Antifungal Activity against Fusarium oxysporum of Botanical End-Products: An Integration of Chemical Composition and Antifungal Activity Datasets to Identify Antifungal Bioactives
Abstract
:1. Introduction
2. Results and Discussion
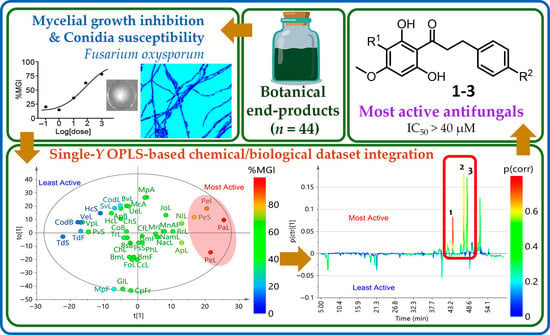
2.1. Antifungal Activity of Test Botanical Extracts
2.2. Integration of Chemical and Activity Datasets: Indirect Recognition of Bioactives
2.3. Isolation and Identification of Antifungal Metabolites
3. Materials and Methods
3.1. Preparation of Botanical Extracts
3.2. In-Vitro Assays against F. oxysporum
3.2.1. Mycelial Growth Inhibition Assay
3.2.2. Conidial Susceptibility Assay
3.3. High-Performance Liquid Chromatography Coupled to Diode Array Detector
3.4. Statistical Analysis
3.5. Purification and Identification of Most-Active Compounds by Semipreparative-Scale HPLC
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garagounis, C.; Delkis, N.; Papadopoulou, K.K. Unraveling the roles of plant specialized metabolites: Using synthetic biology to design molecular biosensors. New Phytol. 2021, 231, 1338–1352. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.; Kazachkova, Y.; Aharoni, A. Catch-22 in specialized metabolism: Balancing defense and growth. J. Exp. Bot. 2021, 72, 6027–6041. [Google Scholar] [CrossRef] [PubMed]
- Zeilinger, S.; Gupta, V.K.; Dahms, T.E.S.; Silva, R.N.; Singh, H.B.; Upadhyay, R.S.; Gomes, E.V.; Tsui, C.K.-M.; Nayak, S.C. Friends or foes? Emerging insights from fungal interactions with plants. FEMS Microbiol. Rev. 2016, 40, 182–207. [Google Scholar] [CrossRef] [Green Version]
- Baayen, R.P.; O’Donnell, K.; Bonants, P.J.; Cigelnik, E.; Kroon, L.P.; Roebroeck, E.J.; Waalwijk, C. Gene genealogies and AFLP analyses in the Fusarium oxysporum complex identify monophyletic and nonmonophyletic formae speciales causing wilt and rot disease. Phytopathology 2000, 90, 891–900. [Google Scholar] [CrossRef] [Green Version]
- Rubio-Reque, G.L.; Baltodano-Sánchez, F.D.M.; Liliana, I.; Wilson-Krugg, J.H.; Muñoz-Ríos, M.A. In vitro resistance of Rhizoctonia solani and Fusarium oxysporum to the fungicides Benzomil 500, Rhizolex-T and Homai-WP. Rev. Biológica Univ. Trujillo 2008, 28. [Google Scholar]
- Thottempudi, V.; Shreeve, J.M. Synthesis and promising properties of a new family of high-Density energetic salts of 5-Nitro-3-trinitromethyl-1H-1,2,4-triazole and 5,5′-bis(trinitromethyl)-3,3′-azo-1H-1,2,4-triazole. J. Am. Chem. Soc. 2011, 133, 19982–19992. [Google Scholar] [CrossRef] [PubMed]
- Pagani, A.P.S.; Dianese, A.C.; Café-Filho, A.C. Management of wheat blast with synthetic fungicides, partial resistance and silicate and phosphite minerals. Phytoparasitica 2014, 42, 609–617. [Google Scholar] [CrossRef]
- Gu, K.X.; Song, X.S.; Xiao, X.M.; Duan, X.X.; Wang, J.X.; Duan, Y.B.; Hou, Y.P.; Zhou, M.G. Aβ2-Tubulin dsRNA derived from Fusarium asiaticum confers plant resistance to multiple phytopathogens and reduces fungicide resistance. Pestic. Biochem. Physiol. 2019, 153, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, V.; Vashisht, D.; Cletus, J.; Sakthivel, N. Plant β-1,3-Glucanases: Their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 2012, 34, 1983–1990. [Google Scholar] [CrossRef]
