MALDI Mass Spectrometry Imaging: A Potential Game-Changer in a Modern Microbiology
Abstract
:1. Modern Microbiology and Current Limitations
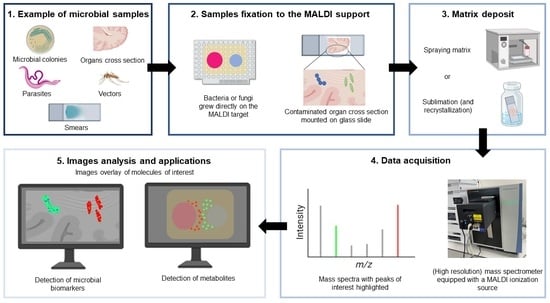
2. Mass Spectrometry Imaging: A Picture Is Worth a Thousand Words
3. Where Are We with MALDI Mass Spectrometry Imaging in Microbiology?
3.1. Microbial and Host-Microbes’ Interactions
| Class | Main Objective | Organisms | Analyzer | Lateral Resolution | Year | References |
|---|---|---|---|---|---|---|
| Bacteria | Biofilm formation | Bacillus | TOF | NA | 2015 | [72] |
| TOF/TOF | 250 µm | 2016 | [84] | |||
| Pseudomonas | TOF/TOF | 100 µm | 2014 | [85] | ||
| Pseudomonas Staphylococcus | TOF | 50 µm | 2016 | [86] | ||
| Listeria | TOF | 100 µm | 2018 | [87] | ||
| Biomarker identification | Mycobacterium | LTQ-orbitrap | 50 µm | 2018 | [88] | |
| Drug effect | Pseudomonas | TOF | 500 µm | 2015 | [70] | |
| Host-microbes’ interactions | Intracellular microbial communities of Bathymodiolus | Q-orbitrap | 3 µm | 2020 | [89] | |
| Pseudonocardia | LTQ-orbitrap | 75 µm | 2017 | [90] | ||
| Gut microbiota | NA | 50 µm | 2022 | [91] | ||
| Gut microbiota | TOF/TOF | NA | 2012 | [67] | ||
| Streptomyces | LTQ-orbitrap | NA | 2011 | [92] | ||
| Francisella | FT-ICR | 75 µm | 2017 | [93] | ||
| Escherichia Pseudomonas | TOF/TOF | NA | 2020 | [94] | ||
| Microbial interactions | Pseudomonas Escherichia Staphylococcus | Q-orbitrap | 50 µm | 2019 | [76] | |
| Bacillus Streptomyces | TOF/TOF | NA | 2012 | [80] | ||
| Lysobacter Bacillus Pseudomonas Streptomyces Staphylococcus Mycobacterium | TOF | 200–800 µm | 2012 | [64] | ||
| Lysobacter | TOF/TOF | 50 µm | 2015 | [61] | ||
| Bacillus Staphylococcus | TOF | 200–350 µm | 2011 | [66] | ||
| Paenibacillus Bacillus | TOF/TOF | 300 µm | 2019 | [63] | ||
| Bacillus | TOF | NA | 2010 | [65] | ||
| Pseudomonas | TOF | 400 µm | 2016 | [62] | ||
| Pseudomonas Aspergillus | TOF/TOF | 400–600 µm | 2012 | [68] | ||
| Bacillus Streptomyces | TOF/TOF | NA | 2009 | [60] | ||
| Paenibacillus | FT-ICR | NA | 2013 | [73] | ||
| Sample preparation | Myxobacteria | TOF/TOF | 350 µm | 2015 | [71] | |
| Bacillus | TOF/TOF | 200 µm | 2016 | [95] | ||
| Spatial distribution | Microbial mat | FT-ICR | 25 µm | 2020 | [96] | |
| Fungi | Microbial interactions | Trichoderma Rhizoctonia | TOF/TOF | NA | 2016 | [74] |
| Host-microbes’ interactions | Aspergillus | FT-ICR | 50 µm | 2020 | [78] | |
| Aspergillus | TOF/TOF | 500 µm | 2019 | [79] | ||
| Sample preparation | Aspergillus | TOF/TOF | 35 µm | 2014 | [97] | |
| Parasites & Vectors | Chemical characterization | Schistosoma | LTQ | 50 µm | 2014 | [98] |
| Q-orbitrap | 5 µm | 2020 | [99] | |||
| Anopheles | Q-orbitrap | 12 µm | 2015 | [100] | ||
| Drug distribution | Schistosoma | Q-orbitrap | 5–9 µm | 2021 | [101] | |
| Fasciola | Q-orbitrap | 10 µm | 2022 | [102] | ||
| Q-orbitrap | 10 µm | 2020 | [103] | |||
| Host-microbes’ interactions | Schistosoma | Q-orbitrap | 10 µm | 2022 | [104] | |
