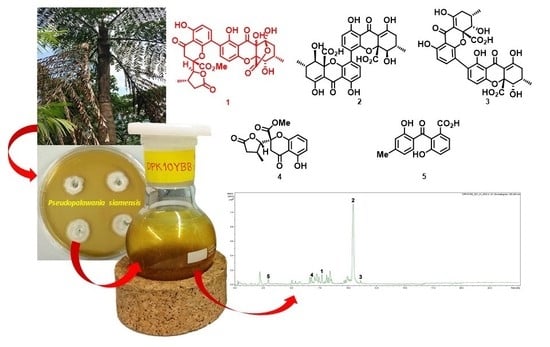
Polyketide-Derived Secondary Metabolites from a Dothideomycetes Fungus, Pseudopalawania siamensis gen. et sp. nov., (Muyocopronales) with Antimicrobial and Cytotoxic Activities
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection, Specimen Examination and Isolation of Fungi
2.2. DNA Extraction, PCR Amplification and Sequencing
2.3. Phylogenetic Analysis
2.4. General Information of Chromatography and Spectral Methods
2.5. Fermentation and Extraction
2.6. Isolation of Compounds 1–5
2.7. Spectral Data
Pseudopalawanone (1)
2.8. Antimicrobial Activity and Cytotoxicity Assays
3. Results and Discussion
3.1. Phylogenetic Analysis
3.2. Taxonomy
3.2.1. Pseudopalawania Mapook and K.D. Hyde, gen. nov.
3.2.2. Pseudopalawania siamensis Mapook and K.D. Hyde, sp. nov.
3.3. Structure Elucidation of the New Compound
3.4. Biological Activity of Compounds 1–5
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bills, G.F.; Gloer, J.B. Biologically Active Secondary Metabolites from the Fungi. Microbiol. Spectr. 2016, 4, 1–32. [Google Scholar] [CrossRef] [PubMed]
- De Silva, D.D.; Rapior, S.; Fons, F.; Bahkali, A.H.; Hyde, K.D. Medicinal mushrooms in supportive cancer therapies: An approach to anti-cancer effects and putative mechanisms of action. Fungal Divers. 2012, 55, 1–35. [Google Scholar] [CrossRef]
- Sandargo, B.; Chepkirui, C.; Cheng, T.; Chaverra-Munoz, L.; Thongbai, B.; Stadler, M.; Hüttel, S. Biological and chemical diversity go hand in hand: Basidomycota as source of new pharmaceuticals and agrochemicals. Biotechnol. Adv. 2019, 37, 107344. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Xu, J.; Rapior, S.; Jeewon, R.; Lumyong, S.; Niego, A.G.T.; Abeywickrama, P.D.; Aluthmuhandiram, J.V.S.; Brahamanage, R.S.; Brooks, S.; et al. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 2019, 97, 1–136. [Google Scholar] [CrossRef] [Green Version]
- Chomcheon, P.; Sriubolmas, N.; Wiyakrutta, S.; Ngamrojanavanich, N.; Chaichit, N.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. Cyclopentenones, Scaffolds for organic syntheses produced by the endophytic fungus mitosporic Dothideomycete sp. LRUB20. J. Nat. Prod. 2006, 69, 1351–1353. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.S.; Ko, W.; Kim, J.W.; Jeong, M.-H.; Ko, S.-K.; Hur, J.-S.; Oh, H.; Jang, J.-H.; Ahn, J.S. Bioactive α-pyrone derivatives from the endolichenic fungus Dothideomycetes sp. EL003334. J. Nat. Prod. 2018, 81, 1084–1088. [Google Scholar] [CrossRef]
- Wu, B.; Wiese, J.; Labes, A.; Kramer, A.; Schmaljohann, R.; Imhoff, J.F. Lindgomycin, an unusual antibiotic polyketide from a marine fungus of the Lindgomycetaceae. Mar. Drugs 2015, 13, 4617–4632. [Google Scholar] [CrossRef] [Green Version]
- Rupcic, Z.; Chepkirui, C.; Hernández-Restrepo, M.; Crous, P.W.; Luangsa-ard, J.J.; Stadler, M. New nematicidal and antimicrobial secondary metabolites from a new species in the new genus, Pseudobambusicola thailandica. MycoKeys 2018, 33, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Phukhamsakda, C.; Macabeo, A.P.G.; Yuyama, K.T.; Hyde, K.D.; Stadler, M. Biofilm inhibitory abscisic acid derivatives from the plant-associated Dothideomycete fungus, Roussoella sp. Molecules 2018, 23, 2190. [Google Scholar] [CrossRef] [Green Version]
- Phukhamsakda, C.; Macabeo, A.P.G.; Huch, V.; Cheng, T.; Hyde, K.D.; Stadler, M. Sparticolins A–G, biologically active oxidized spirodioxynaphthalene derivatives from the ascomycete Sparticola junci. J. Nat. Prod. 2019, 82, 2878–2885. [Google Scholar] [CrossRef]
- Macabeo, A.P.G.; Pilapil, L.A.E.; Garcia, K.Y.M.; Quimque, M.T.J.; Phukhamsakda, C.; Cruz, A.J.C.; Hyde, K.D.; Stadler, M. Alpha-Glucosidase- and lipase-inhibitory phenalenones from a new species of Pseudolophiostoma originating from Thailand. Molecules 2020, 25, 965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chomnunti, P.; Hongsanan, S.; Aguirre-Hudson, B.; Tian, Q.; Peršoh, D.; Dhami, M.K.; Alias, A.S.; Xu, J.; Liu, X.; Stadler, M.; et al. The sooty moulds. Fungal Divers. 2014, 66, 1–36. [Google Scholar] [CrossRef]
- Crous, P.W.; Gams, W.; Stalpers, J.A.; Robert, V.; Stegehuis, G. MycoBank: An online initiative to launch mycology into the 21st century. Stud. Mycol. 2004, 50, 19–22. [Google Scholar]
- Vilgalys, R.; Hester, M. rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. 1990, 18, 315–322. [Google Scholar]
- Rehner, S.A. Primers for Elongation Factor 1-Alpha (EF1-Alpha). 2001. Available online: http://ocid.nacse.org/research/deephyphae/EF1primer.pdf (accessed on 6 April 2020).
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Mapook, A.; Hyde, K.D.; McKenzie, E.H.C.; Jones, E.B.G.; Bhat, D.J.; Jeewon, R.; Stadler, M.; Samarakoon, M.C.; Malaithong, M.; Tanunchai, B.; et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed). Fungal Divers. 2020, in press. [Google Scholar] [CrossRef]
- Mapook, A.; Boonmee, S.; Ariyawansa, H.A.; Tibpromma, S.; Campesori, E.; Jones, E.B.G.; Bahkali, A.H.; Hyde, K.D. Taxonomic and phylogenetic placement of Nodulosphaeria. Mycol. Prog. 2016, 15, 34. [Google Scholar] [CrossRef]
- Hongsanan, S.; Sánchez-Ramírez, S.; Crous, P.W.; Ariyawansa, H.A.; Zhao, R.L.; Hyde, K.D. The evolution of fungal epiphytes. Mycosphere 2016, 7, 1690–1712. [Google Scholar] [CrossRef]
- Crous, P.W.; Luangsa-ard, J.J.; Wingfield, M.J.; Carnegie, A.J.; Hernández-Restrepo, M.; Lombard, L.; Roux, J.; Barreto, R.W.; Baseia, I.G.; Cano-Lira, J.F.; et al. Fungal planet description sheets: 785–867. Persoonia 2018, 41, 238–417. [Google Scholar] [CrossRef]
- Hernández-Restrepo, M.; Bezerra, J.D.P.; Tan, Y.P.; Wiederhold, N.; Crous, P.W.; Guarro, J.; Gené, J. Re-evaluation of Mycoleptodiscus species and morphologically similar fungi. Persoonia 2019, 42, 205–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mapook, A.; Hyde, K.D.; Dai, D.-Q.; Li, J.; Jones, E.B.G.; Bahkali, A.H.; Boonmee, S. Muyocopronales, ord. nov., (Dothideomycetes, Ascomycota) and a reappraisal of Muyocopron species from northern Thailand. Phytotaxa 2016, 265, 225–237. [Google Scholar] [CrossRef]
- Mapook, A.; Hyde, K.D.; Hongsanan, S.; Phukhamsakda, C.; Li, J.F.; Boonmee, S. Palawaniaceae fam. nov., a new family (Dothideomycetes, Ascomycota) to accommodate Palawania species and their evolutionary time estimates. Mycosphere 2016, 7, 1732–1745. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v14: Tree Figure Drawing Tool. 2014. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 6 April 2020).
