycorrhiza 28:343–356
Cummings NJ et al (2016) Diversity of root-endophytic Trichoderma from Malaysian Borneo. Mycol Progr 15(5):50
de Souza Sebastianes FL et al (2013) Species diversity of culturable endophytic fungi from Brazilian mangrove forests. Curr Genet 59:153–166
Dong A et al (2004) Diversity of soil fungi in Liangshui natural reseve, Xiaoxing’ anling Forest Region. J Northwest Forest Univ 32:8–10 (in Chinese)
Druzhinina IS et al (2004) Hypocrea flaviconidia, a new species from Costa Rica with yellow conidia. Stud Mycol 50:401–407
Druzhinina IS et al (2005) An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet Biol 42:813–828
Druzhinina IS et al (2006) The first 100 Trichoderma species characterized by molecular data. Mycoscience 47:55–64
Druzhinina IS et al (2012) Molecular phylogeny and species delimitation in the section Longibrachiatum of Trichoderma. Fungal Genet Biol 49:358–368
El-Hassan SA et al (2013) Use of Trichoderma hamatum for biocontrol of lentil vascular wilt disease: efficacy, mechanisms of interaction and future prospects. J Plant Protect Res 53:12–26
Friedl MA, Druzhinina IS (2011) Taxon-specific metagenomics of Trichoderma reveals a narrow community of opportunistic species that regulate each other’s development. Microbiology 158:69–83
Gajera HP et al (2015) Trichoderma viride induces pathogenesis related defense response against rot pathogen infection in groundnut (Arachis hypogaea L.). Infect Genet Evol 34:314–325
Gams W, Bissett J (1998) Morphology and identification of Trichoderma. Trichoderma and Gliocladium. Taylor and Francis, London, pp 3–34
Garrido-Benavent I et al (2020) Differential colonization and succession of microbial communities in rock and soil substrates on a maritime antarctic glacier forefield. Front Microbiol 11:126
Gazis R et al (2011) Species delimitation in fungal endophyte diversity studies and its implications in ecological and biogeographic inferences. Mol Ecol 20:3001–3013
Geml J et al (2014) Large-scale fungal diversity assessment in the Andean Yungas forests reveals strong community turnover among forest types along an altitudinal gradient. Mol Ecol 23:2452–2472
Gherbawy Y et al (2004) Trichoderma populations from alkaline agricultural soil in the Nile valley, Egypt, consist of only two species. Mycol Progr 3:211–218
Gomes T et al (2018) Endophytic and epiphytic phyllosphere fungal communities are shaped by different environmental factors in a mediterranean ecosystem. Microb Ecol 76:668–679
Harman GE (2000) Myths and dogmas of biocontrol—changes in perceptions derived from research on Trichoderma harzianum T-22. Plant Dis 84:377–393
Harman GE et al (2004) Trichoderma species—Opportunistic, avirulent plant symbionts. Nat Rev Microbiol 2:43–56
Hoyos-Carvajal L et al (2009) Genetic and metabolic biodiversity of Trichoderma from Colombia and adjacent neotropic regions. Fungal Genet Biol 46:615–631
Jaklitsch WM (2009) European species of Hypocrea Part I. The green-spored species. Stud Mycol 63:1–91
Jaklitsch WM, Voglmayr H (2015) Biodiversity of Trichoderma (Hypocreaceae) in Southern Europe and Macaronesia. Stud Mycol 80:1–87
Jaklitsch WM et al (2005) Hypocrea voglmayrii sp nov from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 97:1365–1378
Jaklitsch WM et al (2013) Disentangling the Trichoderma viridescens complex. Persoonia 31:112–146
Jiang Y et al (2016) Trichoderma Biodiversity of Agricultural Fields in East China Reveals a Gradient Distribution of Species. PLoS ONE 11(8):e0160613
Kamala T et al (2015) Phylogeny and Taxonomical Investigation of Trichoderma spp. from Indian Region of Indo-Burma Biodiversity Hot Spot Region with Special Reference to Manipur. Biomed Research Int 2015:21
Kashyap PL et al (2017) Trichoderma for climate resilient agriculture. World J Microbiol Biotechnol 33:155
Kopchinskiy A et al (2005) TrichoBLAST: a multilocus database for Trichoderma and Hypocrea identifications. Mycol Res 109:658–660
Kubicek CP et al (2003) Genetic and metabolic diversity of Trichoderma: a case study on South-East Asian isolates. Fungal Genet Biol 38:310–319
Kubicek CP et al (2008) Fungal genus Hypocrea/Trichoderma: from barcodes to biodiversity. J Zhejiang Univ Sci B 9:753–763
Kullnig C et al (2000) Molecular identification of Trichoderma species from Russia, Siberia and the Himalaya. Mycol Res 104:1117–1125
Larkin MA et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Margalef R (1958) Information theory in ecology. General Syst 3:36–71