| Parasitic nematodes | TOF & Q-orbitrap | 25–8 µm | 2021 | [105] | ||
| Protozoa | Method improvement | Paramecium | NA | 1.4 µm | 2016 | [106] |
| Host-microbes’ interactions | Leishmania | FT-ICR | 50 µm | 2021 | [107] | |
| Viruses | Biomarker identification | MDV | TOF/TOF | NA | 2019 | [108] |
| Parvovirus | TOF/TOF | 50 µm | 2022 | [109] | ||
| HPV | TOF | NA | 2011 | [110] | ||
| Microbial interactions | EhV201 | FT-ICR | 100 µm | 2019 | [77] |
3.2. Biofilms and Microbial Mats Formation
3.3. Chemical Characterization and Visualization
3.4. Drug Distribution and Effect
3.5. Biomarker Identification and Diagnostics
4. How Could It Be Applied to Overcome Current Limitations?
5. Outlooks and Future Challenges for MALDI-Imaging in Microbiology
5.1. Pragmatic Aspects
5.2. The Importance of Standardizing Sample Preparation
5.3. Hardware and Bioinformatic Infrastructure: A Subject of Matter
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feucherolles, M. MALDI-TOF-Enabled Subtyping and Antimicrobial Resistance Screening of the Food- and Waterborne Pathogen Campylobacter. Ph.D. Thesis, University of Luxembourg, Luxembourg, 2022. [Google Scholar]
- Bertelli, C.; Greub, G. Rapid Bacterial Genome Sequencing: Methods and Applications in Clinical Microbiology. Clin. Microbiol. Infect. 2013, 19, 803–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavezzo, E.; Barzon, L.; Toppo, S.; Palù, G. Third Generation Sequencing Technologies Applied to Diagnostic Microbiology: Benefits and Challenges in Applications and Data Analysis. Expert Rev. Mol. Diagn. 2016, 16, 1011–1023. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.; Oliveira, J.; Sousa, M. Bioinformatics and Computational Tools for Next-Generation Sequencing Analysis in Clinical Genetics. J. Clin. Med. 2020, 9, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF Mass Spectrometry: An Emerging Technology for Microbial Identification and Diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chabriere, E.; Bassène, H.; Drancourt, M.; Sokhna, C. MALDI-TOF-MS and Point of Care Are Disruptive Diagnostic Tools in Africa. New Microbes New Infect. 2018, 26, S83–S88. [Google Scholar] [CrossRef] [PubMed]
- Carbonnelle, E.; Mesquita, C.; Bille, E.; Day, N.; Dauphin, B.; Beretti, J.L.; Ferroni, A.; Gutmann, L.; Nassif, X. MALDI-TOF Mass Spectrometry Tools for Bacterial Identification in Clinical Microbiology Laboratory. Clin. Biochem. 2011, 44, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Rotcheewaphan, S.; Lemon, J.K.; Desai, U.U.; Henderson, C.M.; Zelazny, A.M. Rapid One-Step Protein Extraction Method for the Identification of Mycobacteria Using MALDI-TOF MS. Diagn. Microbiol. Infect. Dis. 2019, 94, 355–360. [Google Scholar] [CrossRef]
- Normand, A.C.; Becker, P.; Gabriel, F.; Cassagne, C.; Accoceberry, I.; Gari-Toussaint, M.; Hasseine, L.; De Geyter, D.; Pierard, D.; Surmont, I.; et al. Validation of a New Web Application for Identification of Fungi by Use of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2017, 55, 2661–2670. [Google Scholar] [CrossRef] [Green Version]
- Iles, R.K.; Iles, J.K.; Zmuidinaite, R.; Roberts, M. A How to Guide: Clinical Population Test Development and Authorization of MALDI-ToF Mass Spectrometry-Based Screening Tests for Viral Infections. Viruses 2022, 14, 1958. [Google Scholar] [CrossRef]
- Camarasa, C.G.; Cobo, F. Application of MALDI-TOF Mass Spectrometry in Clinical Virology. In The Use of Mass Spectrometry Technology (MALDI-TOF) in Clinical Microbiology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 167–180. ISBN 9780128144527. [Google Scholar]
- Iles, R.K.; Zmuidinaite, R.; Iles, J.K.; Carnell, G.; Sampson, A.; Heeney, J.L. A Clinical MALDI-ToF Mass Spectrometry Assay for SARS-CoV-2: Rational Design and Multi-Disciplinary Team Work. Diagnostics 2020, 10, 746. [Google Scholar] [CrossRef]
- El Hamzaoui, B.; Laroche, M.; Almeras, L.; Bérenger, J.M.; Raoult, D.; Parola, P. Detection of Bartonella Spp. in Fleas by MALDI-TOF MS. PLoS Negl. Trop. Dis. 2018, 12, e0006189. [Google Scholar] [CrossRef] [PubMed]
- Boucheikhchoukh, M.; Laroche, M.; Aouadi, A.; Dib, L.; Benakhla, A.; Raoult, D.; Parola, P. MALDI-TOF MS Identification of Ticks of Domestic and Wild Animals in Algeria and Molecular Detection of Associated Microorganisms. Comp. Immunol. Microbiol. Infect. Dis. 2018, 57, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Sy, I.; Margardt, L.; Ngbede, E.O.; Adah, M.I.; Yusuf, S.T.; Keiser, J.; Rehner, J.; Utzinger, J.; Poppert, S.; Becker, S.L. Identification of Adult Fasciola Spp. Using Matrix-Assisted Laser/Desorption Ionization Time-of-Flight (Maldi-Tof) Mass Spectrometry. Microorganisms 2021, 9, 82. [Google Scholar] [CrossRef]
- Wendel, T.; Feucherolles, M.; Rehner, J.; Keiser, J.; Poppert, S.; Utzinger, J.; Becker, S.L.; Sy, I. Evaluating Different Storage Media for Identification of Taenia saginata Proglottids Using MALDI-TOF Mass Spectrometry. Microorganisms 2021, 9, 2006. [Google Scholar] [CrossRef] [PubMed]