- Stenroos, S.; Laukka, T.; Huhtinen, S.; Döbbeler, P.; Myllys, L.; Syrjänen, K.; Hyvönen, J. Multiple origins of symbioses between ascomycetes and bryophytes suggested by a five-gene phylogeny. Cladistics 2010, 26, 281–300. [Google Scholar] [CrossRef]
- Li, L.; Pan, H.; Liu, W.; Chen, M.Y.; Zhong, C.H. First report of Alternaria alternata causing postharvest rot of kiwifruit in China. Plant Dis. 2017, 101, 1046. [Google Scholar] [CrossRef]
- Alves, J.L.; Woudenberg, J.H.C.; Duarte, L.L.; Crous, P.W.; Barreto, R.W. Reappraisal of the genus Alternariaster (Dothideomycetes). Persoonia 2013, 31, 77–85. [Google Scholar] [CrossRef] [Green Version]
- Cheewangkoon, R.; Groenewald, J.Z.; Summerell, B.A.; Hyde, K.D.; To-Anun, C.; Crous, P.W. Myrtaceae, a cache of fungal biodiversity. Persoonia 2009, 23, 55–85. [Google Scholar] [CrossRef] [Green Version]
- Crous, P.W.; Wingfield, M.J.; Schumacher, R.K.; Summerell, B.A.; Giraldo, A.; Gené, J.; Guarro, J.; Wanasinghe, D.N.; Hyde, K.D.; Camporesi, E.; et al. Fungal planet description sheets: 281-319. Persoonia 2014, 33, 212–289. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Hofmann, T.A.; Kirschner, R.; Piepenbring, M. Phylogenetic relationships and new records of Asterinaceae (Dothideomycetes) from Panama. Fungal Divers. 2010, 43, 39–53. [Google Scholar] [CrossRef]
- Dai, D.; Bhat, D.J.; Liu, J.; Chukeatirote, E.; Zhao, R.; Hyde, K.D. Bambusicola, a new genus from bamboo with asexual and sexual morphs. Cryptogam. Mycol. 2012, 33, 363–379. [Google Scholar] [CrossRef]
- Liu, J.-K.; Phookamsak, R.; Doilom, M.; Wikee, S.; Li, Y.-M.; Ariyawansha, H.; Boonmee, S.; Chomnunti, P.; Dai, D.-Q.; Bhat, J.D.; et al. Towards a natural classification of Botryosphaeriales. Fungal Divers. 2012, 57, 149–210. [Google Scholar] [CrossRef]
- Schoch, C.L.; Shoemaker, R.A.; Seifert, K.A.; Hambleton, S.; Spatafora, J.W.; Crous, P.W. A Multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 2006, 98, 1041–1052. [Google Scholar] [CrossRef]
- Beimforde, C.; Feldberg, K.; Nylinder, S.; Rikkinen, J.; Tuovila, H.; Dörfelt, H.; Gube, M.; Jackson, D.J.; Reitner, J.; Seyfullah, L.J.; et al. Estimating the phanerozoic history of the Ascomycota lineages: Combining fossil and molecular data. Mol. Phylogenetics Evol. 2014, 78, 386–398. [Google Scholar] [CrossRef] [PubMed]
- Lutzoni, F.; Pagel, M.; Reeb, V. Major fungal lineages are derived from lichen symbiotic ancestors. Nature 2001, 411, 937–940. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Crous, P.W.; Groenewald, J.Z.; Boehm, E.W.A.; Burgess, T.I.; de Gruyter, J.; de Hoog, G.S.; Dixon, L.J.; Grube, M.; Gueidan, C.; et al. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 2009, 64, 1–15. [Google Scholar] [CrossRef]
- Cai, L.; Hyde, K.D. Ascorhombispora aquatica gen. et sp. nov. from a freshwater habitat in China, and its phylogenetic placement based on molecular data. Cryptogam. Mycol. 2007, 28, 291–300. [Google Scholar]
- Hongsanan, S.; Chomnunti, P.; Crous, P.W.; Chukeatirote, E.; Hyde, K.D. Introducing Chaetothyriothecium, a new genus of Microthyriales. Phytotaxa 2014, 161, 157–164. [Google Scholar] [CrossRef] [Green Version]
- Hyde, K.D.; Jones, E.B.G.; Liu, J.-K.; Ariyawansa, H.; Boehm, E.; Boonmee, S.; Braun, U.; Chomnunti, P.; Crous, P.W.; Dai, D.-Q.; et al. Families of Dothideomycetes. Fungal Divers. 2013, 63, 1–313. [Google Scholar] [CrossRef]
- Tibell, L. Tholurna dissimilis and generic delimitations in Caliciaceae inferred from nuclear ITS and LSU rDNA phylogenies (Lecanorales, Lichenized Ascomycetes). Mycol. Res. 2003, 107, 1403–1418. [Google Scholar] [CrossRef]
- Muggia, L.; Hafellner, J.; Wirtz, N.; Hawksworth, D.L.; Grube, M. The sterile microfilamentous lichenized fungi Cystocoleus ebeneus and Racodium rupestre are relatives of plant pathogens and clinically important Dothidealean fungi. Mycol. Res. 2008, 112, 50–56. [Google Scholar] [CrossRef]
- Verkley, G.J.M.; Dukik, K.; Renfurm, R.; Göker, M.; Stielow, J.B. Novel genera and species of Coniothyrium-like fungi in Montagnulaceae (Ascomycota). Persoonia 2014, 32, 25–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariyawansa, H.A.; Camporesi, E.; Thambugala, K.M.; Mapook, A.; Kang, J.-C.; Alias, S.A.; Chukeatirote, E.; Thines, M.; McKenzie, E.H.C.; Hyde, K.D. Confusion surrounding Didymosphaeria —Phylogenetic and morphological evidence suggest Didymosphaeriaceae is not a distinct family. Phytotaxa 2014, 176, 102–119. [Google Scholar] [CrossRef] [Green Version]
- Spatafora, J.W.; Sung, G.-H.; Johnson, D.; Hesse, C.; O’Rourke, B.; Serdani, M.; Spotts, R.; Lutzoni, F.; Hofstetter, V.; Miadlikowska, J.; et al. A five-gene phylogeny of Pezizomycotina. Mycologia 2006, 98, 1018–1028. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Abreu, V.P.; Bazzicalupo, A.; Thilini Chethana, K.W.; Clericuzio, M.; Dayarathne, M.C.; Dissanayake, A.J.; Ekanayaka, A.H.; He, M.-Q.; et al. Fungal diversity notes 603–708: Taxonomic and phylogenetic notes on genera and species. Fungal Divers. 2017, 87, 1–235. [Google Scholar] [CrossRef]
- Pang, K.-L.; Hyde, K.D.; Alias, S.A.; Suetrong, S.; Guo, S.-Y.; Idid, R.; Gareth Jones, E.B. Dyfrolomycetaceae, a new family in the Dothideomycetes, Ascomycota. Cryptogam. Mycol. 2013, 34, 223–232. [Google Scholar] [CrossRef]
- Suetrong, S.; Schoch, C.L.; Spatafora, J.W.; Kohlmeyer, J.; Volkmann-Kohlmeyer, B.; Sakayaroj, J.; Phongpaichit, S.; Tanaka, K.; Hirayama, K.; Jones, E.B.G. Molecular systematics of the marine Dothideomycetes. Stud. Mycol. 2009, 64, 155–173S6. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Phookamsak, R.; Ariyawansa, H.A.; Boonmee, S.; Zhao, Q.; et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Zhang, J.-F.; Liu, J.-K.; Hyde, K.D.; Chen, Y.-Y.; Liu, Y.-X.; Liu, Z.-Y. Two new species of Dyfrolomyces (Dyfrolomycetaceae, Dothideomycetes) from Karst landforms. Phytotaxa 2017, 313, 267–277. [Google Scholar] [CrossRef]
