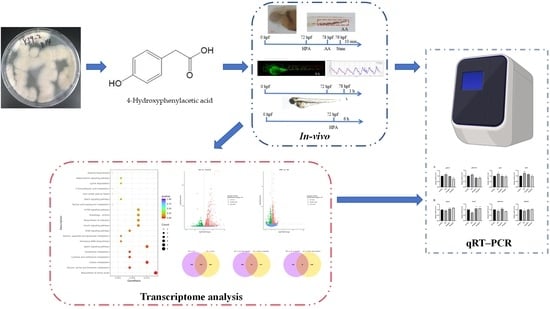
Marine-Fungus-Derived Natural Compound 4-Hydroxyphenylacetic Acid Induces Autophagy to Exert Antithrombotic Effects in Zebrafish
Abstract
:1. Introduction
2. Results
2.1. Safe Concentration of HPA for Zebrafish
2.2. Antithrombotic Activity of HPA
2.3. Functional Classification and Annotation of The Transcriptome
2.4. qRT–PCR Analysis
3. Discussion
4. Experimental Methods
4.1. General Experimental Procedures
4.2. Fungal Material
4.3. Fermentation, Extraction, and Isolation
4.4. Safety Concentration Test
4.5. Bioassay Protocols
4.5.1. Zebrafish Maintenance
4.5.2. Chemical Treatment
4.5.3. Caudal Thrombus Assay
4.5.4. Cardiac Erythrocyte Assay
4.5.5. Caudal Blood Flow Velocity and Heart Rate Assay
4.5.6. Inflammatory Cell Assay
4.6. Transcriptome Analysis
4.7. qRT–PCR Assay
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Barzkar, N.; Jahromi, S.T.; Vianello, F. Marine microbial fibrinolytic enzymes: An overview of source, production, biochemical properties and thrombolytic activity. Mar. Drugs 2022, 20, 46. [Google Scholar] [CrossRef] [PubMed]
- Heron, M. Deaths: Leading causes for 2019. Natl. Vital Stat. Rep. 2021, 70, 1–114. [Google Scholar] [PubMed]
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. GBD-NHLBI-JACC global burden of cardiovascular diseases writing group. Global burden of cardiovascular diseases and risk factors, 1990–2019: Update from the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fang, C.; Yao, M.; Wu, D.; Chen, M.; Guo, T.; Mo, J. Research progress of NF-κB signaling pathway and thrombosis. Front. Immunol. 2023, 14, 1257988. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.N.; Xie, C.Y. Advancing investigate of deep vein thrombosis for inflammation and signaling pathway. Chin. J. Immunol. 2020, 36, 113–118. [Google Scholar]
- Su, X.L.; Su, W.; Wang, Y.; Wang, Y.H.; Ming, X.; Kong, Y. The pyrrolidinoindoline alkaloid Psm2 inhibits platelet aggregation and thrombus formation by affecting PI3K/Akt signaling. Acta Pharmacol. Sin. 2016, 37, 1208–1217. [Google Scholar] [CrossRef] [PubMed]
- Mu, F.; Jiang, Y.; Ao, F.; Wu, H.; You, Q.; Chen, Z. RapaLink-1 plays an antithrombotic role in antiphospholipid syndrome by improving autophagy both in vivo and vitro. Biochem. Biophys. Res. Commun. 2020, 525, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, M.; Oota, E.; Soeda, T.; Masumo, K.; Tomita, Y.; Kato, T.; Imanishi, T. Emergency cardiac surgery and heparin resistance in a patient with essential thrombocythemia. JA Clin. Rep. 2016, 2, 35. [Google Scholar] [CrossRef] [PubMed]
- Stupnisek, M.; Franjic, S.; Drmic, D.; Hrelec, M.; Kolenc, D.; Radic, B.; Bojic, D.; Vcev, A.; Seiwerth, S.; Sikiric, P. Pentadecapeptide BPC 157 reduces bleeding time and thrombocytopenia after amputation in rats treated with heparin, warfarin or aspirin. Thromb. Res. 2012, 129, 652–659. [Google Scholar] [CrossRef]
- Barbosa, F.; Pinto, E.; Kijjoa, A.; Pinto, M.; Sousa, E. Targeting antimicrobial drug resistance with marine natural products. Int. J. Antimicrob. Agents 2020, 56, 106005. [Google Scholar] [CrossRef]
- Ma, H.G.; Liu, Q.; Zhu, G.L.; Liu, H.S.; Zhu, W.M. Marine natural products sourced from marine-derived Penicillium fungi. J. Asian Nat. Prod. Res. 2016, 18, 92–115. [Google Scholar] [CrossRef]