- Feucherolles, M.; Poppert, S.; Utzinger, J.; Becker, S.L. MALDI-TOF Mass Spectrometry as a Diagnostic Tool in Human and Veterinary Helminthology: A Systematic Review. Parasites Vectors 2019, 12, 245. [Google Scholar] [CrossRef]
- Feucherolles, M.; Cauchie, H.; Penny, C. MALDI-TOF Mass Spectrometry and Specific Biomarkers: Potential New Key for Swift Identification of Antimicrobial Resistance in Foodborne Pathogens. Microorganisms 2019, 7, 593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feucherolles, M.; Nennig, M.; Becker, S.L.; Martiny, D.; Losch, S.; Penny, C.; Cauchie, H.M.; Ragimbeau, C. Investigation of MALDI - TOF Mass Spectrometry for Assessing the Molecular Diversity of Campylobacter jejuni and Comparison with MLST and CgMLST: A Luxembourg One-Health Study. Diagnostics 2021, 11, 1949. [Google Scholar] [CrossRef]
- Godmer, A.; Benzerara, Y.; Normand, A.C.; Veziris, N.; Gallah, S.; Eckert, C.; Morand, P.; Piarroux, R.; Aubry, A. Revisiting Species Identification within the Enterobacter cloacae Complex by Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry. Microbiol. Spectr. 2021, 9, e00661-21. [Google Scholar] [CrossRef]
- Saleeb, P.G.; Drake, S.K.; Murray, P.R.; Zelazny, A.M. Identification of Mycobacteria in Solid-Culture Media by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2011, 49, 1790–1794. [Google Scholar] [CrossRef] [Green Version]
- Neuschlova, M.; Vladarova, M.; Kompanikova, J.; Sadlonova, V.; Novakova, E. Identification of Mycobacterium Species by MALDI-TOF Mass Spectrometry. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2017; Volume 1021, pp. 37–42. [Google Scholar]
- Body, B.A.; Beard, M.A.; Slechta, E.S.; Hanson, K.E.; Barker, A.P.; Babady, N.E.; McMillen, T.; Tang, Y.W.; Brown-Elliott, B.A.; Iakhiaeva, E.; et al. Evaluation of the Vitek MS v3.0 Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry System for Identification of Mycobacterium and Nocardia Species. J. Clin. Microbiol. 2018, 56, e00237-18. [Google Scholar] [CrossRef]
- Solntceva, V.; Kostrzewa, M.; Larrouy-Maumus, G. Detection of Species-Specific Lipids by Routine MALDI TOF Mass Spectrometry to Unlock the Challenges of Microbial Identification and Antimicrobial Susceptibility Testing. Front. Cell. Infect. Microbiol. 2021, 10, 621452. [Google Scholar] [CrossRef] [PubMed]
- Mahé, P.; Arsac, M.; Chatellier, S.; Monnin, V.; Perrot, N.; Mailler, S.; Girard, V.; Ramjeet, M.; Surre, J.; Lacroix, B.; et al. Automatic Identification of Mixed Bacterial Species Fingerprints in a MALDI-TOF Mass-Spectrum. Bioinformatics 2014, 30, 1280–1286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mörtelmaier, C.; Panda, S.; Robertson, I.; Krell, M.; Christodoulou, M.; Reichardt, N.; Mulder, I. Identification Performance of MALDI-ToF-MS upon Mono- and Bi-Microbial Cultures Is Cell Number and Culture Proportion Dependent. Anal. Bioanal. Chem. 2019, 411, 7027–7038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Lin, Y.; Qiao, L. Direct MALDI-TOF MS Identification of Bacterial Mixtures. Anal. Chem. 2018, 90, 10400–10408. [Google Scholar] [CrossRef]
- Griffin, P.M.; Price, G.R.; Schooneveldt, J.M.; Schlebusch, S.; Tilse, M.H.; Urbanski, T.; Hamilton, B.; Ventera, D. Use of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry to Identify Vancomycin-Resistant Enterococci and Investigate the Epidemiology of an Outbreak. J. Clin. Microbiol. 2012, 50, 2918–2931. [Google Scholar] [CrossRef] [Green Version]
- Brackmann, M.; Leib, S.L.; Tonolla, M.; Schürch, N.; Wittwer, M. Antimicrobial Resistance Classification Using MALDI-TOF-MS Is Not That Easy: Lessons from Vancomycin-Resistant Enterococcus faecium. Clin. Microbiol. Infect. 2020, 26, 391–393. [Google Scholar] [CrossRef] [Green Version]
- Sandrin, T.R.; Demirev, P.A. Characterization of Microbial Mixtures by Mass Spectrometry. Mass Spectrom. Rev. 2018, 37, 321–349. [Google Scholar] [CrossRef]
- Welker, M.; van Belkum, A.; Girard, V.; Charrier, J.-P.; Pincus, D. An Update on the, Routine Application of Maldi-Tof Ms, in Clinical Microbiology. Expert Rev. Proteom. 2019, 16, 695–710. [Google Scholar] [CrossRef]
- Schubert, S.; Kostrzewa, M. MALDI-TOF MS in the Microbiology Laboratory: Current Trends. Curr. Issues Mol. Biol. 2017, 23, 17–20. [Google Scholar] [CrossRef] [Green Version]
- Porter, J.E.; Sanchez, L.M. MALDI Mass Spectrometry Imaging for Microbiology. In MALDI Mass Spectrometry Imaging: From Fundamentals to Spatial Omics; Porta Siegel, T., Ed.; Royal Society of Chemistry: London, UK, 2021; pp. 291–320. ISBN 978-1839162411. [Google Scholar]
- Dunham, S.J.B.; Ellis, J.F.; Li, B.; Sweedler, J.V. Mass Spectrometry Imaging of Complex Microbial Communities. Acc. Chem. Res. 2017, 50, 96–104. [Google Scholar] [CrossRef]
- Watrous, J.D.; Dorrestein, P.C. Imaging Mass Spectrometry in Microbiology. Nat. Rev. Microbiol. 2011, 9, 683–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ercibengoa, M.; Alonso, M. Future Applications of MALDI-TOF Mass Spectrometry in Clinical Microbiology; Elsevier Inc.: Amsterdam, The Netherlands, 2018; ISBN 9780128144510. [Google Scholar]