- Papendorf, M.G. Leptodiscus africanus sp. nov. Trans. Brit. Mycol. Soc. 1967, 50, 687–690. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egidi, E.; de Hoog, G.S.; Isola, D.; Onofri, S.; Quaedvlieg, W.; de Vries, M.; Verkley, G.J.M.; Stielow, J.B.; Zucconi, L.; Selbmann, L. Phylogeny and taxonomy of meristematic rock-inhabiting black fungi in the Dothideomycetes based on multi-locus phylogenies. Fungal Divers. 2014, 65, 127–165. [Google Scholar] [CrossRef]
- Ertz, D.; Diederich, P. Dismantling melaspileaceae: A first phylogenetic study of Buelliella, Hemigrapha, Karschia, Labrocarpon and Melaspilea. Fungal Divers. 2015, 71, 141–164. [Google Scholar] [CrossRef]
- Tanaka, K.; Hirayama, K.; Yonezawa, H.; Sato, G.; Toriyabe, A.; Kudo, H.; Hashimoto, A.; Matsumura, M.; Harada, Y.; Kurihara, Y.; et al. Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud. Mycol. 2015, 82, 75–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wang, H.K.; Fournier, J.; Crous, P.W.; Jeewon, R.; Pointing, S.B.; Hyde, K.D. Towards a phylogenetic clarification of Lophiostoma/Massarina and morphologically similar genera in the Pleosporales. Fungal Divers. 2009, 38, 225–251. [Google Scholar]
- Madrid, H.; Gené, J.; Cano, J.; Guarro, J. A new species of Leptodiscella from Spanish soil. Mycol. Prog. 2012, 11, 535–541. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Phukhamsakda, C.; Thambugala, K.M.; Bulgakov, T.S.; Wanasinghe, D.N.; Perera, R.H.; Mapook, A.; Camporesi, E.; Kang, J.-C.; Gareth Jones, E.B.; et al. Revision and phylogeny of Leptosphaeriaceae. Fungal Divers. 2015, 74, 19–51. [Google Scholar] [CrossRef]
- de Gruyter, J.; Woudenberg, J.H.C.; Aveskamp, M.M.; Verkley, G.J.M.; Groenewald, J.Z.; Crous, P.W. Redisposition of Phoma-like anamorphs in Pleosporales. Stud. Mycol. 2013, 75, 1–36. [Google Scholar] [CrossRef] [Green Version]
- Chomnunti, P.; Schoch, C.L.; Aguirre–Hudson, B.; Ko-Ko, T.W.; Hongsanan, S.; Jones, E.B.G.; Kodsueb, R.; Phookamsak, R.; Chukeatirote, E.; Bahkali, A.H.; et al. Capnodiaceae. Fungal Divers. 2011, 51, 103–134. [Google Scholar] [CrossRef]
- Aveskamp, M.M.; de Gruyter, J.; Woudenberg, J.H.C.; Verkley, G.J.M.; Crous, P.W. Highlights of the Didymellaceae: A polyphasic approach to characterise Phoma and related Pleosporalean genera. Stud. Mycol. 2010, 65, 1–60. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Hyde, K.D.; Jayasiri, S.C.; Buyck, B.; Chethana, K.W.T.; Dai, D.Q.; Dai, Y.C.; Daranagama, D.A.; Jayawardena, R.S.; Lücking, R.; et al. Fungal diversity notes 111–252—Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015, 75, 27–274. [Google Scholar] [CrossRef]
- Liu, J.K.; Hyde, K.D.; Jones, E.B.G.; Ariyawansa, H.A.; Bhat, D.J.; Boonmee, S.; Maharachchikumbura, S.S.N.; McKenzie, E.H.C.; Phookamsak, R.; Phukhamsakda, C.; et al. Fungal diversity notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 2015, 72, 1–197. [Google Scholar] [CrossRef]
- Quaedvlieg, W.; Verkley, G.J.M.; Shin, H.-D.; Barreto, R.W.; Alfenas, A.C.; Swart, W.J.; Groenewald, J.Z.; Crous, P.W. Sizing up Septoria. Stud. Mycol. 2013, 75, 307–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senwanna, C.; Hongsanan, S.; Phookamsak, R.; Tibpromma, S.; Cheewangkoon, R.; Hyde, K.D. Muyocopron heveae sp. nov. and M. dipterocarpi appears to have host-jumped to rubber. Mycol. Prog. 2019, 18, 741–752. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Jeewon, R.; Phillips, A.J.L.; Bhat, D.J.; Wanasinghe, D.N.; Liu, J.K.; Lu, Y.Z.; et al. Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 2019, 10, 1–186. [Google Scholar] [CrossRef]
- Tibpromma, S.; McKenzie, E.H.C.; Karunarathna, S.C.; Xu, J.; Hyde, K.D.; Hu, D.M. Muyocopron garethjonesii sp. nov. (Muyocopronales, Dothideomycetes) on Pandanus sp. Mycosphere 2016, 7, 1480–1489. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; Bhat, J.D.; Mortimer, P.E.; Xu, J.; Promputtha, I.; Doilom, M.; Yang, J.-B.; Tang, A.M.C.; Karunarathna, S.C. Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from southern Thailand. MycoKeys 2018, 33, 25–67. [Google Scholar] [CrossRef]
- Boehm, E.W.A.; Schoch, C.L.; Spatafora, J.W. On the Evolution of the Hysteriaceae and Mytilinidiaceae (Pleosporomycetidae, Dothideomycetes, Ascomycota) using four nuclear genes. Mycol. Res. 2009, 113, 461–479. [Google Scholar] [CrossRef]
- Ferrer, A.; Miller, A.N.; Shearer, C.A. Minutisphaera and Natipusilla: Two new genera of freshwater Dothideomycetes. Mycologia 2011, 103, 411–423. [Google Scholar] [CrossRef] [Green Version]
- Crous, P.W.; Wingfield, M.J.; Guarro, J.; Cheewangkoon, R.; van der Bank, M.; Swart, W.J.; Stchigel, A.M.; Cano-Lira, J.F.; Roux, J.; Madrid, H.; et al. Fungal planet description sheets: 154–213. Persoonia 2013, 31, 188–296. [Google Scholar] [CrossRef]
- Mapook, A.; Boonmee, S.; Liu, J.-K.; Jones, E.B.G.; Bahkali, A.H.; Hyde, K.D. Taxonomic and phylogenetic placement of Phaeodimeriella (Pseudoperisporiaceae, Pleosporales). Cryptogam. Mycol. 2016, 37, 157–176. [Google Scholar] [CrossRef]
- Lumbsch, H.T.; Schmitt, I.; Lindemuth, R.; Miller, A.; Mangold, A.; Fernandez, F.; Huhndorf, S. Performance of four ribosomal DNA regions to infer higher-level phylogenetic relationships of inoperculate Euascomycetes (Leotiomyceta). Mol. Phylogenet. Evol. 2005, 34, 512–524. [Google Scholar] [CrossRef]
