Abstract
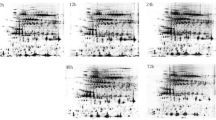
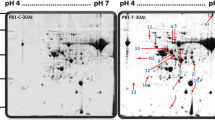
Fungal diseases are crucial factors in reducing chickpea production. Ascochyta Blight is caused by the necrotrophic fungi Didymella rabiei and is one of the most destructive diseases in most world areas. Therefore, a completely randomized factorial design with five replications was applied to evaluate the effects of Ascochyta Blight disease fungi on the chickpea plant. The chlorophyll index, the activities of superoxide dismutase isoforms, and evolutionary analyses were performed to get further insights. Also, 2D electrophoresis of chickpea leaf proteins, gene ontology, and protein-protein interactions analysis was performed. The results did not show any significant effect of A. rabiei infection on the wet weight of chickpea seedlings. Chlorophyll index levels significantly decreased with A. rabiei infection in both chickpea lines. Electrophoretic analysis of superoxide dismutase on 8% polyacrylamide gel revealed three isoforms. The activities of superoxide dismutase isoforms significantly increased with A. rabiei disease. Identification of proteins was performed according to their isoelectric points and approximate molecular weights. Leaf proteome analysis of chickpea lines showed that the expression of eight reproducible spots changed significantly under A. rabiei disease condition. Candidate proteins were components of defense and regulation systems. High expression of Dual specificity protein, Peptide methionine sulfoxide reductase B2, and chloroplastic phosphatase 1B (the proteins involved in the defense system) reveals their essential functions under A. rabiei infection. Increased activities of superoxide dismutase enzymes and proteins involved in the defense system can reduce A. rabiei infection effects on chickpea seedlings.








Similar content being viewed by others
Notes
b-ZIP Transcription factor TGACGTCA Motif-Binding Prote-in.
REFERENCES
Bagheban, F., Mohraminajad, S., Lotfi, R., Bandehhagh, A., and Karbalaei, H., Study of catalase activity and photosynthetic efficiency of maize lines under Fusarium contamination (Fusarium verticillioides), J. Appl. Res. Plant Prot., 2019, vol. 8, pp. 13–23.https://doi.org/10.1002/elps.1150080203
Blum, H., Beier, H., and Gross, H.J., Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels, Electrophoresis, 1987, vol. 8, pp. 93–99. https://doi.org/10.1002/elps.1150080203
Bradford, M.M., A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, Anal. Biochem., 1976, vol. 72, pp. 248–254. https://doi.org/10.1006/abio.1976.9999
Chen, Z., Silva, H., and Klessig, D.F., Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid, Science, 1993, vol. 262, pp. 1883–1886. https://doi.org/10.1126/science.8266079
Chen, W., Coyne, C., Peever, T., and Muehlbauer, F., Characterization of chickpea differentials for pathogenicity assay of ascochyta blight and identification of chickpea accessions resistant to Didymella rabiei, Plant Pathol., 2004, vol. 53, pp. 759–769. https://doi.org/10.1111/j.1365-3059.2004.01103.x
Chen, Y.-Y., Li, M.-Y., Wu, X.-J., Huang, Y., Ma, J., and Xiong, A.-S., Genome-wide analysis of basic helix–loop–helix family transcription factors and their role in responses to abiotic stress in carrot, Mol. Breed., 2015, vol. 35, pp. 1–12. https://doi.org/10.1007/s11032-015-0319-0
Chivasa, S., Tomé, D.F., Hamilton, J.M., and Slabas, A.R., Proteomic analysis of extracellular ATP-regulated proteins identifies ATP synthase beta-subunit as a novel plant cell death regulator, Mol. Cell Proteomics, 2011, vol. 10, p. M110.003905. https://doi.org/10.1074/mcp.M110.003905
Damerval, C., Zivy, M., and Granier, F., Two-Dimensional Electrophoresis in Plant Biology, Laboratoire de génétique des systèmes végétaux, 1988.