- Li, H.; Li, Z. The Exploration of Microbial Natural Products and Metabolic Interaction Guided by Mass Spectrometry Imaging. Bioengineering 2022, 9, 707. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Tang, W.; Li, B. Mass Spectrometry Imaging and Its Potential in Food Microbiology. Int. J. Food Microbiol. 2022, 371, 109675. [Google Scholar] [CrossRef] [PubMed]
- Croxatta, A.; Prod’hom, G.; Greub, G. Applications of MALDI-TOF Mass Spectrometry in Clinical Diagnostic Microbiology. FEMS Microbiol. Rev. 2012, 36, 380–407. [Google Scholar] [CrossRef]
- Palmer, A.; Trede, D.; Alexandrov, T. Where Imaging Mass Spectrometry Stands: Here Are the Numbers. Metabolomics 2016, 12, 107. [Google Scholar] [CrossRef]
- Caprioli, R.M. Imaging Mass Spectrometry: A Perspective. J. Biomol. Tech. 2019, 30, 7–11. [Google Scholar] [CrossRef] [Green Version]
- Norris, J.L.; Caprioli, R.M. Analysis of Tissue Specimens by Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry in Biological and Clinical Research. Chem. Rev. 2013, 113, 2309–2342. [Google Scholar] [CrossRef] [Green Version]
- Buchberger, A.R.; DeLaney, K.; Johnson, J.; Li, L. Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights. Anal. Chem. 2018, 90, 240–265. [Google Scholar] [CrossRef]
- Aichler, M.; Walch, A. MALDI Imaging Mass Spectrometry: Current Frontiers and Perspectives in Pathology Research and Practice. Lab. Investig. 2015, 95, 422–431. [Google Scholar] [CrossRef] [Green Version]
- Heijs, B.; Tolner, E.A.; Bovée, J.V.M.G.; Van Den Maagdenberg, A.M.J.M.; McDonnell, L.A. Brain Region-Specific Dynamics of On-Tissue Protein Digestion Using MALDI Mass Spectrometry Imaging. J. Proteome Res. 2015, 14, 5348–5354. [Google Scholar] [CrossRef]
- Thiele, H.; Heldmann, S.; Trede, D.; Strehlow, J.; Wirtz, S.; Dreher, W.; Berger, J.; Oetjen, J.; Kobarg, J.H.; Fischer, B.; et al. 2D and 3D MALDI-Imaging: Conceptual Strategies for Visualization and Data Mining. Biochim. Biophys. Acta Proteins Proteom. 2014, 1844, 117–137. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Pan, Y.; Xiong, C.; Han, J.; Wang, X.; Chen, J.; Nie, Z. Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging (MALDI MSI) for in Situ Analysis of Endogenous Small Molecules in Biological Samples. TrAC Trends Anal. Chem. 2022, 157, 116809. [Google Scholar] [CrossRef]
- Ščupáková, K.; Balluff, B.; Tressler, C.; Adelaja, T.; Heeren, R.M.A.; Glunde, K.; Ertaylan, G. Cellular Resolution in Clinical MALDI Mass Spectrometry Imaging: The Latest Advancements and Current Challenges. Clin. Chem. Lab. Med. 2020, 58, 914–929. [Google Scholar] [CrossRef] [PubMed]
- Gemoll, T.; Roblick, U.J.; Habermann, J.K. MALDI Mass Spectrometry Imaging in Oncology (Review). Mol. Med. Rep. 2011, 4, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- Salzet, M.; Mériaux, C.; Franck, J.; Wistorski, M.; Fournier, I. MALDI Imaging Technology Application in Neurosciences: From History to Perspectives. In Expression Profiling in Neuroscience; Springer Science: Berlin/Heidelberg, Germany, 2012; Volume 64, pp. 181–223. ISBN 9781617794483. [Google Scholar]
- Sun, N.; Wu, Y.; Nanba, K.; Sbiera, S.; Kircher, S.; Kunzke, T.; Aichler, M.; Berezowska, S.; Reibetanz, J.; Rainey, W.E.; et al. High-Resolution Tissue Mass Spectrometry Imaging Reveals a Refined Functional Anatomy of the Human Adult Adrenal Gland. Endocrinology 2018, 159, 1511–1524. [Google Scholar] [CrossRef] [PubMed]
- Schubert, K.O.; Weiland, F.; Baune, B.T.; Hoffmann, P. The Use of MALDI-MSI in the Investigation of Psychiatric and Neurodegenerative Disorders: A Review. Proteomics 2016, 16, 1747–1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, J.; Sugiura, Y.; Yuki, D.; Hayasaka, T.; Goto-Inoue, N.; Zaima, N.; Kunii, Y.; Wada, A.; Yang, Q.; Nishiura, K.; et al. Abnormal Phospholipids Distribution in the Prefrontal Cortex from a Patient with Schizophrenia Revealed by Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry. Anal. Bioanal. Chem. 2011, 400, 1933–1943. [Google Scholar] [CrossRef] [Green Version]
- Mittal, P.; Condina, M.R.; Klingler-Hoffmann, M.; Kaur, G.; Oehler, M.K.; Sieber, O.M.; Palmieri, M.; Kommoss, S.; Brucker, S.; McDonnell, M.D.; et al. Cancer Tissue Classification Using Supervised Machine Learning Applied to Maldi Mass Spectrometry Imaging. Cancers. 2021, 13, 5388. [Google Scholar] [CrossRef]
- Nishidate, M.; Hayashi, M.; Aikawa, H.; Tanaka, K.; Nakada, N.; Miura, S.; Ryu, S.; Higashi, T.; Ikarashi, Y.; Fujiwara, Y.; et al. Applications of MALDI Mass Spectrometry Imaging for Pharmacokinetic Studies during Drug Development. Drug Metab. Pharmacokinet. 2019, 34, 209–216. [Google Scholar] [CrossRef]
- Handler, A.M.; Pommergaard Pedersen, G.; Troensegaard Nielsen, K.; Janfelt, C.; Just Pedersen, A.; Clench, M.R. Quantitative MALDI Mass Spectrometry Imaging for Exploring Cutaneous Drug Delivery of Tofacitinib in Human Skin. Eur. J. Pharm. Biopharm. 2021, 159, 1–10. [Google Scholar] [CrossRef]
- Tobias, F.; Hummon, A.B. Considerations for MALDI-Based Quantitative Mass Spectrometry Imaging Studies. J. Proteome Res. 2020, 19, 3620–3630. [Google Scholar] [CrossRef]