- James, T.Y.; Kauff, F.; Schoch, C.L.; Matheny, P.B.; Hofstetter, V.; Cox, C.J.; Celio, G.; Gueidan, C.; Fraker, E.; Miadlikowska, J.; et al. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 2006, 443, 818–822. [Google Scholar] [CrossRef]
- Crous, P.W.; Schoch, C.L.; Hyde, K.D.; Wood, A.R.; Gueidan, C.; de Hoog, G.S.; Groenewald, J.Z. Phylogenetic lineages in the Capnodiales. Stud. Mycol. 2009, 64, 17–47. [Google Scholar] [CrossRef] [PubMed]
- Thambugala, K.M.; Hyde, K.D.; Tanaka, K.; Tian, Q.; Wanasinghe, D.N.; Ariyawansa, H.A.; Jayasiri, S.C.; Boonmee, S.; Camporesi, E.; Hashimoto, A.; et al. Towards a natural classification and backbone tree for Lophiostomataceae, Floricolaceae, and Amorosiaceae fam. nov. Fungal Divers. 2015, 74, 199–266. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Thambugala, K.M.; Manamgoda, D.S.; Jayawardena, R.; Camporesi, E.; Boonmee, S.; Wanasinghe, D.N.; Phookamsak, R.; Hongsanan, S.; Singtripop, C.; et al. Towards a natural classification and backbone tree for Pleosporaceae. Fungal Divers. 2015, 71, 85–139. [Google Scholar] [CrossRef]
- Tian, Q.; Liu, J.K.; Hyde, K.D.; Wanasinghe, D.N.; Boonmee, S.; Jayasiri, S.C.; Luo, Z.L.; Taylor, J.E.; Phillips, A.J.L.; Bhat, D.J.; et al. Phylogenetic relationships and morphological reappraisal of Melanommataceae (Pleosporales). Fungal Divers. 2015, 74, 267–324. [Google Scholar] [CrossRef]
- Braun, U.; Crous, P.W.; Groenewald, J.Z.; Scheuer, C. Pseudovirgaria, a fungicolous hyphomycete genus. IMA Fungus 2011, 2, 65–69. [Google Scholar] [CrossRef] [Green Version]
- Arzanlou, M.; Groenewald, J.Z.; Gams, W.; Braun, U.; Shin, H.-D.; Crous, P.W. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Stud. Mycol. 2007, 58, 57–93. [Google Scholar] [CrossRef]
- Verkley, G.J.M.; Quaedvlieg, W.; Shin, H.-D.; Crous, P.W. A new approach to species delimitation in Septoria. Stud. Mycol. 2013, 75, 213–305. [Google Scholar] [CrossRef] [Green Version]
- Winton, L.M.; Stone, J.K.; Hansen, E.M.; Shoemaker, R.A. The systematic position of Phaeocryptopus gaeumannii. Mycologia 2007, 99, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Batzer, J.C.; Arias, M.M.D.; Harrington, T.C.; Gleason, M.L.; Groenewald, J.Z.; Crous, P.W. Four species of Zygophiala (Schizothyriaceae, Capnodiales) are associated with the sooty blotch and flyspeck complex on apple. Mycologia 2008, 100, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Dong, Y.; Phookamsak, R.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Liu, N.G.; Abeywickrama, P.D.; Mapook, A.; Wei, D.; et al. Fungal diversity notes 1151–1273: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020, 100, 5–277. [Google Scholar] [CrossRef] [Green Version]
- Samerpitak, K.; Van der Linde, E.; Choi, H.-J.; Gerrits van den Ende, A.H.G.; Machouart, M.; Gueidan, C.; de Hoog, G.S. Taxonomy of Ochroconis, genus including opportunistic pathogens on humans and animals. Fungal Divers. 2014, 65, 89–126. [Google Scholar] [CrossRef]
- Kruys, A.; Eriksson, O.E.; Wedin, M. Phylogenetic relationships of coprophilous Pleosporales (Dothideomycetes, Ascomycota), and the classification of some bitunicate taxa of unknown position. Mycol. Res. 2006, 110, 527–536. [Google Scholar] [CrossRef]
- Ismail, S.I.; Batzer, J.C.; Harrington, T.C.; Crous, P.W.; Lavrov, D.V.; Li, H.; Gleason, M.L. Ancestral state reconstruction infers phytopathogenic origins of sooty blotch and flyspeck fungi on apple. Mycologia 2016, 108, 292–302. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Schoch, C.L.; Fournier, J.; Crous, P.W.; de Gruyter, J.; Woudenberg, J.H.C.; Hirayama, K.; Tanaka, K.; Pointing, S.B.; Spatafora, J.W.; et al. Multi-locus phylogeny of Pleosporales: A taxonomic, ecological and evolutionary re-evaluation. Stud. Mycol. 2009, 64, 85–102. [Google Scholar] [CrossRef]
- Crous, P.W.; Schubert, K.; Braun, U.; de Hoog, G.S.; Hocking, A.D.; Shin, H.-D.; Groenewald, J.Z. Opportunistic, human-pathogenic species in the Herpotrichiellaceae are phenotypically similar to saprobic or phytopathogenic species in the Venturiaceae. Stud. Mycol. 2007, 58, 185–217. [Google Scholar] [CrossRef]
- Hongsanan, S.; Tian, Q.; Bahkali, A.H.; Yang, J.-B.; Mckenzie, E.H.C.; Chomnunti, P.; Hyde, K.D. Zeloasperisporiales ord. nov., and two new species of Zeloasperisporium. Cryptogam. Mycol. 2015, 36, 301–317. [Google Scholar] [CrossRef]
- Kuephadungphan, W.; Macabeo, A.P.G.; Luangsa-ard, J.J. Studies on the biologically active secondary metabolites of the new spider parasitic fungus Gibellula gamsii. Mycol. Prog. 2019, 18, 135–146. [Google Scholar] [CrossRef] [Green Version]
- Macabeo, A.P.G.; Cruz, A.J.C.; Narmani, A.; Arzanlou, M.; Babai-Ahari, A.; Pilapil, L.A.E.; Garcia, K.Y.M.; Huch, V.; Stadler, M. Tetrasubstituted α-pyrone derivatives from the endophytic fungus, Neurospora udagawae. Phytochem. Lett. 2020, 35, 147–151. [Google Scholar] [CrossRef]
- Wu, H.; Hyde, K.D. Re-appraisal of Scolecopeltidium. Mycotaxon 2013, 125, 1–10. [Google Scholar] [CrossRef]
- Wu, H.X.; Schoch, C.L.; Boonmee, S.; Bahkali, A.H.; Chomnunti, P.; Hyde, K.D. A Reappraisal of Microthyriaceae. Fungal Divers. 2011, 51, 189–248. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Jaklitsch, W.M.; Voglmayr, H.; Hyde, K.D. Epitypification, morphology, and phylogeny of Tothia fuscella. Mycotaxon 2011, 118, 203–211. [Google Scholar] [CrossRef]
- Wu, H.; Li, Y.; Chen, H.; Hyde, K.D. Studies on Microthyriaceae: Some excluded genera. Mycotaxon 2010, 113, 147–156. [Google Scholar] [CrossRef]
- Wu, H.; Hyde, K.D.; Chen, H. Studies on Microthyriaceae: Placement of Actinomyxa, Asteritea, Cirsosina, Polystomellina and Stegothyrium. Cryptog. Mycol. 2011, 32, 3–12. [Google Scholar] [CrossRef]