Dangl, J.L., Dietrich, R.A., and Richberg, M.H., Death don’t have no mercy: cell death programs in plant-microbe interactions, Plant Cell, 1996, vol. 8, p. 1793. https://doi.org/10.1105%2Ftpc.8.10.1793
Danon, A., Rotari, V.I., Gordon, A., Mailhac, N., and Gallois, P., Ultraviolet-C overexposure induces programmed cell death in Arabidopsis, which is mediated by caspase-like activities and which can be suppressed by caspase inhibitors, p35 and Defender against apoptotic death, J. Biol. Chem., 2004, vol. 279, pp. 779–787. https://doi.org/10.1074/jbc.M304468200
Ding, C., Gao, J., Zhang, S., Jiang, N., Su, D., Huang, X., and Zhang, Z., The basic/helix-loop-helix transcription factor family gene RcbHLH112 is a susceptibility gene in gray mould resistance of rose (Rosa chinensis), Int. J. Mol. Sci., 2023, vol. 24. https://doi.org/10.3390/ijms242216305
Farahani, S., Maleki, M., Ford, R., Mehrabi, R., Kanouni, H., Kema, G.H.J., Naji, A.M., and Talebi, R., Genome-wide association mapping for isolate-specific resistance to Ascochyta rabiei in chickpea (Cicer arietinum L.), Physiol. Mol. Plant Pathol., 2022, vol. 121, p. 101883. https://doi.org/10.1016/j.pmpp.2022.101883
Foresto, E., Carezzano, M.E., Giordano, W., and Bogino, P., Ascochyta blight in chickpea: an update, J. Fungi (Basel), 2023, vol. 9. https://doi.org/10.3390/jof9020203
Ge, Y., Cai, Y.M., Bonneau, L., Rotari, V., Danon, A., McKenzie, E.A., McLellan, H., Mach, L., and Gallois, P., Inhibition of cathepsin B by caspase-3 inhibitors blocks programmed cell death in Arabidopsis, Cell Death Differ., 20163, vol. 23, pp. 1493–1501. https://doi.org/10.1038/cdd.2016.34
Gilroy, E.M., Hein, I., Van Der Hoorn, R., Boevink, P.C., Venter, E., McLellan, H., Kaffarnik, F., Hrubikova, K., Shaw, J., Holeva, M., et al., Involvement of cathepsin B in the plant disease resistance hypersensitive response, Plant J., 2007, vol. 52, pp. 1–13. https://doi.org/10.1111/j.1365-313X.2007.03226.x
Gupta, S., Dong, Y., Dijkwel, P.P., Mueller-Roeber, B., and Gechev, T.S., Genome-wide analysis of ROS antioxidant genes in resurrection species suggest an involvement of distinct ROS detoxification systems during desiccation, Int. J. Mol. Sci., 20193, vol. 20, p. 3101. https://doi.org/10.3390/ijms20123101
Heang, D. and Sassa, H., Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice, PLoS One, 2012, vol. 7, p. e31325. https://doi.org/10.1371/journal.pone.0031325
Houssin, C., Eynard, N., Shechter, E., and Ghazi, A.J.B.e.B.A.-B., Effect of osmotic pressure on membrane energy-linked functions in Escherichia coli, Biochim. Biophys. Acta, Bioenerg., 1991, vol. 1056, pp. 76–84. https://doi.org/10.1016/S0005-2728(05)80075-1
Hu, X., Yang, L., Ren, M., Liu, L., Fu, J., and Cui, H., TGA factors promote plant root growth by modulating redox homeostasis or response, J. Integr. Plant Biol., 2022, vol. 64, pp. 1543–1559. https://doi.org/10.1111/jipb.13310
Hu, D., Guo, Q., Zhang, Y., and Chen, F., Maize methionine sulfoxide reductase genes ZmMSRA2 and ZmMSRA5.1 involved in the tolerance to osmotic or salinity stress in Arabidopsis and maize, Plant Mol. Biol. Rep., 2023, vol. 41, pp. 118–133. https://doi.org/10.1007/s11105-022-01354-6
Ishidoh, K.-i., Kinoshita, H., Ihara, F., and Nihira, T., Efficient and versatile transformation systems in entomopathogenic fungus Lecanicillium species, Curr. Genet., 2014, vol. 60, pp. 99–108. https://doi.org/10.1007/s00294-013-0399-5
Jia, Z.-C., Das, D., Zhang, Y., Fernie, A.R., Liu, Y.-G., Chen, M., and Zhang, J., Plant serine/arginine-rich proteins: versatile players in RNA processing, Planta, 2023, vol. 257, p. 109. https://doi.org/10.1007/s00425-023-04132-0
Jiang, N., Wang, L., Lan, Y., Liu, H., Zhang, X., He, W., Wu, M., Yan, H., and Xiang, Y., Genome-wide identification of the Carya illinoinensis bZIP transcription factor and the potential function of S1-bZIPs in abiotic stresses, Tree Genet. Genomes, 2023, vol. 19, p. 47. https://doi.org/10.1007/s11295-023-01622-w
