Abstract
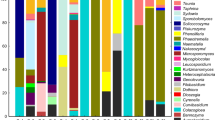
The Mauritian waters harbour micro-organisms that have successfully adapted to their environment; however, the mycobiota associated with algae and sponges have been poorly studied and are awaiting to be identified and exploited. This study focusses on the fungi associated with the sponge Haliclona sp. and two brown algae Turbinaria conoides and Sargassum portierianum from Mauritius. Samples of Haliclona sp., Turbinaria conoides and Sargassum portierianum were collected in the North of Mauritius, and associated fungi were isolated after surface sterilization and plating on artificial media supplemented with seawater. A total of 17 morphologically different isolates were recovered from Haliclona sp., 20 isolates were obtained from Turbinaria conoides, and 16 isolates were from Sargassum portierianum. The selected sponge-associated fungi have been identified as Acremonium sp., Periconia byssoides, Pestalotiopsis sp. Endophytes from Turbinaria conoides were identified as Aspergillus penicilloides, Acremonium sp. and Cordyceps memorabilis, whereas those from Sargassum portierianum were identified as Pseudopithomyces maydicus, Acremonium sclerotigenum and Curvularia lunata. The ITS regions were suitable for identifying fungi up to the genus level. However, species level identification was problematic and should be used with caution.








Similar content being viewed by others
References
Pang KL, Overy DP, Jones EBG et al (2016) ‘Marine fungi’ and ‘marine-derived fungi’ in natural product chemistry research: Toward a new consensual definition. Fungal Biology Reviews 30(4)
Tadych M, White JF (2019) Endophytic microbes. In: Schmidt TM (ed) Encyclopedia of Microbiology. Academic Press, New Brunswick, pp 123–136
Van Soest RWM, Boury-Esnault N, Hooper JNA et al (2019) World Porifera Database. http://www.marinespecies.org/porifera. Accessed 16 August 2018
Bovio E, Garzoli L, Poli A et al (2019) Marine Fungi from the Sponge Grantia compressa: Biodiversity, Chemodiversity, and Biotechnological Potential. Marine Drugs 17(4):220. https://doi.org/10.3390/md17040220
Kossuga MH, Romminger S, Xavier C et al (2012) Evaluating methods for the isolation of marine-derived fungal strains and production of bioactive secondary metabolites. Rev Bras Farmacogn 22(2). https://doi.org/10.1590/S0102-695X2011005000222
Dayarathne MC, Jones EBG, Maharachchikumbura SSN et al (2020) Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 11(1):1–188
Guiry MD, Guiry GM (2019) AlgaeBase. https://www.algaebase.org. Accessed 14 August 2018
Bramhachari PV, Anju S, Sheela GM et al (2019) Secondary Metabolites from Marine Endophytic Fungi: Emphasis on Recent Advances in Natural Product Research. In: Singh BP (ed) Advances in Endophytic Fungal Research, Fungal Biology. Springer Nature, Switzerland, pp 339–350
Kamat S, Kumari M, Taritla S, Jayabaskaran C (2020) Endophytic Fungi of Marine Alga from Konkan Coast, India- A Rich Source of Bioactive Material. Frontiers in Marine Science 7(31)
Wang XF, Wang MM, Zhao Y, Li CL, Li W (2018) Diversity of culturable fungi associated with marine macroalgae from coast of Qingdao. China Mycosystema 37(3):281–293
Jeewon R, Ittoo J, Mahadeb D, Jaufeerally-Fakim Y et al (2013) DNA based identification and phylogenetic characterization of endophytes and saprobic fungi from Antidesma madagascariense, a medicinal plant in Mauritius. Journal of Mycology 781914:1–10
Thompson JD, Higgings DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Swofford DL (2002) PAUP* Version 4.0 b10 Phylogenetic Acalysis Uisng Parsimony (*and other methods. Sinauer, Sunderland
Zhou J, Diao X, Wang T et al (2018) Phylogenetic diversity and antioxidant activities of culturable fungal endophytes associated with the mangrove species Rhizophora stylosa and R. mucronata in the South China Sea. PLoS ONE 13(6)