- Kuvarina, A.E.; Gavryushina, I.A.; Sykonnikov, M.A.; Efimenko, T.A.; Markelova, N.N.; Bilanenko, E.N.; Bondarenko, S.A.; Kokaeva, L.Y.; Timofeeva, A.V.; Serebryakova, M.V.; et al. Exploring peptaibol’s profile, antifungal, and antitumor activity of Emericellipsin A of Emericellopsis Species from Soda and Saline Soils. Molecules 2022, 27, 1736. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Saha, S. The genus Simplicillium and Emericellopsis: A review of phytochemistry and pharmacology. Biotechnol. Appl. Biochem. 2022, 69, 2229–2239. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.R.; Yang, B.; Li, H.M.; Fu, C.Y.; Liu, Y.H.; Li, Y.Q. Research about the secondary metabolites of soft coral-derivedepiphytic fungus Emericellopsis sp. SCSIO41202 from the South China Sea. Acta Med. Sin. 2022, 35, 7–12. [Google Scholar]
- Inostroza, A.; Lara, L.; Paz, C.; Perez, A.; Galleguillos, F.; Hernandez, V.; Becerra, J.; González-Rocha, G.; Silva, M. Antibiotic activity of Emerimicin IV isolated from Emericellopsis minima from Talcahuano Bay, Chile. Nat. Prod. Res. 2018, 32, 1361–1364. [Google Scholar] [CrossRef]
- Baranova, A.A.; Georgieva, M.L.; Bilanenko, E.N.; Andreev, Y.A.; Rogozhin, E.A.; Sadykova, V.S. Antimicrobial potential of alkalophilic micromycetes Emericellopsis alkalina. Appl. Biochem. Microbiol. 2017, 53, 703–710. [Google Scholar] [CrossRef]
- Kuvarina, A.E.; Gavryushina, I.A.; Kulko, A.B.; Ivanov, I.A.; Rogozhin, E.A.; Georgieva, M.L.; Sadykova, V.S. The Emericellipsins A-E from an alkalophilic fungus Emericellopsis alkalina show potent activity against multidrug-resistant pathogenic fungi. J. Fungi 2021, 7, 153. [Google Scholar] [CrossRef] [PubMed]
- Abraúl, M.; Alves, A.; Hilário, S.; Melo, T.; Conde, T.; Domingues, M.R.; Rey, F. Evaluation of lipid extracts from the marine fungi Emericellopsis cladophorae and Zalerion maritima as a source of anti-Inflammatory, antioxidant and antibacterial compounds. Mar. Drugs 2023, 21, 199. [Google Scholar] [CrossRef] [PubMed]
- Santoriello, C.; Zon, L.I. Hooked! Modeling human disease in zebrafish. J. Clin. Investig. 2012, 122, 2337–2343. [Google Scholar] [CrossRef] [PubMed]
- Zon, L.I.; Peterson, R.T. In vivo drug discovery in the zebrafish. Nat. Rev. Drug Discov. 2005, 4, 35–44. [Google Scholar] [CrossRef]
- Chico, T.J.; Ingham, P.W.; Crossman, D.C. Modeling cardiovascular disease in the zebrafish. Trends Cardiovasc. Med. 2008, 18, 150–155. [Google Scholar] [PubMed]
- Zhu, X.Y.; Liu, H.C.; Guo, S.Y.; Xia, B.; Song, R.S.; Lao, Q.C.; Xuan, Y.X.; Li, C.Q. A zebrafish thrombosis model for assessing antithrombotic drugs. Zebrafish 2016, 13, 335–344. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Li, M.C.; Li, Y.Y.; Yan, H.Y.; Han, L.W.; Huang, X.; Li, S.W.; Wang, P.; Shi, Y.P.; Wang, L.; et al. Screening of antithrombotic active Fractions and chemical constituents in Sparganii Rhizoma based on zebrafish model. Chin. J. Hosp. Pharm. 2019, 39, 1439–1443. [Google Scholar]
- Zhang, Y.; Zhu, X.Y.; Guo, S.Y.; Li, C.Q. Phenylhydrazine-induced modelling of zebrafish thrombosis. Lab. Anim. Comp. Med. 2015, 35, 27–31. [Google Scholar]
- Chen, Y.S. Studies on metabolic products of Hypochnus sasakii Shirai: Isolation of p-hydroxyphenylacetic acid and its physiological activity. Agric. Biol. Chem. 1958, 22, 136–142. [Google Scholar]
- Ohtani, K.; Fujioka, S.; Kawano, T.; Shimada, A.; Kimura, Y. Nematicidal activities of 4-hydroxyphenylacetic acid and oidiolactone D produced by the fungus Oidiodendron sp. Z. Naturforsch. C. J. Biosci. 2011, 66, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Flubacher, N.; Baltenweck, R.; Hugueney, P.; Fischer, J.; Thines, E.; Riemann, M.; Nick, P.; Khattab, I.M. The fungal metabolite 4-hydroxyphenylacetic acid from Neofusicoccum parvum modulates defence responses in grapevine. Plant Cell Environ. 2023, 46, 3575–3591. [Google Scholar] [CrossRef] [PubMed]