Jukanti, A.K., Gaur, P.M., Gowda, C., and Chibbar, R.N., Nutritional quality and health benefits of chickpea (Cicer arietinum L.): a review, Br. J. Nutr., 2012, vol. 108, pp. S11–S26. https://doi.org/10.1017/S0007114512000797
Kasote, D.M., Katyare, S.S., Hegde, M.V., and Bae, H., Significance of antioxidant potential of plants and its relevance to therapeutic applications, Int. J. Biol. Sci., 2015, vol. 11, pp. 982–991. https://doi.org/10.7150/ijbs.12096
Kaur, K., Grewal, S.K., Singh, S., Rani, U., and Bhardwaj, R.D., Timing and intensity of upregulated defensive enzymes is a key factor determining resistance in chickpea to Ascochyta rabiei, Physiol. Mol. Plant Pathol., 2021, vol. 114, p. 101645. https://doi.org/10.1016/j.pmpp.2021.101645
Kavousi, H.R., Marashi, H., and Bagheri, A., Expression of phenylpropanoid pathway genes in chickpea defense against race 3 of Ascochyta rabiei, Plant Pathol. J., 2009, vol. 8.
Khaledi, N., Taheri, P., and Falahati-Rastegar, M., Evaluation of resistance and the role of some defense responses in wheat cultivars to Fusarium head blight, J. Plant Prot. Res., 2017, vol. 57. https://doi.org/10.1515/jppr-2017-0054
Kosová, K., Vítámvás, P., Urban, M.O., and Prášil, I.T., Plant proteome responses to salinity stress–comparison of glycophytes and halophytes, Funct. Plant Biol., 2013, vol. 40, pp. 775–786. https://doi.org/10.1071/FP12375
Kufel, J., Diachenko, N., and Golisz, A., Alternative splicing as a key player in the fine-tuning of the immunity response in Arabidopsis, Mol. Plant Pathol., 2022, vol. 23, pp. 1226–1238. https://doi.org/10.1111/mpp.13228
Kumar, R., Tiwari, R.K., Jeevalatha, A., Siddappa, S., Shah, M.A., Sharma, S., Sagar, V., Kumar, M., and Chakrabarti, S.K., Potato apical leaf curl disease: current status and perspectives on a disease caused by tomato leaf curl New Delhi virus, J. Plant Dis. Prot., 2021, vol. 128, pp. 897–911. https://doi.org/10.1007/s41348-021-00463-w
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K., MEGA X: molecular evolutionary genetics analysis across computing platforms, Mol. Biol. Evol., 2018, vol. 35, pp. 1547–1549. https://doi.org/10.1093/molbev/msy096
Ladizinsky, G. and Adler, A., The origin of chickpea Cicer arietinum L., Euphytica, 1976, vol. 25, pp. 211–217. https://doi.org/10.1007/BF00041547
Laugier, E., Tarrago, L., Vieira Dos Santos, C., Eymery, F., Havaux, M., and Rey, P., Arabidopsis thaliana plastidial methionine sulfoxide reductases B, MSRBs, account for most leaf peptide MSR activity and are essential for growth under environmental constraints through a role in the preservation of photosystem antennae, Plant J., 2010, vol. 61, pp. 271–282. https://doi.org/10.1111/j.1365-313X.2009.04053.x
Liu, W., Zhao, C., Liu, L., Huang, D., Ma, C., Li, R., and Huang, L., Genome-wide identification of the TGA gene family in kiwifruit (Actinidia chinensis spp.) and revealing its roles in response to Pseudomonas syringae pv. actinidiae (Psa) infection, Int. J. Biol. Macromol., 2022, vol. 222, pp. 101–113. https://doi.org/10.1016/j.ijbiomac.2022.09.154
Luo, Z., Hu, X., Wu, Z., Liu, X., Wu, C., and Zeng, Q., Identification and expression profiling of TGA transcription factor genes in sugarcane reveals the roles in response to Sporisorium scitamineum infection, Agriculture, 2022, vol. 12. https://doi.org/10.3390/agriculture12101644
Makonya, G.M., Ogola, J.B.O., Gabier, H., Rafudeen, M.S., Muasya, A.M., Crespo, O., Maseko, S., Valentine, A.J., Ottosen, C.-O., Rosenqvist, E., and Chimphango, S.B.M., Proteome changes and associated physiological roles in chickpea (Cicer arietinum) tolerance to heat stress under field conditions, Funct. Plant Biol., 2022, vol. 49, pp. 13–24.