- Wu, H.; Tian, Q.; Li, W.; Hyde, K.D. A reappraisal of Microthyriaceae. Phytotaxa 2014, 176, 201–212. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Li, Y.-P.; Li, X.-X.; Lu, Z.-H.; Zheng, Q.-H.; Liu, Q.-Y. Isolation of 4,4′-bond secalonic acid D from the marine-derived fungus Penicillium oxalicum with inhibitory property against hepatocellular carcinoma. J. Antibiot. 2019, 72, 34–44. [Google Scholar] [CrossRef]
- Bao, J.; Sun, Y.-L.; Zhang, X.-Y.; Han, Z.; Gao, H.-C.; He, F.; Qian, P.-Y.; Qi, S.-H. Antifouling and antibacterial polyketides from marine gorgonian coral-associated fungus Penicillium sp. SCSGAF 0023. J. Antibiot. 2013, 66, 219–223. [Google Scholar] [CrossRef] [Green Version]
- El-Elimat, T.; Figueroa, M.; Raja, H.A.; Graf, T.N.; Swanson, S.M.; Falkinham, J.O.; Wani, M.C.; Pearce, C.J.; Oberlies, N.H. Biosynthetically distinct cytotoxic polyketides from Setophoma terrestris. Eur. J. Org. Chem. 2015, 2015, 109–121. [Google Scholar] [CrossRef] [Green Version]
- Tan, S.; Yang, B.; Liu, J.; Xun, T.; Liu, Y.; Zhou, X. Penicillixanthone A, a marine-derived dual-coreceptor antagonist as anti-HIV-1 agent. Nat. Prod. Res. 2019, 33, 1467–1471. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Krohn, K.; Zia-Ullah Flörke, U.; Pescitelli, G.; Di Bari, L.; Antus, S.; Kurtán, T.; Rheinheimer, J.; Draeger, S. New mono- and dimeric members of the secalonic acid family: Blennolides A-G isolated from the fungus Blennoria sp. Chemistry 2008, 14, 4913–4923. [Google Scholar] [CrossRef]
- Wittine, K.; Saftić, L.; Peršurić, Ž.; Kraljević Pavelić, S. Novel antiretroviral structures from marine organisms. Molecules 2019, 24, 3486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asai, T.; Otsuki, S.; Sakurai, H.; Yamashita, K.; Ozeki, T.; Oshima, Y. Benzophenones from an endophytic fungus, Graphiopsis chlorocephala, from Paeonia lactiflora cultivated in the presence of an NAD+-dependent HDAC inhibitor. Org. Lett. 2013, 15, 2058–2061. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Norphanphoun, C.; Chen, J.; Dissanayakem, A.J.; Doilom, M.; Hongsanan, S.; Jayawardena, R.S.; Jeewon, R.; Perera, R.H.; Thongbai, B.; et al. Thailand’s amazing diversity—An estimated 55–96% of fungi in northern Thailand are novel. Fungal Divers. 2018, 93, 215–239. [Google Scholar] [CrossRef]






| Taxa | Strain No. 1 | GenBank Accession Numbers 2 | References | ||||
|---|---|---|---|---|---|---|---|
| LSU | SSU | RPB2 | ITS | TEF | |||
| Acrospermum adeanum | M133 | EU940104 | EU940031 | EU940320 | EU940180 | - | Stenroos et al. [26] |
| Acrospermum compressum | M151 | EU940084 | EU940012 | EU940301 | EU940161 | - | Stenroos et al. [26] |
| Acrospermum gramineum | M152 | EU940085 | EU940013 | EU940302 | EU940162 | - | Stenroos et al. [26] |
| Alternaria alternata | KFRD-18 | KX609781 | KX609769 | - | KX346897 | KY094931 | Li et al. [27] |
| Alternariaster bidentis | CBS 134021 | KC609341 | - | KC609347 | KC609333 | - | Alves et al. [28] |
| Antennariella placitae | CBS:124785 | GQ303299 | - | - | MH863403 | - | Cheewangkoon et al. [29] |
| Arxiella dolichandrae | CBS 138853 | KP004477 | - | - | KP004449 | - | Crous et al. [30] |
| Arxiella terrestris | CBS 268.65 | MH870201 | - | - | MH858565 | - | Vu et al. [31] |
| Asterina fuchsiae | TH590 | GU586216 | GU586210 | - | - | - | Hofmann et al. [32] |
| Asterina phenacis | TH589 | GU586217 | GU586211 | - | - | - | Hofmann et al. [32] |
| Bambusicola massarinia | MFLUCC 11-0389 | JX442037 | JX442041 | KU940169 | JX442033 | - | Dai et al. [33] |
| Bambusicola splendida | MFLUCC 11-0439 | JX442038 | JX442042 | - | JX442034 | - | Dai et al. [33] |
| Botryosphaeria agaves | MFLUCC 11-0125 | JX646808 | JX646825 | - | JX646791 | JX646856 | Liu et al. [34] |
| Botryosphaeria tsugae | AFTOL-ID 1586 | DQ767655 | - | DQ767644 | - | DQ677914 | Schoch et al. [35] |
| Calicium salicinum | CBS 100898 | KF157982 | KF157970 | KF157998 | - | - | Beimforde et al. [36] |
| Calicium viride | 10-VII-1997 (DUKE) | AF356670 | AF356669 | AY641031 | - | - | Lutzoni et al. [37] |
| Camarosporium quaternatum | CBS 483.95 | GU301806 | GU296141 | GU357761 | KY929149 | GU349044 | Schoch et al. [38] |
| Capnodium salicinum | AFTOL-ID 937 | DQ678050 | DQ677997 | - | - | DQ677889 | Schoch et al. [37] |
| Caryospora minima | - | EU196550 | EU196551 | - | - | - | Cai and Hyde [39] |
| Chaetothyriothecium elegans | CPC 21375 | KF268420 | - | - | - | - | Hongsanan et al. [40] |
| Corynespora cassiicola | CBS 100822 | GU301808 | GU296144 | GU371742 | - | GU349052 | Schoch et al. [38] |
| Corynespora smithii | CABI 5649b | GU323201 | - | GU371783 | - | GU349018 | Schoch et al. [38] |
| Cucurbitaria berberidis | MFLUCC 11-0387 | KC506796 | KC506800 | - | - | - | Hyde et al. [41] |
| Cyphelium inquinans | Tibell 22283 (UPS) | AY453639 | U86695 | - | AY450584 | - | Tibell [42] |
| Cyphelium tigillare | Tibell 22343 (UPS) | AY453641 | AF241545 | - | AY452497 | - | Tibell [42] |
| Cystocoleus ebeneus | L161 | EU048578 | EU048571 | - | - | - | Muggia et al. [43] |
| Didymella exigua | CBS 183.55 | JX681089 | EU754056 | GU371764 | MH857436 | KR184187 | Verkley et al. [44] |
| Didymosphaeria rubi-ulmifolii | MFLUCC 14-0023 | KJ436586 | KJ436588 | - | - | - | Ariyawansa et al. [45] |
| Dothiora cannabinae | AFTOL ID 1359 | DQ470984 | DQ479933 | DQ470936 | - | DQ471107 | Spatafora et al. [46] |
| Dyfrolomyces phetchaburiensis | MFLUCC 15-0951 | MF615402 | MF615403 | - | - | - | Hyde et al. [47] |
| Dyfrolomyces rhizophorae | BCC15481 | - | KF160009 | - | - | - | Pang et al. [48] |
| Dyfrolomyces rhizophorae | JK 5456A | GU479799 | - | - | - | GU479860 | Suetrong et al. [49] |