- Roepstorff, P. MALDI-TOF Mass Spectrometry in Protein Chemistry. Proteom. Funct. Genom. 2000, 88, 81–97. [Google Scholar] [CrossRef]
- Yang, J.Y.; Phelan, V.V.; Simkovsky, R.; Watrous, J.D.; Trial, R.M.; Fleming, T.C.; Wenter, R.; Moore, B.S.; Golden, S.S.; Pogliano, K.; et al. Primer on Agar-Based Microbial Imaging Mass Spectrometry. J. Bacteriol. 2012, 194, 6023–6028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.L.; Xu, Y.; Straight, P.; Dorrestein, P.C. Translating Metabolic Exchange with Imaging Mass Spectrometry. Nat. Chem. Biol. 2009, 5, 885–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Bruijn, I.; Cheng, X.; de Jager, V.; Expósito, R.G.; Watrous, J.; Patel, N.; Postma, J.; Dorrestein, P.C.; Kobayashi, D.; Raaijmakers, J.M. Comparative Genomics and Metabolic Profiling of the Genus Lysobacter. BMC Genom. 2015, 16, 991. [Google Scholar] [CrossRef] [Green Version]
- Michelsen, C.F.; Khademi, S.M.H.; Johansen, H.K.; Ingmer, H.; Dorrestein, P.C.; Jelsbak, L. Evolution of Metabolic Divergence in Pseudomonas aeruginosa during Long-Term Infection Facilitates a Proto-Cooperative Interspecies Interaction. ISME J. 2016, 10, 1330–1336. [Google Scholar] [CrossRef] [Green Version]
- Luzzatto-Knaan, T.; Melnik, A.V.; Dorrestein, P.C. Mass Spectrometry Uncovers the Role of Surfactin as an Interspecies Recruitment Factor. ACS Chem. Biol. 2019, 14, 459–467. [Google Scholar] [CrossRef]
- Gonzalez, D.J.; Xu, Y.; Yang, Y.L.; Esquenazi, E.; Liu, W.T.; Edlund, A.; Duong, T.; Du, L.; Molnár, I.; Gerwick, W.H.; et al. Observing the Invisible through Imaging Mass Spectrometry, a Window into the Metabolic Exchange Patterns of Microbes. J. Proteom. 2012, 75, 5069–5076. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.T.; Yang, Y.L.; Xu, Y.; Lamsa, A.; Haste, N.M.; Yang, J.Y.; Ng, J.; Gonzalez, D.; Ellermeier, C.D.; Straight, P.D.; et al. Imaging Mass Spectrometry of Intraspecies Metabolic Exchange Revealed the Cannibalistic Factors of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2010, 107, 16286–16290. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, D.J.; Haste, N.M.; Hollands, A.; Fleming, T.C.; Hamby, M.; Pogliano, K.; Nizet, V.; Dorrestein, P.C. Microbial Competition between Bacillus subtilis and Staphylococcus aureus Monitored by Imaging Mass Spectrometry. Microbiology 2011, 157, 2485–2492. [Google Scholar] [CrossRef]
- Rath, C.M.; Alexandrov, T.; Higginbottom, S.K.; Song, J.; Milla, M.E.; Fischbach, M.A.; Sonnenburg, J.L.; Dorrestein, P.C. Molecular Analysis of Model Gut Microbiotas by Imaging Mass Spectrometry and Nanodesorption Electrospray Ionization Reveals Dietary Metabolite Transformations. Anal. Chem. 2012, 84, 9259–9267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moree, W.J.; Phelan, V.V.; Wu, C.H.; Bandeira, N.; Cornett, D.S.; Duggan, B.M.; Dorrestein, P.C. Interkingdom Metabolic Transformations Captured by Microbial Imaging Mass Spectrometry. Proc. Natl. Acad. Sci. USA 2012, 109, 13811–13816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, J.; Dorrestein, P.C. Emerging Mass Spectrometry Techniques for the Direct Analysis of Microbial Colonies. Curr. Opin. Microbiol. 2014, 19, 120–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phelan, V.V.; Fang, J.; Dorrestein, P.C. Mass Spectrometry Analysis of Pseudomonas aeruginosa Treated with Azithromycin. J. Am. Soc. Mass Spectrom. 2015, 26, 873–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, T.; Dorrestein, P.C. Homogeneous Matrix Deposition on Dried Agar for MALDI Imaging Mass Spectrometry of Microbial Cultures. J. Am. Soc. Mass Spectrom. 2015, 26, 1959–1962. [Google Scholar] [CrossRef] [Green Version]
- Bleich, R.; Watrous, J.D.; Dorrestein, P.C.; Bowers, A.A.; Shank, E.A. Thiopeptide Antibiotics Stimulate Biofilm Formation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2015, 112, 3086–3091. [Google Scholar] [CrossRef] [Green Version]
- Schleyer, G.; Shahaf, N.; Ziv, C.; Dong, Y.; Meoded, R.A.; Helfrich, E.J.N.; Schatz, D.; Rosenwasser, S.; Rogachev, I.; Aharoni, A.; et al. In Plaque-Mass Spectrometry Imaging of a Bloom-Forming Alga during Viral Infection Reveals a Metabolic Shift towards Odd-Chain Fatty Acid Lipids. Nat. Microbiol. 2019, 4, 527–538. [Google Scholar] [CrossRef]
- Juříková, T.; Luptáková, D.; Kofroňová, O.; Škríba, A.; Novák, J.; Marešová, H.; Palyzová, A.; Petřík, M.; Havlíček, V.; Benada, O. Bringing Sem and Msi Closer than Ever before: Visualizing Aspergillus and Pseudomonas Infection in the Rat Lungs. J. Fungi 2020, 6, 257. [Google Scholar] [CrossRef]
- Martin, H.C.; Ibáñez, R.; Nothias, L.F.; Boya, P.C.A.; Reinert, L.K.; Rollins-Smith, L.A.; Dorrestein, P.C.; Gutiérrez, M. Viscosin-like Lipopeptides from Frog Skin Bacteria Inhibit Aspergillus fumigatus and Batrachochytrium dendrobatidis Detected by Imaging Mass Spectrometry and Molecular Networking. Sci. Rep. 2019, 9, 3017. [Google Scholar] [CrossRef]