Mi, H., Ebert, D., Muruganujan, A., Mills, C., Albou, L.-P., Mushayamaha, T., and Thomas, P.D., PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API, Nucleic Acids Res., 2021, vol. 49, pp. D394–D403. https://doi.org/10.1093/nar/gkaa1106
Moharramnejad, S. and Valizadeh, M., A key response of grain yield and superoxide dismutase in maize (Zea mays L.) to water deficit stress, J. Plant Physiol. Breed., 2019, vol. 9, pp. 77–84.
Mohsenzadeh Golfazani, M., Taghvaei, M.M., Samizadeh Lahiji, H., Ashery, S., and Raza, A., Investigation of proteins’ interaction network and the expression pattern of genes involved in the ABA biogenesis and antioxidant system under methanol spray in drought-stressed rapeseed, 3 Biotech, 2022, vol. 12, p. 217. https://doi.org/10.1007/s13205-022-03290-4
Mostafaie, A., Theoretical and Practical Guide Protein Electrophoresis in Gel, Press yadavaran, 2003.
Nalçacı, N., Kafadar, F.N., Özkan, A., Turan, A., Başbuğa, S., Anay, A., Mart, D., Öğut, E., Sarpkaya, K., Atik, O., and Can, C., Epiphytotics of chickpea Ascochyta blight in Turkey as influenced by climatic factors, J. Plant Dis. Prot., 2021, vol. 128, pp. 1121–1128. https://doi.org/10.1007/s41348-021-00458-7
Negash Tufa, E., Distribution of Ascochyta Blight (Didymella rabiei) and Evaluation of Chickpea (Cicer rietinum L.) Varieties to the Disease in East Shewa, Central Ethiopia, Haramaya University, 2021.
Nizam, S., Singh, K., and Verma, P.K., Expression of the fluorescent proteins DsRed and EGFP to visualize early events of colonization of the chickpea blight fungus Ascochyta rabiei, Curr. Genet., 2010, vol. 56, pp. 391–399. https://doi.org/10.1007/s00294-010-0305-3
Noshi, M., Mori, D., Tanabe, N., Maruta, T., and Shigeoka, S., Arabidopsis clade IV TGA transcription factors, TGA10 and TGA9, are involved in ROS-mediated responses to bacterial PAMP flg22, Plant Sci., 2016, vol. 252, pp. 12–21. https://doi.org/10.1016/j.plantsci.2016.06.019
Pandey, A.K. and Basandrai, A.K., Will Macrophomina phaseolina spread in legumes due to climate change? A critical review of current knowledge, J. Plant Dis. Prot., 2021, vol. 128, pp. 9–18. https://doi.org/10.1007/s41348-020-00374-2
Pasandideh Arjmand, M., Samizadeh Lahiji, H., Mohsenzadeh Golfazani, M., and Biglouei, M.H., Evaluation of protein’s interaction and the regulatory network of some drought-responsive genes in Canola under drought and re-watering conditions, Physiol. Mol. Biol. Plants, 2023, vol. 29, pp. 1085–1102. https://doi.org/10.1007/s12298-023-01345-1
Rossignol, M., Peltier, J.B., Mock, H.P., Matros, A., Maldonado, A.M., and Jorrín, J.V., Plant proteome analysis: a 2004–2006 update, Proteomics, 2006, vol. 6, pp. 5529–5548. https://doi.org/10.1002/pmic.200600260
Sanjog, T.T., Feroz, K., and Suman, P.S.K., Cathepsin B-like protease from chili pepper revealed by “in silico” approach, Plant Omics, 2016. https://doi.org/10.3316/informit.027795688541121
Shah, J.A., Iqbal, A., Mahmood, M.T., Aslam, M., Abbas, M., and Ahmad, I., Screening of elite chickpea germplasm against Ascochyta blight under controlled conditions, Pak. J. Agric. Res., 2021, vol. 34, pp. 774–780. https://doi.org/10.17582/journal.pjar/2021/34.4.774.780