| Dyfrolomyces thailandica | MFLU 16-1173 | KX611366 | KX611367 | - | - | - | Hyde et al. [50] |
| Dyfrolomyces thamplaensis | MFLUCC 15-0635 | KX925435 | KX925436 | - | - | KY814763 | Zhang et al. [51] |
| Dyfrolomyces tiomanensis | NTOU3636 | KC692156 | KC692155 | - | - | KC692157 | Pang et al. [48] |
| Elsinoe fawcettii | CPC 18535 | JN940382 | JN940559 | - | KX887207 | KX886853 | Schoch et al. [52] |
| Elsinoe verbenae | CPC 18561 | JN940391 | JN940562 | - | KX887298 | KX886942 | Schoch et al. [53] |
| Extremus antarcticus | CCFEE 5312 | KF310020 | - | KF310086 | KF309979 | - | Egidi et al. [54] |
| Gonatophragmium triuniae | CBS 138901 | KP004479 | - | - | KP004451 | - | Crous et al. [30] |
| Helicascus nypae | BCC 36751 | GU479788 | GU479754 | GU479826 | - | GU479854 | Suetrong et al. [49] |
| Julella avicenniae | BCC 20173 | GU371822 | GU371830 | GU371786 | - | GU371815 | Schoch et al. [38] |
| Karschia cezannei | Cezanne-Eichler B26 | KP456152 | - | - | - | - | Ertz and Diederich [55] |
| Katumotoa bambusicola | KT 1517a | AB524595 | AB524454 | AB539095 | NR_154103 | AB539108 | Tanaka et al. [56] |
| Labrocarpon canariense | Ertz 16907 (BR) | KP456157 | - | - | - | - | Ertz and Diederich [55] |
| Lentithecium fluviatile | CBS 123090 | FJ795450 | FJ795492 | FJ795467 | - | - | Zhang et al. [57] |
| Leptodiscella africana | CBS 400.65 | MH870275 | - | - | MH858635 | - | Vu et al. [31] |
| Leptodiscella brevicatenata | FMR 10885 | FR821311 | - | - | FR821312 | - | Madrid et al. [58] |
| Leptodiscella chlamydospora | MUCL 28859 | FN869567 | - | - | FR745398 | - | Madrid et al. [58] |
| Leptodiscella rintelii | CBS 144927 | LR025181 | - | - | LR025180 | - | Papendorf [52] |
| Leptosphaeria doliolum | MFLUCC 15-1875 | KT454719 | KT454734 | - | KT454727 | - | Ariyawansa et al. [59] |
| Leptosphaerulina australis | CBS 317.83 | EU754166 | GU296160 | GU371790 | MH861604 | GU349070 | de Gruyter et al. [60] |
| Leptoxyphium cacuminum | MFLUCC10-0049 | JN832602 | JN832587 | - | - | - | Chomnunti et al. [61] |
| Lophiotrema nucula | CBS 627 86 | GU301837 | GU296167 | GU371792 | LC194497 | GU349073 | Schoch et al. [38] |
| Lophium mytilinum | AFTOL-ID 1609 | DQ678081 | DQ678030 | DQ677979 | - | DQ677926 | Schoch et al. [35] |
| Massarina bambusina | H 4321 | AB807536 | AB797246 | - | LC014578 | AB808511 | Tanaka et al. [56] |
| Massarina eburnea | CBS 473.64 | GU301840 | GU296170 | GU371732 | - | GU349040 | Schoch et al. [38] |
| Melanomma pulvis-pyrius | CBS 371 75 | GU301845 | FJ201989 | GU371798 | - | GU349019 | Schoch et al. [38] |
| Melaspileopsiscf. diplasiospora | Ertz 16247 (BR) | KP456164 | - | - | - | - | Ertz and Diederich [55] |
| Melomastia maolanensis | GZCC 16-0102 | KY111905 | KY111906 | - | - | KY814762 | Zhang et al. [51] |
| Microsphaeropsis olivacea | CBS 233 77 | GU237988 | - | KT389643 | MH861055 | - | Aveskamp et al. [62] |
| Microthyrium buxicola | MFLUCC 15-0213 | KT306552 | KT306550 | - | - | - | Ariyawansa et al. [63] |
| Microthyrium microscopicum | CBS 115976 | GU301846 | GU296175 | GU371734 | - | GU349042 | Schoch et al. [44] |
| Multiseptospora thailandica | MFLUCC 11-0183 | KP744490 | KP753955 | - | KP744447 | KU705657 | Liu et al. [64] |
| Murispora rubicunda | IFRD 2017 | FJ795507 | GU456308 | - | - | GU456289 | Zhang et al. [57] |
| Muyocopron alcornii | BRIP 43897 | MK487708 | - | MK492712 | MK487735 | MK495956 | Hernández-Restrepo et al. [22] |
| Muyocopron atromaculans | MUCL 34983 | MK487709 | - | MK492713 | MK487736 | MK495957 | Hernández-Restrepo et al. [22] |
| Muyocopron castanopsis | MFLUCC 10-0042 | - | JQ036225 | - | - | - | Mapook et al. [23] |
| Muyocopron castanopsis | MFLUCC 14-1108 | KU726965 | KU726968 | KY225778 | MT137784 | MT136753 | Mapook et al. [23] |
| Muyocopron chromolaenae | MFLUCC 17-1513 | MT137876 | MT137881 | MT136761 | MT137777 | MT136756 | Mapook et al. [24] |
| Muyocopron chromolaenicola | MFLUCC 17-1470 | MT137877 | MT137882 | - | MT137778 | MT136757 | Mapook et al. [24] |
| Muyocopron coloratum | CBS 720.95 | MK487710 | - | MK492714 | NR_160197 | MK495958 | Hernández-Restrepo et al. [22] |
| Muyocopron dipterocarpi | MFLUCC 14-1103 | KU726966 | KU726969 | KY225779 | MT137785 | MT136754 | Mapook et al. [23] |
| Muyocopron dipterocarpi | MFLUCC 17-0075 | MH986833 | MH986829 | - | MH986837 | - | Senwanna et al. [65] |
| Muyocopron dipterocarpi | MFLUCC 17-0354 | MH986834 | MH986830 | - | MH986838 | - | Senwanna et al. [65] |
| Muyocopron dipterocarpi | MFLUCC 17-0356 | MH986835 | MH986831 | - | MH986839 | - | Senwanna et al. [66] |
| Muyocopron dipterocarpi | MFLUCC 18-0470 | MK348001 | MK347890 | - | MK347783 | - | Jayasiri et al. [67] |
| Muyocopron garethjonesii | MFLU 16-2664 | KY070274 | KY070275 | - | - | - | Tibpromma et al. [68] |
| Muyocopron geniculatum | CBS 721.95 | MK487711 | - | MK492715 | MK487737 | MK495959 | Hernández-Restrepo et al. [22] |
| Muyocopron heveae | MFLUCC 17-0066 | MH986832 | MH986828 | - | MH986836 | - | Senwanna et al. [66] |
| Muyocopron laterale | CBS 141029 | MK487712 | - | MK492716 | MK487738 | MK495960 | Hernández-Restrepo et al. [22] |
| Muyocopron laterale | IMI 324533 | MK487713 | - | MK492717 | MK487739 | MK495961 | Hernández-Restrepo et al. [22] |
| Muyocopron laterale | CBS 719.95 | MK487714 | - | MK492718 | MK487740 | MK495962 | Hernández-Restrepo et al. [22] |
| Muyocopron laterale | CBS 141033 | MK487715 | - | MK492719 | MK487741 | MK495963 | Hernández-Restrepo et al. [22] |
| Muyocopron laterale | URM 7802 | MK487716 | - | MK492720 | MK487742 | MK495964 | Hernández-Restrepo et al. [22] |