- de Alencar, E.R.; Faroni, L.R.D.A.; de Fátima Ferreira Soares, N.; da Silva, W.A.; da Silva Carvalho, M.C. Efficacy of ozone as a fungicidal and detoxifying agent of aflatoxins in peanuts. J. Sci. Food Agric. 2012, 92, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Palareti, G.; Legnani, C.; Cosmi, B.; Antonucci, E.; Erba, N.; Poli, D.; Testa, S.; Tosetto, A. Comparison between different D-Dimer cutoff values to assess the individual risk of recurrent venous thromboembolism: Analysis of results obtained in the DULCIS study. Int. J. Lab. Hematol. 2016, 38, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Naeini, A.; Ziglari, T.; Shokri, H.; Khosravi, A.R. Assessment of growth-inhibiting effect of some plant essential oils on different Fusarium isolates. J. Mycol. Med. 2010, 20, 174–178. [Google Scholar] [CrossRef]
- Seepe, H.A.; Nxumalo, W.; Amoo, S.O. Natural products from medicinal plants against phytopathogenic Fusarium species: Current research endeavours, challenges and prospects. Molecules 2021, 26, 6539. [Google Scholar] [CrossRef]
- Shahidi, F.; Varatharajan, V.; Oh, W.Y.; Peng, H. Phenolic compounds in agri-Food by-Products, their bioavailability and health effects. J. Food Bioact. 2019, 5, 57–119. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, S.; Thakur, S.; Jaitak, V.; Bhardwaj, P. Gene and metabolite profiling reveals flowering and survival strategies in Himalayan Rhododendron arboreum. Gene 2019, 690, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dória, G.A.A.; Silva, W.J.; Carvalho, G.A.; Alves, P.B.; Cavalcanti, S.C.H. A study of the larvicidal activity of two Croton species from northeastern Brazil against Aedes aegypti. Pharm. Biol. 2010, 48, 615–620. [Google Scholar] [CrossRef]
- Álvarez-Caballero, J.M.; Cuca-Suárez, L.E.; Coy-Barrera, E. Bio-Guided fractionation of ethanol extract of leaves of Esenbeckia alata Kunt (Rutaceae) led to the isolation of two cytotoxic quinoline alkaloids: Evidence of selectivity against leukemia cells. Biomolecules 2019, 9, 585. [Google Scholar] [CrossRef] [Green Version]
- Granato, D.; Nunes, D.S.; Barba, F.J. An integrated strategy between food chemistry, biology, nutrition, pharmacology, and statistics in the development of functional foods: A proposal. Trends Food Sci. Technol. 2017, 62, 13–22. [Google Scholar] [CrossRef]
- Kellogg, J.J.; Todd, D.A.; Egan, J.M.; Raja, H.A.; Oberlies, N.H.; Kvalheim, O.M.; Cech, N.B. Biochemometrics for natural products research: Comparison of data analysis approaches and application to identification of bioactive compounds. J. Nat. Prod. 2016, 79, 376–386. [Google Scholar] [CrossRef] [Green Version]
- Salehi, B.; Zakaria, Z.A.; Gyawali, R.; Ibrahim, S.A.; Rajkovic, J.; Shinwari, Z.K.; Khan, T.; Sharifi-Rad, J.; Ozleyen, A.; Turkdonmez, E.; et al. Piper species: A comprehensive review on their phytochemistry, biological activities and applications. Molecules 2019, 24, 1364. [Google Scholar] [CrossRef] [Green Version]
- Tangarife-Castaño, V.; Correa-Royero, J.B.; Roa-Linares, V.C.; Pino-Benitez, N.; Betancur-Galvis, L.A.; Durán, D.C.; Stashenko, E.E.; Mesa-Arango, A.C. Anti-dermatophyte, anti-Fusarium and cytotoxic activity of essential oils and plant extracts of Piper genus. J. Essent. Oil Res. 2014, 26, 221–227. [Google Scholar] [CrossRef]
- Muñoz Castellanos, L.; Amaya Olivas, N.; Ayala-Soto, J.; De La Contreras, C.M.O.; Zermeño Ortega, M.; Sandoval Salas, F.; Hernández-Ochoa, L. In vitro and in vivo antifungal activity of clove (Eugenia caryophyllata) and pepper (Piper nigrum L.) essential oils and functional extracts against Fusarium oxysporum and Aspergillus niger in tomato (Solanum lycopersicum L.). Int. J. Microbiol. 2020, 2020, 1–8. [Google Scholar] [CrossRef]