Singh, R., Kumar, K., Purayannur, S., Chen, W., and Verma, P.K., Ascochyta rabiei: a threat to global chickpea production, Mol. Plant Pathol., 2022, vol. 23, pp. 1241–1261. https://doi.org/10.1111/mpp.13235
Sowders, J.M. and Tanaka, K., A histochemical reporter system to study extracellular ATP response in plants, Front. Plant Sci., 2023, vol. 14, p. 1183335. https://doi.org/10.3389/fpls.2023.1183335
Su, W., Raza, A., Gao, A., Jia, Z., Zhang, Y., Hussain, M.A., Mehmood, S.S., Cheng, Y., Lv, Y., and Zou, X., Genome-wide analysis and expression profile of superoxide dismutase (SOD) gene family in rapeseed (Brassica napus L.) under different hormones and abiotic stress conditions, Antioxidants, 2021, vol. 10, p. 1182. https://doi.org/10.3390/antiox10081182
Szklarczyk, D., Gable, A.L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., Simonovic, M., Doncheva, N.T., Morris, J.H., and Bork, P., STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets, Nucleic Acids Res., 2019, vol. 47, pp. D607–D613. https://doi.org/10.1093/nar/gky1131
Taghvaei, M.M., Lahiji, H.S., and Golfazani, M.M., Evaluation of expression changes, proteins interaction network, and microRNAs targeting catalase and superoxide dismutase genes under cold stress in rapeseed (Brassica napus L.), OCL, 2022, vol, 29.https://doi.org/10.1051/ocl/2021051
Thakur, R., Sharma, S., Devi, R., Sirari, A., Tiwari, R.K., Lal, M.K., and Kumar, R., Exploring the molecular basis of resistance to Botrytis cinerea in chickpea genotypes through biochemical and morphological markers, PeerJ, 2023, vol. 11, p. e15560. https://doi.org/10.7717/peerj.15560
Thompson, J.D., Higgins, D.G., and Gibson, T.J., CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice, Nucleic Acids Res., 1994, vol. 22, pp. 4673–4680. https://doi.org/10.1093/nar/22.22.4673
Tivoli, B. and Banniza, S., Comparison of the epidemiology of ascochyta blights on grain legumes, in Ascochyta Blights of Grain Legumes, Springer, 2007, pp. 59–76. https://doi.org/10.1007/978-1-4020-6065-6_7
Vilela, B., Pagès, M., and Lumbreras, V., Regulation of MAPK signaling and cell death by MAPK phosphatase MKP2, Plant Signal. Behav., 2010, vol. 5, pp. 1497–1500. https://doi.org/10.4161/psb.5.11.13645
Wang, F., Lin, R., Feng, J., Qiu, D., Chen, W., and Xu, S., Wheat bHLH transcription factor gene, TabHLH060, enhances susceptibility of transgenic Arabidopsis thaliana to Pseudomonas syringae, Physiol. Mol. Plant Pathol., 2015a, vol. 90, pp. 123–130. https://doi.org/10.1016/j.pmpp.2015.04.007
Wang, J., Hu, Z., Zhao, T., Yang, Y., Chen, T., Yang, M., Yu, W., and Zhang, B., Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum), BMC Genomics, 2015b, vol. 16, p. 39. https://doi.org/10.1186/s12864-015-1249-2
Wang, G., Fu, X., Zhao, W., Zhang, M., and Chen, F., Ectopic expression of maize plastidic methionine sulfoxide reductase ZmMSRB1 enhances salinity stress tolerance in Arabidopsis thaliana, Plant Mol. Biol. Rep., 2022, vol. 40, pp. 284–295. https://doi.org/10.1007/s11105-021-01320-8
Wen, J., Jiang, F., Liu, M., Zhou, R., Sun, M., Shi, X., Zhu, Z., and Wu, Z., Identification and expression analysis of cathepsin B-like protease 2 genes in tomato at abiotic stresses especially at high temperature, Sci. Hortic., 2021, vol. 277, p. 109799. https://doi.org/10.1016/j.scienta.2020.109799