- Barger, S.R.; Hoefler, B.C.; Cubillos-Ruiz, A.; Russell, W.K.; Russell, D.H.; Straight, P.D. Imaging Secondary Metabolism of Streptomyces sp. Mg1 during Cellular Lysis and Colony Degradation of Competing Bacillus subtilis. Int. J. Gen. Mol. Microbiol. 2012, 102, 435–445. [Google Scholar] [CrossRef]
- Debois, D.; Ongena, M.; Cawoy, H.; De Pauw, E. MALDI-FTICR MS Imaging as a Powerful Tool to Identify Paenibacillus Antibiotics Involved in the Inhibition of Plant Pathogens. J. Am. Soc. Mass Spectrom. 2013, 24, 1202–1213. [Google Scholar] [CrossRef] [PubMed]
- Holzlechner, M.; Reitschmidt, S.; Gruber, S.; Zeilinger, S.; Marchetti-Deschmann, M. Visualizing Fungal Metabolites during Mycoparasitic Interaction by MALDI Mass Spectrometry Imaging. Proteomics 2016, 16, 1742–1746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debois, D.; Ongena, M.; Cawoy, H.; De Pauw, E. In Situ Analysis of Bacterial Lipopeptide Antibiotics by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2016; Volume 1401, pp. 161–173. [Google Scholar]
- Brockmann, E.U.; Steil, D.; Bauwens, A.; Soltwisch, J.; Dreisewerd, K. Advanced Methods for MALDI-MS Imaging of the Chemical Communication in Microbial Communities. Anal. Chem. 2019, 91, 15081–15089. [Google Scholar] [CrossRef] [PubMed]
- Brockmann, E.U.; Potthoff, A.; Tortorella, S.; Soltwisch, J.; Dreisewerd, K. Infrared MALDI Mass Spectrometry with Laser-Induced Postionization for Imaging of Bacterial Colonies. J. Am. Soc. Mass Spectrom. 2021, 32, 1053–1064. [Google Scholar] [CrossRef]
- Niehaus, M.; Soltwisch, J.; Belov, M.E.; Dreisewerd, K. Transmission-Mode MALDI-2 Mass Spectrometry Imaging of Cells and Tissues at Subcellular Resolution. Nat. Methods 2019, 16, 925–931. [Google Scholar] [CrossRef]
- Karas, M.; Glückmann, M.; Schäfer, J. Ionization in Matrix-Assisted Laser Desorption/Ionization: Singly Charged Molecular Ions Are the Lucky Survivors. J. Mass Spectrom. 2000, 35, 1–12. [Google Scholar] [CrossRef]
- Si, T.; Li, B.; Zhang, K.; Xu, Y.; Zhao, H.; Sweedler, J.V. Characterization of Bacillus subtilis Colony Biofilms via Mass Spectrometry and Fluorescence Imaging. J. Proteome Res. 2016, 15, 1955–1962. [Google Scholar] [CrossRef]
- Lanni, E.J.; Masyuko, R.N.; Driscoll, C.M.; Aerts, J.T.; Shrout, J.D.; Bohn, P.W.; Sweedler, J.V. MALDI-Guided SIMS: Multiscale Imaging of Metabolites in Bacterial Biofilms. Anal. Chem. 2014, 86, 9139–9145. [Google Scholar] [CrossRef] [Green Version]
- Wakeman, C.A.; Moore, J.L.; Noto, M.J.; Zhang, Y.; Singleton, M.D.; Prentice, B.M.; Gilston, B.A.; Doster, R.S.; Gaddy, J.A.; Chazin, W.J.; et al. The Innate Immune Protein Calprotectin Promotes Pseudomonas aeruginosa and Staphylococcus aureus Interaction. Nat. Commun. 2016, 7, 11951. [Google Scholar] [CrossRef] [Green Version]
- Santos, T.; Théron, L.; Chambon, C.; Viala, D.; Centeno, D.; Esbelin, J.; Hébraud, M. MALDI Mass Spectrometry Imaging and in Situ Microproteomics of Listeria monocytogenes Biofilms. J. Proteomics 2018, 187, 152–160. [Google Scholar] [CrossRef]
- Blanc, L.; Lenaerts, A.; Dartois, V.; Prideaux, B. Visualization of Mycobacterial Biomarkers and Tuberculosis Drugs in Infected Tissue by MALDI-MS Imaging. Anal. Chem. 2018, 90, 6275–6282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geier, B.; Sogin, E.M.; Michellod, D.; Janda, M.; Kompauer, M.; Spengler, B.; Dubilier, N.; Liebeke, M. Spatial Metabolomics of in Situ Host–Microbe Interactions at the Micrometre Scale. Nat. Microbiol. 2020, 5, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Gemperline, E.; Horn, H.A.; Delaney, K.; Currie, C.R.; Li, L. Imaging with Mass Spectrometry of Bacteria on the Exoskeleton of Fungus-Growing Ants. ACS Chem. Biol. 2017, 12, 1980–1985. [Google Scholar] [CrossRef]
- Hulme, H.; Meikle, L.M.; Strittmatter, N.; Swales, J.; Hamm, G.; Brown, S.L.; Milling, S.; Macdonald, A.S.; Goodwin, R.J.A.; Burchmore, R.; et al. Mapping the Influence of the Gut Microbiota on Small Molecules across the Microbiome Gut Brain Axis. J. Am. Soc. Mass Spectrom. 2022, 33, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Schoenian, I.; Spiteller, M.; Ghaste, M.; Wirth, R.; Herz, H.; Spiteller, D. Chemical Basis of the Synergism and Antagonism in Microbial Communities in the Nests of Leaf-Cutting Ants. Proc. Natl. Acad. Sci. USA 2011, 108, 1955–1960. [Google Scholar] [CrossRef] [Green Version]
- Scott, A.J.; Post, J.M.; Lerner, R.; Ellis, S.R.; Lieberman, J.; Shirey, K.A.; Heeren, R.M.A.; Bindila, L.; Ernst, R.K. Host-Based Lipid Inflammation Drives Pathogenesis in Francisella Infection. Proc. Natl. Acad. Sci. USA 2017, 114, 12596–12601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Chandler, C.E.; Jackson, S.N.; Woods, A.S.; Goodlett, D.R.; Ernst, R.K.; Scott, A.J. On-Tissue Derivatization of Lipopolysaccharide for Detection of Lipid A Using MALDI-MSI. Anal. Chem. 2020, 92, 13667–13671. [Google Scholar] [CrossRef]
- Li, B.; Comi, T.J.; Si, T.; Dunham, S.J.B.; Sweedler, J.V. A One-Step Matrix Application Method for MALDI Mass Spectrometry Imaging of Bacterial Colony Biofilms. J. Mass Spectrom. 2016, 51, 1030–1035. [Google Scholar] [CrossRef] [Green Version]