- Xiang, C.-P.; Shi, Y.-N.; Liu, F.-F.; Li, H.-Z.; Zhang, Y.-J.; Yang, C.-R.; Xu, M. A survey of the chemical compounds of Piper spp. (Piperaceae) and their biological activities. Nat. Prod. Commun. 2016, 11, 1403–1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, R.; Darin, K.; Nath, P.; Deb, P. An overview of various Piper species for their biological activities. Int. J. Pharma Res. Rev. 2014, 3, 67–75. [Google Scholar]
- Gutiérrez, Y.; Montes, R.; Scull, R.; Sánchez, A.; Cos, P.; Monzote, L.; Setzer, W.N. Chemodiversity associated with cytotoxicity and antimicrobial activity of Piper aduncum var. ossanum. Chem. Biodivers. 2016, 13, 1715–1719. [Google Scholar] [CrossRef]
- Navickiene, H.M.D.; Morandim, A.D.A.; Alécio, A.C.; Regasini, L.O.; Bergamo, D.C.B.; Telascrea, M.; Cavalheiro, A.J.; Lopes, M.N.; Bolzani, V.; Furlan, V.S.B.M.; et al. Composition and antifungal activity of essential oils from Piper aduncum, Piper arboreum and Piper tuberculatum. Quim. Nova 2006, 29, 467–470. [Google Scholar] [CrossRef] [Green Version]
- Piton, L.P.; Turchen, L.M.; Butnariu, A.R.; Pereira, M.J.B. Natural insecticide based-Leaves extract of Piper aduncum (Piperaceae) in the control of stink bug brown soybean. Ciência Rural. 2014, 44, 1915–1920. [Google Scholar] [CrossRef] [Green Version]
- Yanisia, D.; Oriela, P.; Benedicto, M. Effect of four essential oils on Fusarium spp. Rev. Protección Veg. 2013, 28, 232–235. [Google Scholar]
- Polhit, A.; Pinto, A.; Mause, R. Piper aduncum L.: Pluripotente plant and important phytochemical substance source. Rev. Fitos 2006, 2, 7–18. [Google Scholar]
- Mannai, S.; Horrigue-Raouani, N.; M’Hamdi, N. Effect of six fungicides against Fusarium oxysporum and F. solani associated with peach seedlings decline in tunisian nurseries. Annu. Res. Rev. Biol. 2018, 26, 1–11. [Google Scholar] [CrossRef]
- Kumar, S.; Singh, D.; Yadav, J.K.; Kumar, S.; Verma, S.K. Efficacy of plant extracts, bioagents and fungicides against Fusarium udum causing pigeonpea wilt. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 2652–2660. [Google Scholar] [CrossRef] [Green Version]
- Mamza, W.S.; Zarafi, A.B.; Alabi, O. Fusarium pallidoroseum L. on castor (Ricinus communis) in Samaru, Nigeria. Arch. Phytopathol. Plant. Prot. 2010, 43, 871–874. [Google Scholar] [CrossRef]
- Cordova-Albores, L.; Zapotitla, E.S.; Ríos, M.; Barrera-Necha, L.; Hernández-López, M.; Bautista-Baños, S. Microscopic study of the morphology and metabolic activity of Fusarium oxysporum f. sp. gladioli treated with Jatropha curcas oil and derivatives. J. Microsc. Ultrastruct. 2016, 4, 28–35. [Google Scholar]
- Monzote, L.; Scull, R.; Cos, P.; Setzer, W. Essential oil from Piper aduncum: Chemical analysis, antimicrobial assessment, and literature review. Medicines 2017, 4, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nick, P.; Zdeněk, O. Metabolic Engineering of Wood Formation. In Applied Plant Cell Biology; Springer: Berlin/Heidelberg, 2014; pp. 369–391. [Google Scholar]
- Suárez, K.; Coy, E. Diversity of bioactive naturally-Occurring organic compounds: A singularity expressed by the secondary metabolism plasticity. Rev. Fac. Ciencias Basicas 2016, 12, 252–269. [Google Scholar]
- Mohamed, M.S.M.; Saleh, A.M.; Abdel-Farid, I.B.; El-Naggar, S.A. Growth, hydrolases and ultrastructure of Fusarium oxysporum as affected by phenolic rich extracts from several xerophytic plants. Pestic. Biochem. Physiol. 2017, 141, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhou, X.; Shi, X.; Liu, Y.; Wang, M.; Shang, X.; Gu, D.; Wang, W.; Wu, C. In vitro responses of Fusarium oxysporum f.sp. niveum to phenolic acids in decaying watermelon tissues. Phytochem. Lett. 2014, 8, 171–178. [Google Scholar] [CrossRef]