- Fries, L.; Iwasaki, H. p-Hydroxyphenylacetic acid and other phenolic compounds as growth stimulators of the red alga Porphyra tenera. Plant Sci. Lett. 1976, 6, 299–307. [Google Scholar] [CrossRef]
- An, S.; Yao, Y.; Wu, J.; Hu, H.; Wu, J.; Sun, M.; Li, J.; Zhang, Y.; Li, L.; Qiu, W.; et al. Gut-derived 4-hydroxyphenylacetic acid attenuates sepsis-induced acute kidney injury by upregulating ARC to inhibit necroptosis. Biochim. Biophys. Acta Mol. Basis Dis. 2024, 1870, 166876. [Google Scholar] [CrossRef]
- Liu, Z.; Xi, R.; Zhang, Z.; Li, W.; Liu, Y.; Jin, F.; Wang, X. 4-Hydroxyphenylacetic acid attenuated inflammation and edema via suppressing HIF-1α in seawater aspiration-induced lung injury in rats. Int. J. Mol. Sci. 2014, 15, 12861–12884. [Google Scholar] [CrossRef] [PubMed]
- Takahama, U.; Oniki, T.; Murata, H. The presence of 4-hydroxyphenylacetic acid in human saliva and the possibility of its nitration by salivary nitrite in the stomach. FEBS Lett. 2002, 518, 116–118. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Jiang, Z.; Chang, X.; Xue, H.; Yahefu, W.; Zhang, X. 4-Hydroxyphenylacetic acid prevents acute APAP-induced liver injury by increasing phase II and antioxidant enzymes in mice. Front. Pharmacol. 2018, 9, 653. [Google Scholar] [CrossRef] [PubMed]
- Fries, L. Growth regulating effects of phenylacetic acid and p-hydroxyphenylacetic acid on Fucus spiralis L. (Phaeophyceae, Fucales) in axenic culture. Phycologia 1977, 16, 451–455. [Google Scholar] [CrossRef]
- Vissiennon, C.; Nieber, K.; Kelber, O.; Butterweck, V. Route of administration determines the anxiolytic activity of the flavonols kaempferol, quercetin and myricetin—are they prodrugs? J. Nutr. Biochem. 2012, 23, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Skorochod, I.; Kurdish, I. P-62–Quantum-chemical study of the antioxidant mechanisms of 4-hydroxyphenylacetic acid a metabolite of Bacillus subtilis IMV V-7023 in gas phase. Free Radic. Biol. Med. 2018, 120, S63. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, C.; Zhang, G.; Zhan, H.; Liu, B.; Li, C.; Wang, L.; Wang, H.; Wang, J. Antimicrobial mechanism of 4-hydroxyphenylacetic acid on Listeria monocytogenes membrane and virulence. Biochem. Biophys. Res. Commun. 2021, 572, 145–150. [Google Scholar]
- Ko, H.S.; Jin, R.D.; Krishnan, H.B.; Lee, S.B.; Kim, K.Y. Biocontrol ability of Lysobacter antibioticus HS124 against Phytophthora blight is mediated by the production of 4-hydroxyphenylacetic acid and several lytic enzymes. Curr. Microbiol. 2009, 59, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Sapkota, K.; Park, S.E.; Kim, S.; Kim, S.J. Thrombolytic, anticoagulant and antiplatelet activities of codiase, a bi-functional fibrinolytic enzyme from Codium fragile. Biochimie 2013, 95, 1266–1277. [Google Scholar] [CrossRef] [PubMed]
- Mourão, P.A. Perspective on the use of sulfated polysaccharides from marine organisms as a source of new antithrombotic drugs. Mar. Drugs 2015, 13, 2770–2784. [Google Scholar] [CrossRef] [PubMed]
- Doshi, G.; Nailwal, N. A Review on molecular mechanisms and patents of marine-derived anti-thrombotic agents. Curr. Drug Targets 2021, 22, 318–335. [Google Scholar] [CrossRef] [PubMed]