- Wörmer, L.; Gajendra, N.; Schubotz, F.; Matys, E.D.; Evans, T.W.; Summons, R.E.; Hinrichs, K.U. A Micrometer-Scale Snapshot on Phototroph Spatial Distributions: Mass Spectrometry Imaging of Microbial Mats in Octopus Spring, Yellowstone National Park. Geobiology 2020, 18, 742–759. [Google Scholar] [CrossRef]
- Vergeiner, S.; Schafferer, L.; Haas, H.; Müller, T. Improved MALDI-TOF Microbial Mass Spectrometry Imaging by Application of a Dispersed Solid Matrix. J. Am. Soc. Mass Spectrom. 2014, 25, 1498–1501. [Google Scholar] [CrossRef]
- Ferreira, M.S.; De Oliveira, D.N.; De Oliveira, R.N.; Allegretti, S.M.; Vercesi, A.E.; Catharino, R.R. Mass Spectrometry Imaging: A New Vision in Differentiating Schistosoma mansoni Strains. J. Mass Spectrom. 2014, 49, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Kadesch, P.; Quack, T.; Gerbig, S.; Grevelding, C.G.; Spengler, B. Tissue-and Sex-Specific Lipidomic Analysis of Schistosoma mansoni Using High-Resolution Atmospheric Pressure Scanning Microprobe Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging. PLoS Negl. Trop. Dis. 2020, 14, e0008145. [Google Scholar] [CrossRef]
- Khalil, S.M.; Römpp, A.; Pretzel, J.; Becker, K.; Spengler, B. Phospholipid Topography of Whole-Body Sections of the Anopheles stephensi Mosquito, Characterized by High-Resolution Atmospheric-Pressure Scanning Microprobe Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging. Anal. Chem. 2015, 87, 11309–11316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mokosch, A.S.; Gerbig, S.; Grevelding, C.G.; Haeberlein, S.; Spengler, B. High-Resolution AP-SMALDI MSI as a Tool for Drug Imaging in Schistosoma mansoni. Anal. Bioanal. Chem. 2021, 413, 2755–2766. [Google Scholar] [CrossRef] [PubMed]
- Morawietz, C.M.; Peter Ventura, A.M.; Grevelding, C.G.; Haeberlein, S.; Spengler, B. Spatial Visualization of Drug Uptake and Distribution in Fasciola hepatica Using High-Resolution AP-SMALDI Mass Spectrometry Imaging. Parasitol. Res. 2022, 121, 1145–1153. [Google Scholar] [CrossRef]
- Morawietz, C.M.; Houhou, H.; Puckelwaldt, O.; Hehr, L.; Dreisbach, D.; Mokosch, A.; Roeb, E.; Roderfeld, M.; Spengler, B.; Haeberlein, S. Targeting Kinases in Fasciola hepatica: Anthelminthic Effects and Tissue Distribution of Selected Kinase Inhibitors. Front. Vet. Sci. 2020, 7, 611270. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, K.R.; Peter Ventura, A.; Gerbig, S.; Roderfeld, M.; Quack, T.; Grevelding, C.G.; Roeb, E.; Spengler, B. Changes in the Lipid Profile of Hamster Liver after Schistosoma mansoni Infection, Characterized by Mass Spectrometry Imaging and LC–MS/MS Analysis. Anal. Bioanal. Chem. 2022, 414, 3653–3665. [Google Scholar] [CrossRef]
- Geier, B.; Oetjen, J.; Ruthensteiner, B.; Polikarpov, M.; Gruber-Vodicka, H.R.; Liebeke, M. Connecting Structure and Function from Organisms to Molecules in Small-Animal Symbioses through Chemo-Histo-Tomography. Proc. Natl. Acad. Sci. USA 2021, 118, e2023773118. [Google Scholar] [CrossRef]
- Kompauer, M.; Heiles, S.; Spengler, B. Atmospheric Pressure MALDI Mass Spectrometry Imaging of Tissues and Cells at 1.4-µm Lateral Resolution. Nat. Methods 2016, 14, 90–96. [Google Scholar] [CrossRef]
- Kloehn, J.; Boughton, B.A.; Saunders, E.C.; O’callaghan, S.; Binger, K.J.; McConville, M.J. Identification of Metabolically Quiescent Leishmania Mexicana Parasites in Peripheral and Cured Dermal Granulomas Using Stable Isotope Tracing Imaging Mass Spectrometry. MBio 2021, 12, e00129-21. [Google Scholar] [CrossRef]
- Pauker, V.I.; Bertzbach, L.D.; Hohmann, A.; Kheimar, A.; Teifke, J.P.; Mettenleiter, T.C.; Karger, A.; Kaufer, B.B. Imaging Mass Spectrometry and Proteome Analysis of Marek’s Disease Virus-Induced Tumors. mSphere 2019, 4, e00569-18 . [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rourke, M.B.O.; Roediger, B.R.; Jolly, C.J.; Crossett, B.; Padula, M.P.; Hansbro, P.M. Viral Biomarker Detection and Validation Using MALDI Mass Spectrometry Imaging (MSI). Proteomes 2022, 10, 33. [Google Scholar]
- Schwamborn, K.; Krieg, R.C.; Uhlig, S.; Ikenberg, H.; Wellmann, A. MALDI Imaging as a Specific Diagnostic Tool for Routine Cervical Cytology Specimens. Int. J. Mol. Med. 2011, 27, 417–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniel, N.; Lécuyer, E.; Chassaing, B. Host/Microbiota Interactions in Health and Diseases—Time for Mucosal Microbiology! Mucosal Immunol. 2021, 14, 1006–1016. [Google Scholar] [CrossRef]
- Rich, V.I.; Maier, R.M. Aquatic Environments. Environ. Microbiol. 2015, 111–138. [Google Scholar] [CrossRef]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and Its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, J.P.L.; Bollwein, C.; Schwamborn, K. Mass Spectrometry Imaging Spatial Tissue Analysis toward Personalized Medicine. Life 2022, 12, 1037. [Google Scholar] [CrossRef]
- Bertzbach, L.D.; Kaufer, B.B.; Karger, A. Applications of Mass Spectrometry Imaging in Virus Research. In Advances in Virus Research; Academic Press Inc.: Cambridge, MA, USA, 2021; Volume 109, pp. 31–62. ISBN 9780128230428. [Google Scholar]