| Muyocopron laterale | URM 7801 | MK487717 | - | MK492721 | MK487743 | - | Hernández-Restrepo et al. [22] |
| Muyocopron laterale | CBS 127677 | MK487718 | - | MK492722 | MK487744 | MK495965 | Hernández-Restrepo et al. [22] |
| Muyocopron laterale | CBS 145310 | MK487719 | - | MK492723 | MK487745 | MK495966 | Hernández-Restrepo et al. [22] |
| Muyocopron laterale | CBS 145315 | MK487720 | - | MK492724 | MK487746 | MK495967 | Hernández-Restrepo et al. [22] |
| Muyocopron laterale | CBS 145313 | MK487721 | - | MK492725 | MK487747 | MK495968 | Hernández-Restrepo et al. [22] |
| Muyocopron laterale | CBS 145309 | MK487722 | - | MK492726 | MK487748 | MK495969 | Hernández-Restrepo et al. [22] |
| Muyocopron laterale | CBS 145314 | MK487723 | - | MK492727 | MK487749 | MK495970 | Hernández-Restrepo et al. [22] |
| Muyocopron laterale | CBS 145311 | MK487724 | - | MK492728 | MK487750 | - | Hernández-Restrepo et al. [22] |
| Muyocopron laterale | CBS 145312 | MK487725 | - | MK492729 | MK487751 | MK495971 | Hernández-Restrepo et al. [22] |
| Muyocopron laterale | CBS 145316 | MK487726 | - | MK492730 | MK487752 | MK495972 | Hernández-Restrepo et al. [22] |
| Muyocopron laterale | FMR13797 | MK874616 | - | MK875802 | MK874615 | MK875803 | Hernández-Restrepo et al. [22] |
| Muyocopron lithocarpi | MFLUCC 10-0041 | JQ036230 | JQ036226 | - | - | - | Mapook et al. [23] |
| Muyocopron lithocarpi | MFLUCC 14-1106 | KU726967 | KU726970 | KY225780 | MT137786 | MT136755 | Mapook et al. [23] |
| Muyocopron lithocarpi | MFLU 18-2087 | MK347930 | MK347821 | - | MK347716 | - | Jayasiri et al. [66] |
| Muyocopron lithocarpi | MFLU 18-2088 | MK347931 | MK347822 | - | MK347717 | - | Jayasiri et al. [66] |
| Muyocopron lithocarpi | MFLUCC 16-0962 | MK348034 | MK347923 | - | - | - | Jayasiri et al. [66] |
| Muyocopron lithocarpi | MFLUCC 17-1465 | MT137878 | MT137883 | - | MT137779 | MT136758 | Mapook et al. [24] |
| Muyocopron lithocarpi | MFLUCC 17-1466 | MT137879 | MT137884 | - | MT137780 | MT136759 | Mapook et al. [24] |
| Muyocopron lithocarpi | MFLUCC 17-1500 | MT137880 | MT137885 | MT136762 | MT137781 | MT136760 | Mapook et al. [24] |
| Muyocopron zamiae | CBS 203.71 | MK487727 | - | MK492731 | - | MK495973 | Hernández-Restrepo et al. [22] |
| Mycoleptodiscus endophytica | MFLUCC 17-0545 | MG646946 | MG646978 | - | MG646961 | MG646985 | Tibpromma et al. [69] |
| Mycoleptodiscus suttonii | CBS 276.72 | MK487728 | - | MK492732 | MK487753 | MK495974 | Hernández-Restrepo et al. [22] |
| Mycoleptodiscus suttonii | CBS 141030 | MK487729 | - | MK492733 | - | MK495975 | Hernández-Restrepo et al. [22] |
| Mycoleptodiscus terrestris | CBS 231.53 | MK487730 | - | MK492734 | MK487754 | MK495976 | Hernández-Restrepo et al. [22] |
| Mycoleptodiscus terrestris | IMI 159038 | MK487731 | - | MK492735 | MK487755 | MK495977 | Hernández-Restrepo et al. [22] |
| Myriangium duriaei | CBS 260.36 | NG_027579 | AF242266 | KT216528 | MH855793 | - | Schoch et al. [35] |
| Myriangium hispanicum | CBS 247.33 | GU301854 | GU296180 | GU371744 | MH855426 | GU349055 | Schoch et al. [38] |
| Mytilinidion rhenanum | CBS 135.34 | FJ161175 | FJ161136 | FJ161115 | - | FJ161092 | Boehm et al. [70] |
| Natipusilla decorospora | AF236 1a | HM196369 | HM196376 | - | - | - | Ferrer et al. [71] |
| Natipusilla naponensis | AF217 1a | HM196371 | HM196378 | - | - | - | Ferrer et al. [71] |
| Neocochlearomyces chromolaenae | BCC 68250 | MK047514 | MK047552 | - | MK047464 | MK047573 | Crous et al. [21] |
| Neocochlearomyces chromolaenae | BCC 68251 | MK047515 | MK047553 | - | MK047465 | MK047574 | Crous et al. [21] |
| Neocochlearomyces chromolaenae | BCC 68252 | MK047516 | MK047554 | - | MK047466 | MK047575 | Crous et al. [21] |
| Neocylindroseptoria pistaciae | CBS 471.69 | KF251656 | - | KF252161 | KF251152 | KF253112 | Quaedvlieg et al. [65] |
| Neomycoleptodiscus venezuelense | CBS 100519 | MK487732 | - | MK492736 | MK487756 | MK495978 | Hernández-Restrepo et al. [22] |
| Palawania thailandensis | MFLUCC 14-1121 | KY086493 | KY086495 | KY086496 | MT137787 | - | Mapook et al. [24] |
| Palawania thailandensis | MFLU 16-1871 | KY086494 | - | - | MT137788 | - | Mapook et al. [24] |
| Paramycoleptodiscus albizziae | CPC 27552 | MH878220 | - | - | - | - | Vu et. al. [31] |
| Paramycoleptodiscus albizziae | CBS 141320 | KX228330 | - | MK492737 | KX228279 | MK495979 | Crous et. al. [72] |
| Phaeodimeriella cissampeli | MFLU 16-0558 | KU746806 | KU746808 | KU746810 | - | KU746812 | Mapook et. al. [73] |
| Phaeodimeriella dilleniae | MFLU 14-0013 | KU746805 | KU746807 | KU746809 | - | KU746811 | Mapook et. al. [73] |
| Phaeotrichum benjaminii | CBS 541.72 | AY004340 | AY016348 | GU357788 | MH860561 | DQ677892 | Lumbsch et. al. [74] |
| Physcia aipolia | AFTOL-ID 84 | DQ782904.1 | DQ782876 | DQ782862 | DQ782836 | DQ782892 | James et. al. [75] |
| Piedraia hortae | CBS 480.64 | GU214466 | - | KF902289 | GU214647 | - | Crous et. al. [76] |
| Platystomum crataegi | MFLUCC 14-0925 | KT026109 | KT026113 | - | NG_063580 | KT026121 | Thambugala et. al. [77] |
| Pleomassaria siparia | AFTOL-ID 1600 | DQ678078 | DQ678027 | DQ677976 | - | DQ677923 | Schoch et. al. [35] |
| Pleospora herbarum | IT 956 | KP334709 | KP334729 | KP334733 | KP334719 | KP334731 | Ariyawansa et. al. [78] |
| Preussia funiculata | CBS 659.74 | GU301864 | GU296187 | GU371799 | - | GU349032 | Schoch et. al. [38] |
| Pseudomassariosphaeria bromicola | IT-1333 | KT305994 | KT305996 | - | KT305998 | KT305999 | Ariyawansa et. al. [63] |
| Pseudopalawania siamensis | MFLUCC 17-1476a | - | MT137789 | - | MT137782 | MT136752 | This study |