- Haque, N.; Parveen, S.; Tang, T.; Wei, J.; Huang, Z. Marine natural products in clinical use. Mar. Drugs 2022, 20, 528. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.P.; Zhang, Y.G.; Li, H.N.; Kong, H.T.; Zhang, S.S.; Zhang, X.M.; Li, X.B.; Liu, K.C.; Han, L.W.; Tian, Q.P. Discovery and identification of antithrombotic chemical markers in Gardenia Fructus by herbal metabolomics and zebrafish model. J. Ethnopharmacol. 2020, 253, 112679. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zhao, X.; Liu, H.; Wang, Y.; Zhao, C.; Zhao, T.; Zhao, B.; Wang, Y. Identification of a quality marker (Q-Marker) of danhong injection by the zebrafish thrombosis model. Molecules 2017, 22, 1443. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, P.; Zhang, S.; Zhang, X.; Wang, L.; Zhang, Y.; Li, X.; Liu, K. Study on the mechanism of the danggui-chuanxiong herb pair on treating thrombus through network pharmacology and zebrafish models. ACS Omega 2021, 6, 14677–14691. [Google Scholar] [CrossRef]
- Zhu, H.; Lan, C.; Zhao, D.; Wang, N.; Du, D.; Luo, H.; Lu, H.; Peng, Z.; Wang, Y.; Qiao, Z.; et al. Wuliangye Baijiu but not ethanol reduces cardiovascular disease risks in a zebrafish thrombosis model. NPJ Sci. Food 2022, 6, 55. [Google Scholar] [CrossRef] [PubMed]
- Stark, K.; Massberg, S. Interplay between inflammation and thrombosis in cardiovascular pathology. Nat. Rev. Cardiol. 2021, 18, 666–682. [Google Scholar] [CrossRef] [PubMed]
- Schrijvers, D.M.; De Meyer, G.R.Y.; Martinet, W. Autophagy in atherosclerosis: A potential drug target for plaque stabilization. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2787–2791. [Google Scholar] [CrossRef] [PubMed]
- Denton, D.; Xu, T.; Kumar, S. Autophagy as a pro-death pathway. Immunol. Cell Biol. 2015, 93, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Martinet, W.; De Meyer, G.R.Y. Autophagy in atherosclerosis: A cell survival and death phenomenon with therapeutic potential. Circ. Res. 2009, 104, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Cosemans, J.M.; Angelillo-Scherrer, A.; Mattheij, N.J.; Heemskerk, J.W. The effects of arterial flow on platelet activation, thrombus growth, and stabilization. Cardiovasc. Res. 2013, 99, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Koupenova, M.; Kehrel, B.E.; Corkrey, H.A.; Freedman, J.E. Thrombosis and platelets: An update. Eur. Heart J. 2017, 38, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Ampofo, E.; Später, T.; Müller, I.; Eichler, H.; Menger, M.D.; Laschke, M.W. The marine-derived kinase inhibitor fascaplysin exerts anti-thrombotic activity. Mar. Drugs 2015, 13, 6774–6791. [Google Scholar] [CrossRef]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K pathway in human disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Liu, M.; Xu, R.; Wang, G.; Li, J.; Chen, P.; Ma, W.; Mwangi, J.; Lu, Q.; Duan, Z.; et al. The 14-3-3ζ-c-Src-integrin-β3 complex is vital for platelet activation. Blood 2020, 136, 974–988. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Tian, Z.Z.; Ma, X.L.; Zuo, X.; Ya, F.L.; Yang, Y. Fruitflow, a water-soluble tomato concentrate, inhibits platelet activation, aggregation and thrombosis by regulating the signaling pathway of PI3K/Akt and MAPKs. J. Sun Yat-Sen Univ. Med. Sci. 2020, 41, 243–250. [Google Scholar]
- Aslan, J.E.; Tormoen, G.W.; Loren, C.P.; Pang, J.; McCarty, O.J. S6K1 and mTOR regulate Rac1-driven platelet activation and aggregation. Blood 2011, 118, 3129–3136. [Google Scholar] [CrossRef]