Xin, J., Li, C., Ning, K., Qin, Y., Shang, J.-X., and Sun, Y., AtPFA-DSP3, an atypical dual-specificity protein tyrosine phosphatase, affects salt stress response by modulating MPK3 and MPK6 activity, Plant, Cell Environ., 2021, vol. 44, pp. 1534–1548. https://doi.org/10.1111/pce.14002
Xu, R., Zhou, J., Zheng, E., Yang, Y., Li, D., Chen, Y., Yan, C., Chen, J., and Wang, X., Systemic acquired resistance plays a major role in bacterial blight resistance in a progeny of somatic hybrids of cultivated rice (Oryza sativa L.) and wild rice (Oryza meyeriana L.), J. Plant Dis. Prot., 2023, vol. 128, pp. 1023–1040. https://doi.org/10.1007/s41348-021-00457-8
Xu, S., Zhang, Z., Jing, B., Gannon, P., Ding, J., Xu, F., Li, X., and Zhang, Y., Transportin-SR is required for proper splicing of resistance genes and plant immunity, PLoS Genet., 2011, vol. 7, p. e1002159. https://doi.org/10.1371/journal.pgen.1002159
Yasuda, M., Ishikawa, A., Jikumaru, Y., Seki, M., Umezawa, T., Asami, T., Maruyama-Nakashita, A., Kudo, T., Shinozaki, K., and Yoshida, S., Antagonistic interaction between systemic acquired resistance and the abscisic acid–mediated abiotic stress response in Arabidopsis, Plant Cell, 2008, vol. 20, pp. 1678–1692. https://doi.org/10.1105/tpc.107.054296
Zhu, J., Ding, P., Li, Q., Gao, Y., Chen, F., and Xia, G., Molecular characterization and expression profile of methionine sulfoxide reductase gene family in maize (Zea mays) under abiotic stresses, Gene, 2021, vol. 562, pp. 159–168. https://doi.org/10.1016/j.gene.2015.02.066
Zuo, Z.-F., Lee, H.-Y., and Kang, H.-G., Basic helix-loop-helix transcription factors: regulators for plant growth development and abiotic stress responses, Int. J. Mol. Sci., 2023, vol. 24. https://doi.org/10.3390/ijms24021419
Funding
This work was supported by ongoing institutional funding. No additional grants to carry out or direct this particular research were obtained.
Author information
Authors and Affiliations
Contributions
M.M.G, planned and designed the study, drafted the manuscript, and participated in all the experiments. Y.S. and M.M.T. participated in all experiments and data analysis. H.S.L. and A.M. and A.R. participated in study design and supervised the study. All authors participated in revising and finalizing the manuscript.
Corresponding author
Ethics declarations
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This work does not contain any studies involving human and animal subjects.
CONFLICT OF INTEREST
The authors of this work declare that they have no conflicts of interest.
Additional information
Publisher’s Note.
Pleiades Publishing remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
About this article
Cite this article
Shafiei, Y., Golfazani, M.M., Mostafaie, A. et al. Evaluation of Superoxide Dismutase Isoforms Activity and Defense System-Related Proteins’ Expression in Ascochyta Blight-Infected Chickpea Using 2D Electrophoresis Technique. Biol Bull Russ Acad Sci (2024). https://doi.org/10.1134/S1062359023603336
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1134/S1062359023603336