| Pseudopalawania siamensis | MFLUCC 17-1476b | - | MT137790 | - | MT137783 | - | This study |
| Pseudostrickeria muriformis | MFLUCC 13-0764 | KT934254 | KT934258 | - | - | KT934262 | Tian et. al. [79] |
| Pseudovirgaria grisea | CPC 19134 | JF957614 | - | - | JF957609 | - | Braun et. al. [80] |
| Pseudovirgaria hyperparasitica | CPC 10753 | EU041824 | - | - | EU041767 | - | Arzanlou et. al. [81] |
| Ramularia endophylla | CBS 113265 | KF251833 | - | KP894673 | KF251220 | - | Verkley et. al. [82] |
| Rasutoria pseudotsugae | rapssd | EF114704 | EF114729 | - | EF114687 | - | Winton et al. [83] |
| Rasutoria tsugae | ratstk | EF114705 | EF114730 | GU371809 | EF114688 | - | Winton et al. [83] |
| Salsuginea ramicola | KT 2597.1 | GU479800 | GU479768 | GU479833 | - | GU479861 | Suetrong et al. [49] |
| Schizothyrium pomi | CBS 406.61 | EF134949 | - | KF902384 | - | - | Batzer et al. [84] |
| Setoapiospora thailandica | MFLUCC 17-1426 | MN638847 | MN638851 | - | MN638862 | MN648731 | Hyde et al. [85] |
| Stictographa lentiginosa | Ertz 17570 (BR) | KP456170 | - | - | - | - | Ertz and Diederich [55] |
| Sympoventuria capensis | CBS 120136 | KF156104 | KF156094 | - | KF156039 | - | Samerpitak et al. [86] |
| Teratosphaeria fibrillosa | CBS 121707 | GU323213 | GU296199 | GU357767 | MH863138 | KF903305 | Schoch et al. [38] |
| Trichodelitschia munkii | Kruys 201 (UPS) | DQ384096 | DQ384070 | - | - | - | Kruys et al. [87] |
| Tumidisporashoreae | MFLUCC 14-0574 | KT314074 | KT314076 | - | - | - | Ariyawansa et al. [63] |
| Uwebraunia commune | NC132C1d | - | - | KT216546 | - | - | Ismail et al. [88] |
| Venturia inaequalis | CBS 594.70 | GU301879 | GU296205 | GU357757 | KF156040 | GU349022 | Schoch et al. [38] |
| Xenolophium applanatum | CBS 123127 | GU456330 | GU456313 | GU456355 | - | GU456270 | Zhang et al. [89] |
| Zeloasperisporium hyphopodioides | CBS 218.95 | EU035442 | - | - | - | - | Crous et al. [90] |
| Zeloasperisporium siamense | IFRDCC 2194 | JQ036228 | JQ036223 | - | - | - | Mapook et. al. [73] |
| Zeloasperisporium wrightiae | MFLUCC 15-0225 | KT387737 | KT387738 | - | - | - | Hongsanan et al. [91] |
| No. | δH, m, J (Hz) | δC, m | No. | δH, m, J (Hz) | δC, m |
|---|---|---|---|---|---|
| 1 | - | 160.1, C | 1′ | - | 161.8, C |
| 2 | - | 117.6, C | 2′ | 6.66, d (8.7) | 110.4, CH |
| 3 | 7.82, d (8.6) | 143.8, CH | 3′ | 7.54, d (8.7) | 141.2, CH |
| 4 | 6.77, d (8.6) | 108.3, CH | 4′ | - | 114.0, C |
| 4a | - | 158.3, C | 4a′ | - | 155.6, C |
| 5 | 4.44, d (4.0) | 74.1, CH | 5′ | 4.38, d (2.5) | 88.1, CH |
| 6 | 2.13, m | 30.4, CH | 6′ | 2.65, m | 29.9, CH |
| 7a | 2.36, dd (15.9, 13.6) | 33.8, CH2 | 7′a | 2.18, m | 35.8, CH2 |
| b | 2.12, m | b | 1.99, dd (18.3, 3.1) | ||
| 8 | - | 108.9, C | 8′ | - | 176.5, C |
| 8a | - | 73.6, C | 8a′a | 3.14, d (16.9) | 39.6, CH2 |
| b | 2.98, d (16.9) | ||||
| 9 | - | 194.9, C | 9′ | - | 193.6, C |
| 9a | - | 106.8, C | 9a′ | - | 107.6, C |
| 10a | - | 84.7, C | 10a′ | - | 84.8, C |
| 11 | 1.20, d (6.5) | 14.9, CH3 | 11′ | 1.16, d (7.2) | 20.9, CH3 |
| 12 | - | 176.6, C | 12′ | - | 168.5, C |
| 13 | - | - | 13′ | 3.80, s | 53.7, CH3 |
| 1-OH | 11.35, s | - | 1′-OH | 11.51, s | - |
| Tested Organisms | Strain No. | Minimum Inhibitory Concentration (MIC) [μg/mL] | |||||
|---|---|---|---|---|---|---|---|
| Compounds | Positive Control * | ||||||
| 1 | 2 | 3 | 4 | 5 | |||
| Fungi | |||||||
| Candida albicans | DSM 1665 | - | 66.7 | - | - | - | 66.7 (20 µL N) |
| Cryptococcus neoformans | DSM 15466 | - | - | - | - | - | 66.7 (20 µL N) |
| Mucor hiemalis | DSM 6766 | - | - | 66.7 | - | - | 66.7 (20 µL N) |
| Pichia anomala | DSM 6766 | - | - | - | - | - | 66.7 (20 µL N) |
| Rhodoturula glutinis | DSM 10134 | - | - | - | - | - | 16.7 (20 µL N) |
| Schizosaccharomyces pombe | DSM 70572 | - | - | - | - | - | 33.3 (20 µL N) |
| Bacteria | |||||||
| Bacillus subtilis | DSM 10 | 66.7 | 1.0 | 4.2 | - | - | 8.3 (20 µL O) |
| Chromobacterium violaceum | DSM 30191 | - | - | - | - | - | 1.7 (2 µL O) |
| Escherichia coli | DSM 1116 | - | - | - | - | - | 3.3 (2 µL O) |
| Micrococcus luteus | DSM 1790 | 66.7 | 8.3 | 33.3 | - | - | 0.4 (2 µL O) |
| Mycobacterium smegmatis | ATCC 700084 | - | 66.7 | - | - | - | 3.3 (2 µL K) |
| Pseudomonas aeruginosa | PA14 | - | - | - | - | - | 0.8 (2 µL G) |
| Staphylococcus aureus | DSM 346 | 66.7 | 4.2 | 33.3 | - | - | 0.2 (2 µL O) |
| Cell Lines | IC50 (µM) | |||||
|---|---|---|---|---|---|---|
| Compounds | Epothilone B | |||||
| 1 | 2 | 3 | 4 | 5 | ||
| HeLa cells KB3.1 | 29.7 | 3.9 | 17.2 | - | - | 8.9 × 10−5 |
| Mouse fibroblast L929 | 50.0 | 14.1 | - | - | - | 1.8 × 10−3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mapook, A.; Macabeo, A.P.G.; Thongbai, B.; Hyde, K.D.; Stadler, M. Polyketide-Derived Secondary Metabolites from a Dothideomycetes Fungus, Pseudopalawania siamensis gen. et sp. nov., (Muyocopronales) with Antimicrobial and Cytotoxic Activities. Biomolecules 2020, 10, 569. https://doi.org/10.3390/biom10040569
Mapook A, Macabeo APG, Thongbai B, Hyde KD, Stadler M. Polyketide-Derived Secondary Metabolites from a Dothideomycetes Fungus, Pseudopalawania siamensis gen. et sp. nov., (Muyocopronales) with Antimicrobial and Cytotoxic Activities. Biomolecules. 2020; 10(4):569. https://doi.org/10.3390/biom10040569
Chicago/Turabian StyleMapook, Ausana, Allan Patrick G. Macabeo, Benjarong Thongbai, Kevin D. Hyde, and Marc Stadler. 2020. "Polyketide-Derived Secondary Metabolites from a Dothideomycetes Fungus, Pseudopalawania siamensis gen. et sp. nov., (Muyocopronales) with Antimicrobial and Cytotoxic Activities" Biomolecules 10, no. 4: 569. https://doi.org/10.3390/biom10040569