- Fang, S.; Wan, X.; Zou, X.; Sun, S.; Hao, X.; Liang, C.; Zhang, Z.; Zhang, F.; Sun, B.; Li, H.; et al. Arsenic trioxide induces macrophage autophagy and atheroprotection by regulating ROS-dependent TFEB nuclear translocation and AKT/mTOR pathway. Cell Death Dis. 2021, 12, 88. [Google Scholar] [CrossRef] [PubMed]
- Shao, B.Z.; Han, B.Z.; Zeng, Y.X.; Su, D.F.; Liu, C. The roles of macrophage autophagy in atherosclerosis. Acta Pharmacol. Sin. 2016, 37, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Xu, X.S.; Meng, X.; Li, Y.L.; Li, J.F.; Chen, W.Q. Autophagy of monocytes attenuates the vulnerability of coronary atherosclerotic plaques. Coron. Artery Dis. 2013, 24, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fang, Y.; Huang, Q.; Xu, P.; Lenahan, C.; Lu, J.; Zheng, J.; Dong, X.; Shao, A.; Zhang, J. An updated review of autophagy in ischemic stroke: From mechanisms to therapies. Exp. Neurol. 2021, 340, 113684. [Google Scholar] [CrossRef] [PubMed]
- Lekli, I.; Haines, D.D.; Balla, G.; Tosaki, A. Autophagy: An adaptive physiological countermeasure to cellular senescence and ischaemia/reperfusion-associated cardiac arrhythmias. J. Cell. Mol. Med. 2017, 21, 1058–1072. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.Q.; Ren, C.; Xia, Z.F.; Yao, Y.M. Organelle-specific autophagy in inflammatory diseases: A potential therapeutic target underlying the quality control of multiple organelles. Autophagy 2021, 17, 385–401. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Xie, X.; Luo, X.; Song, Y.L.; Zhao, Y.W.; Huang, W.Z.; Wang, Z.Z.; Xiao, W. Chemical constituents from Rhodiola wallichiana var. cholaensis (I). Chin. Tradit. Herb. Drugs 2015, 46, 3471–3474. [Google Scholar]
- Parkhomchuk, D.; Borodina, T.; Amstislavskiy, V.; Banaru, M.; Hallen, L.; Krobitsch, S.; Lehrach, H.; Soldatov, A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009, 37, e123. [Google Scholar] [CrossRef]










| Gene Name | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| β-actin | ACCACGGCCGAAAGAGAAAT | GATACCGCAAGATTCCATACCC |
| pik3r1 | TTCAGACATCAGCCCACAAGTT | GCCTGACTGTCACTCCATTCG |
| pik3r3b | TGACACCAGTAAACGGCAATG | TGTTTCCACCTTTCCTTAGCG |
| akt1 | GTGAAGGAGAAAGCAACAGGCA | ATGACTTCAGAGCCGTCAGGAA |
| mtor | ACGCTGCACGCACTGATTC | AGAGTCAAATGGTCATAGTCAGGG |
| ulk1b | GTGCCTTCGCAGTGGTTTT | TGCTGTAGGAATACTCGGATGGT |
| eif2ak3 | AACCGTCTGGAGACACCGAT | TGCTGAGGCTTGAGGATACCA |
| becn1 | AGATGGCGTGGCTCGAAAAT | GCACTCCTCACAAAGTGGGT |
| ambra1a | ACGCACTCATCCGTCAATAGG | TTCACCGACGCATTACTGATTTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xin, S.; Zhang, M.; Li, P.; Wang, L.; Zhang, X.; Zhang, S.; Mu, Z.; Lin, H.; Li, X.; Liu, K. Marine-Fungus-Derived Natural Compound 4-Hydroxyphenylacetic Acid Induces Autophagy to Exert Antithrombotic Effects in Zebrafish. Mar. Drugs 2024, 22, 148. https://doi.org/10.3390/md22040148
Xin S, Zhang M, Li P, Wang L, Zhang X, Zhang S, Mu Z, Lin H, Li X, Liu K. Marine-Fungus-Derived Natural Compound 4-Hydroxyphenylacetic Acid Induces Autophagy to Exert Antithrombotic Effects in Zebrafish. Marine Drugs. 2024; 22(4):148. https://doi.org/10.3390/md22040148
Chicago/Turabian StyleXin, Shaoshuai, Mengqi Zhang, Peihai Li, Lizhen Wang, Xuanming Zhang, Shanshan Zhang, Zhenqiang Mu, Houwen Lin, Xiaobin Li, and Kechun Liu. 2024. "Marine-Fungus-Derived Natural Compound 4-Hydroxyphenylacetic Acid Induces Autophagy to Exert Antithrombotic Effects in Zebrafish" Marine Drugs 22, no. 4: 148. https://doi.org/10.3390/md22040148