- Larrouy-Maumus, G.; Puzo, G. Mycobacterial Envelope Lipids Fingerprint from Direct MALDI-TOF MS Analysis of Intact Bacilli. Tuberculosis 2015, 95, 75–85. [Google Scholar] [CrossRef]
- Pizzato, J.; Tang, W.; Bernabeu, S.; Bonnin, R.A.; Bille, E.; Farfour, E.; Guillard, T.; Barraud, O.; Cattoir, V.; Plouzeau, C.; et al. Discrimination of Escherichia coli, Shigella flexneri, and Shigella sonnei Using Lipid Profiling by MALDI-TOF Mass Spectrometry Paired with Machine Learning. Microbiologyopen 2022, 11, e1313. [Google Scholar] [CrossRef]
- Cunsolo, V.; Muccilli, V.; Saletti, R.; Foti, S. Mass Spectrometry in Food Proteomics: A Tutorial. J. Mass Spectrom. 2014, 49, 768–784. [Google Scholar] [CrossRef]
- Calderaro, A.; Buttrini, M.; Montecchini, S.; Rossi, S.; Piccolo, G.; Arcangeletti, M.C.; Medici, M.C.; Chezzi, C.; De Conto, F. MALDI-TOF MS as a New Tool for the Identification of Dientamoeba fragilis. Parasites Vectors 2018, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.Q.; Zheng, P.; Xu, S.T.; Wu, X. Object Detection with Deep Learning: A Review. IEEE Trans. Neural Networks Learn. Syst. 2019, 30, 3212–3232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szulc, J.; Ruman, T. Laser Ablation Remote-Electrospray Ionisation Mass Spectrometry (LARESI MSI) Imaging—New Method for Detection and Spatial Localization of Metabolites and Mycotoxins Produced by Moulds. Toxins 2020, 12, 720. [Google Scholar] [CrossRef]
- Baquer, G.; Sementé, L.; Mahamdi, T.; Correig, X.; Ràfols, P.; García-Altares, M. What Are We Imaging? Software Tools and Experimental Strategies for Annotation and Identification of Small Molecules in Mass Spectrometry Imaging. Mass Spectrom. Rev. 2022; e21794, Early View. [Google Scholar] [CrossRef] [PubMed]
- Zubarev, R.A.; Makarov, A. Orbitrap Mass Spectrometry. Anal. Chem. 2013, 85, 5288–5296. [Google Scholar] [CrossRef] [PubMed]
- Spengler, B. Mass Spectrometry Imaging of Biomolecular Information. Anal. Chem. 2015, 87, 64–82. [Google Scholar] [CrossRef]
- Watrous, J.; Hendricks, N.; Meehan, M.; Dorrestein, P.C. Capturing Bacterial Metabolic Exchange Using Thin Film Desorption Electrospray Ionization-Imaging Mass Spectrometry. Anal. Chem. 2010, 82, 1598–1600. [Google Scholar] [CrossRef] [Green Version]
- Baig, N.F.; Dunham, S.J.B.; Morales-Soto, N.; Shrout, J.D.; Sweedler, J.V.; Bohn, P.W. Multimodal Chemical Imaging of Molecular Messengers in Emerging Pseudomonas aeruginosa Bacterial Communities. Analyst 2015, 140, 6544–6552. [Google Scholar] [CrossRef] [Green Version]
- Dueñas, M.E.; Carlucci, L.; Lee, Y.J. Matrix Recrystallization for MALDI-MS Imaging of Maize Lipids at High-Spatial Resolution. J. Am. Soc. Mass Spectrom. 2016, 27, 1575–1578. [Google Scholar] [CrossRef]
- Angerer, T.B.; Bour, J.; Biagi, J.L.; Moskovets, E.; Frache, G. Evaluation of 6 MALDI-Matrices for 10 Μm Lipid Imaging and On-Tissue MSn with AP-MALDI-Orbitrap. J. Am. Soc. Mass Spectrom. 2022, 33, 760–771. [Google Scholar] [CrossRef]
- Hankin, J.A.; Barkley, R.M.; Murphy, R.C. Sublimation as a Method of Matrix Application for Mass Spectrometric Imaging. J. Am. Soc. Mass Spectrom. 2007, 18, 1646–1652. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Caprioli, R.M. Matrix Sublimation/Recrystallization for Imaging Proteins by Mass Spectrometry at High Spatial Resolution. Anal. Chem. 2011, 83, 5728–5734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grujcic, V.; Taylor, G.T.; Foster, R.A. One Cell at a Time: Advances in Single-Cell Methods and Instrumentation for Discovery in Aquatic Microbiology. Front. Microbiol. 2022, 13, 881088. [Google Scholar] [CrossRef] [PubMed]
- Bednařík, A.; Machálková, M.; Moskovets, E.; Coufalíková, K.; Krásenský, P.; Houška, P.; Kroupa, J.; Navrátilová, J.; Šmarda, J.; Preisler, J. MALDI MS Imaging at Acquisition Rates Exceeding 100 Pixels per Second. J. Am. Soc. Mass Spectrom. 2019, 30, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Egli, A. Digitalization, Clinical Microbiology and Infectious Diseases. Clin. Microbiol. Infect. 2020, 26, 1289–1290. [Google Scholar] [CrossRef]
- Egli, A.; Schrenzel, J.; Greub, G. Digital Microbiology. Clin. Microbiol. Infect. 2020, 26, 1324–1331. [Google Scholar] [CrossRef]
- Abdelmoula, W.M.; Lopez, B.G.C.; Randall, E.C.; Kapur, T.; Sarkaria, J.N.; White, F.M.; Agar, J.N.; Wells, W.M.; Agar, N.Y.R. Peak Learning of Mass Spectrometry Imaging Data Using Artificial Neural Networks. Nat. Commun. 2021, 12, 5544. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feucherolles, M.; Frache, G. MALDI Mass Spectrometry Imaging: A Potential Game-Changer in a Modern Microbiology. Cells 2022, 11, 3900. https://doi.org/10.3390/cells11233900
Feucherolles M, Frache G. MALDI Mass Spectrometry Imaging: A Potential Game-Changer in a Modern Microbiology. Cells. 2022; 11(23):3900. https://doi.org/10.3390/cells11233900
Chicago/Turabian StyleFeucherolles, Maureen, and Gilles Frache. 2022. "MALDI Mass Spectrometry Imaging: A Potential Game-Changer in a Modern Microbiology" Cells 11, no. 23: 3900. https://doi.org/10.3390/cells11233900