- Espinel-Ingroff, A. Germinated and nongerminated conidial suspensions for testing of susceptibilities of Aspergillus spp. to amphotericin B, itraconazole, posaconazole, ravuconazole, and voriconazole. Antimicrob. Agents Chemother. 2001, 45, 605–607. [Google Scholar] [CrossRef] [Green Version]
- Guarro, J.; Llop, C.; Aguilar, C.; Pujol, I. Comparison of in vitro antifungal susceptibilities of conidia and hyphae of filamentous fungi. Antimicrob. Agents Chemother. 1997, 41, 2760–2762. [Google Scholar] [CrossRef] [Green Version]
- Hadacek, F.; Greger, H. Testing of antifungal natural products: Methodologies, comparability of results and assay choice. Phytochem. Anal. 2000, 11, 137–147. [Google Scholar] [CrossRef]
- Shah, F.A.; Ansari, M.A.; Watkins, J.; Phelps, Z.; Cross, J.; Butt, T.M. Influence of commercial fungicides on the germination, growth and virulence of four species of entomopathogenic fungi. Biocontrol Sci. Technol. 2009, 19, 743–753. [Google Scholar] [CrossRef]
- Wiklund, S.; Johansson, E.; Sjöström, L.; Mellerowicz, E.J.; Edlund, U.; Shockcor, J.P.; Gottfries, J.; Moritz, T.; Trygg, J. Visualization of GC/TOF-MS-Based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 2008, 80, 115–122. [Google Scholar] [CrossRef]
- Cloarec, O.; Dumas, M.-E.; Craig, A.; Barton, R.H.; Trygg, J.; Hudson, J.; Blancher, C.; Gauguier, D.; Lindon, J.C.; Holmes, E.; et al. Statistical total correlation spectroscopy: An exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Anal. Chem. 2005, 77, 1282–1289. [Google Scholar] [CrossRef]
- Kew, W.; Goodall, I.; Uhrín, D. Analysis of Scotch Whisky by 1H NMR and chemometrics yields insight into its complex chemistry. Food Chem. 2019, 298. [Google Scholar] [CrossRef]
- Cox, D.G.; Oh, J.; Keasling, A.; Colson, K.L.; Hamann, M.T. The utility of metabolomics in natural product and biomarker characterization. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 3460–3474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Y.; Tianero, M.D.B.; Kwan, J.C.; Wyche, T.P.; Michel, C.R.; Ellis, G.A.; Vazquez-Rivera, E.; Braun, D.R.; Rose, W.E.; Schmidt, E.W.; et al. Structure and biosynthesis of the antibiotic bottromycin D. Org. Lett. 2012, 14, 5050–5053. [Google Scholar] [CrossRef]
- Yuliana, N.D.; Khatib, A.; Choi, Y.H.; Verpoorte, R. Metabolomics for bioactivity assessment of natural products. Phyther. Res. 2011, 25, 157–169. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Wang, X. Serum metabolomics as a novel diagnostic approach for disease: A systematic review. Anal. Bioanal. Chem. 2012, 404, 1239–1245. [Google Scholar] [CrossRef]
- Britton, E.R.; Kellogg, J.J.; Kvalheim, O.M.; Cech, N.B. Biochemometrics to identify synergists and additives from botanical medicines: A case study with Hydrastis canadensis (goldenseal). J. Nat. Prod. 2018, 81, 484–493. [Google Scholar] [CrossRef] [Green Version]
- Caesar, L.K.; Kellogg, J.J.; Kvalheim, O.M.; Cech, N.B. Opportunities and limitations for untargeted mass spectrometry metabolomics to identify biologically active constituents in complex natural product mixtures. J. Nat. Prod. 2019, 82, 469–484. [Google Scholar] [CrossRef]