Suryanarayananand TS Johnson JA (2014) Fungal Endosymbionts of Macroalgae: Need for Enquiries into Diversity and Technological Potential. Oceanography 2(1)
Wang Y, Barth D, Tamminen A, Wiebe MG (2016) Growth of marine fungi on polymeric substrates. BMC Biotechnol 16(3)
Jalili B, Bagheri AS, Soltani J (2020) Identification and salt tolerance evaluation of endophyte fungi isolate from halophyte plants. Int J Environ Sci Technol 17:3459–3466
Ghule S, Sawant IS, Sawant SD et al (2019) Isolation and identification of three new mycoparasites of Erysiphe necator for biological control of grapevine powdery mildew. Australas Plant Pathol 48(4):351–367
Jeewon R, Hyde KD (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7(11):1669–1677
Chi LP, Yang SQ, Li XM et al (2020) A new steroid with 7β,8β-epoxidation from the deep sea fungus Aspergillus penicilloides SD-311
Nazareth SW, Gonsalves V (2014) Halophilic Aspergillus penicillioides from athalassohaline, thalassohaline, and polyhaline environments. Front Microbiol 5(412). https://doi.org/10.3389/fmicb.2014.00412
Wanasinghe DN, Phukhamsakda C, Hyde KD et al (2018) Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Diversity 89:1–236. https://doi.org/10.1007/s13225-018-0395-7
Doilom M, Manawasinghe IS, Jeewon R et al (2017) Can ITS sequence data identify fungal endophytes from cultures? A case study from Rhizophora apiculata. Mycosphere 8(10):1869–1892
Saltamachia SJ, Araújo JPM (2020) Ophiocordyceps desmidiospora, a basal lineage within the ‘Zombie Ant Fungi’ clade. Mycologia
Cheng M, Aung T, Wu M et al (2020) Polar Constituent of the Endophytic Fungus Ophiocordyceps sobolifera. Chem Nat Compd 56:289–291
Herrmann LW, poitevin CG, Schuindt LC, et al (2019) Diversity of fungal endophytes in leaves of Mentha piperita and Mentha canadensis. International Journal of Botany Studies 4(1):44–49
Markovskaja S, Kačergius A (2014) Morphological and molecular characterisation of Periconia pseudobyssoides sp. nov. and closely related P. byssoides. Mycol Prog 13(2): 291–302
Liu J, Chen M, Chen R et al (2020) Three new compounds from the endophytic fungus Periconia sp. F-31. Journal of Chinese Pharmaceutical Science 29(4): 244–251
Marin-Felix Y, Hernández-Restrepo M, Crous PW (2020) Multi-locus phylogeny of the genus Curvularia and description of ten new species. Mycological Progress 19:559–588
Norphanphoun C, Raspé O, Jeewon R et al (2019) Morphological and phylogenetic characterization of novel pestalotioid species associated with mangroves in Thailand. MycoKeys 10(1):531–578. https://doi.org/10.3897/mycokeys.38.28011
Acknowledgements
The authors are grateful to Dr Vishwakalyan Bhoyroo and his team for the provision of samples. The authors also thank the staff of the Faculty of Agriculture and the Faculty of Science of the University of Mauritius for their help.
Funding
This study was supported by the University of Mauritius under grant number Q0200 and the MPhil/PhD scholarship of the Higher Education Commission (HEC).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance Statement
This study aims to isolate and identify selected fast-growing marine fungi associated with sponge and algae from Mauritius. It demonstrates that the ITS rDNA regions, which are commonly used for fungal identification, are useful for genus level identification.
Rights and permissions
About this article
Cite this article
Wong Chin, J.M., Puchooa, D., Bahorun, T. et al. Molecular characterization of marine fungi associated with Haliclona sp. (sponge) and Turbinaria conoides and Sargassum portierianum (brown algae). Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 91, 643–656 (2021). https://doi.org/10.1007/s40011-021-01229-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-021-01229-y