Mukherjee PK et al (2013) Trichoderma research in the genome era. Ann Rev Phytopathol 51:105–129
Naeimi S et al (2011) Species pattern and phylogenetic relationships of Trichoderma strains in rice fields of Southern Caspian Sea, Iran. Cereal Res Commun 39:560–568
Ortega-García JG et al (2015) Effect of Trichoderma asperellum applications and mineral fertilization on growth promotion and the content of phenolic compounds and flavonoids in onions. Sci Hortic 195:8–16
Pielou E (1966) Measurment of diversity in differert types of biological collections. J Theor Biol 13:131–144
Pioli S et al (2018) Community fingerprinting reveals increasing wood-inhabiting fungal diversity in unmanaged Mediterranean forests. For Ecol Manage 408:202–210
Saravanakumar K et al (2016) Biodiversity of Trichoderma Community in the Tidal Flats and Wetland of Southeastern China. PLoS ONE 11(12):e0168020
Shannon C (1948) A mathematical theory of communication. Bell Syst Tech J 27:623–656
Shi Z et al (2014) Diversity and distribution of arbuscular mycorrhizal fungi along altitudinal gradients in Mount Taibai of the Qinling Mountains. Can J Microbiol 60:811–818
Simpson E (1949) Measurement of diversity. Nature 163:688–689
Sun R et al (2012) Trichoderma biodiversity in China. J Appl Genet 53:343–354
Tamura K et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Vargas Gil S et al (2009) Quantitative isolation of biocontrol agents Trichoderma spp., Gliocladium spp. and actinomycetes from soil with culture media. Microbiol Res 164:196–205
Vitti A et al (2016) Trichoderma harzianum T-22 induces systemic resistance in tomato infected by cucumber mosaic virus. Front Plant Sci 7:1520
Wen C et al (1993) Studies on the taxonomy of the genus Trichoderma in southwestern China. Acta Mycol Sin 12:118–130
White TJ, Bruns T, Lee SJ, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc 18(1):315–322
Wu B et al (2013) The biogeography of fungal communities in wetland sediments along the Changjiang River and other sites in China. ISME J 7:1299–1309
Wu Z et al (2017) Diversity and antagonizing effects of Trichoderma spp. in crop rhizosphere soils. J Fungal Res 14:177–182
Wuczkowski M et al (2003) Species pattern and genetic diversity of Trichoderma in a mid-European, primeval floodplain-forest. Microbiol Res 158:125–133
Yu H et al. (2012) Investigation of Trichoderma diversity in Guangxi forset regions. Journal of Northwest Forestry University. 109–113.(in Chinese)
Yu H et al (2010) Investigation on Trichoderma resources in forest soil of Northern Guangxi. Guangxi Agric Sci 41:703–706
Zachow C et al (2009) Fungal diversity in the rhizosphere of endemic plant species of Tenerife (Canary Islands): relationship to vegetation zones and environmental factors. ISME J 3:79–92
Zhang CL et al (2005) Trichoderma biodiversity in China: evidence for a North to South distribution of species in East Asia. FEMS Microbiol Lett 251:251–257
Zhang S et al (2015) Biocontrol potential of a native species of Trichoderma longibrachiatum against Meloidogyne incognita. Appl Soil Ecol 94:21–29
Acknowledgements
We are grateful to Dr. Jianqing Tian (Institute of Microbiology, Chinese Academy of Agricultural Sciences) for the help and direction of analysis in this research.
This work was supported by the National key research and development plan (Chemical fertilizer and pesticide reducing efficiency synergistic technology research and development); Research and demonstration of a new high efficiency biocide (2017YFD0201100–2017YFD0201102); the Beijing National Science Foundation (6192022); the Survey of basic resources of science and technology: A comprehensive survey of biodiversity in the Mongolian Plateau (2019FY102000); the Demonstration of comprehensive prevention and control technology of non-point source pollution in main vegetable producing areas of Huanghuaihai (SQ2018YFD080026).
Author information
Authors and Affiliations
Contributions
JM designed the experiment and writing for the manuscript, ML and ET conducted the experiment, BW and XJ contributed to designing the experiment and writing the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ma, J., Tsegaye, E., Li, M. et al. Biodiversity of Trichoderma from grassland and forest ecosystems in Northern Xinjiang, China. 3 Biotech 10, 362 (2020). https://doi.org/10.1007/s13205-020-02301-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02301-6