- Orjala, J.; Wright, A.D.; Behrends, H.; Folkers, G.; Sticher, O.; Rüegger, H.; Rali, T. Cytotoxic and antibacterial dihydrochalcones from Piper aduncum. J. Nat. Prod. 1994, 57, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Barrero, A.F.; Herrador, M.M.; Arteaga, P.; Rodríguez-García, I.; García-Moreno, M. Resorcinol derivatives and flavonoids of Ononis natrix subspecies ramosissima. J. Nat. Prod. 1997, 60, 65–68. [Google Scholar] [CrossRef]
- Ichino, K. Two flavonoids from two Lindera umbellata varieties. Phytochemistry 1989, 28, 955–956. [Google Scholar] [CrossRef]
- Orjala, J.; Wright, A.D.; Erdelmeier, C.A.J.; Sticher, O.; Rali, T. New monoterpene-Substituted dihydrochalcones from Piper aduncum. Helv. Chim. Acta 1993, 76, 1481–1488. [Google Scholar] [CrossRef]
- Joshi, A.S.; Li, X.-C.; Nimrod, A.C.; ElSohly, H.N.; Walker, L.A.; Clark, A.M. Dihydrochalcones from Piper longicaudatum. Planta Med. 2001, 67, 186–188. [Google Scholar] [CrossRef]
- Funari, C.S.; Gullo, F.P.; Napolitano, A.; Carneiro, R.L.; Mendes-Giannini, M.J.S.; Fusco-Almeida, A.M.; Piacente, S.; Pizza, C.; Silva, D.H.S. Chemical and antifungal investigations of six Lippia species (Verbenaceae) from Brazil. Food Chem. 2012, 135, 2086–2094. [Google Scholar] [CrossRef] [Green Version]
- Álvarez-Caballero, J.M.; Coy-Barrera, E. Chemical and antifungal variability of several accessions of Azadirachta indica A. Juss. from six locations across the Colombian caribbean coast: Identification of antifungal azadirone limonoids. Plants 2019, 8, 555. [Google Scholar] [CrossRef] [Green Version]
- Marentes-Culma, R.; Orduz-Díaz, L.L.; Coy-Barrera, E. Targeted metabolite profiling-based identification of antifungal 5-n-alkylresorcinols occurring in different cereals against Fusarium oxysporum. Molecules 2019, 24, 770. [Google Scholar] [CrossRef] [Green Version]
- Divband, K.; Shokri, H.; Khosravi, A.R. Down-regulatory effect of Thymus vulgaris L. on growth and Tri4 gene expression in Fusarium oxysporum strains. Microb. Pathog. 2017, 104, 1–5. [Google Scholar] [CrossRef]




| # | Plant | BF a | PP b | C c | MGI d (%) | # | Plant | BF a | PP b | C c | MGI d (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Piper aduncum | Pi | L | PaL | 95.26 ± 0.15 A | 24 | Ulex europeus | F | L | UeL | 50.76 ± 0.73 P |
| 2 | Piper elongatum | Pi | L | PeL | 91.31 ± 0.13 B | 25 | Bursera simaruba | Bu | L | BsL | 49.74 ± 0.11 Q |
| 3 | Piper elongatum | Pi | I | PeI | 81.26 ± 0.21 C | 26 | Croton fragrans | E | L | CfL | 48.43 ± 0.09 R |
| 4 | Piper elongatum | Pi | S | PeS | 76.21 ± 0.11 D | 27 | Croton hibiscifolius | E | L | ChL | 47.77 ± 0.23 RS |
| 5 | Anadenanthera peregrina | F | L | ApL | 70.08 ± 0.06 E | 28 | Bursera simaruba | Bu | B | BsB | 47.63 ± 0.22 S |
| 6 | Nectandra longifolia | L | L | NlL | 68.80 ± 0.07 F | 29 | Galipea longiflora | Ru | L | GlL | 46.91 ± 0.30 T |
| 7 | Raphanus raphanistrum | Br | L | RrL | 64.67 ± 0.13 G | 30 | Ficus obtusifolia | Mo | L | FoL | 42.86 ± 0.13 U |
| 8 | Jacaranda obtusifolia | Bi | L | JoL | 62.71 ± 0.34 H | 31 | Croton hibiscifolius | E | S | ChS | 41.96 ± 0.43 V |
| 9 | Nectandra acutifolia | L | L | NacL | 62.66 ± 0.15 H | 32 | Pyrostegia venusta | Bi | S | PvS | 41.03 ± 0.23 W |
| 10 | Protium heptaphyllum | Bu | L | PhL | 60.66 ± 0.47 I | 33 | Baccharis macrantha | A | L | BmL | 40.67 ± 0.13 W |
| 11 | Mimosa pudica | F | A | MpA | 58.48 ± 0.23 J | 34 | Phyllanthus salviifolius | Ph | S | PsS | 39.03 ± 0.05 X |
| 12 | Genista monspessulana | F | F | GmF | 56.61 ± 0.18 K | 35 | Virola peruviana | My | L | VpL | 38.73 ± 0.09 X |
| 13 | Mimosa nigra | F | A + F | MnAF | 55.69 ± 0.02 L | 36 | Trattinnickia rhoifolia | Bu | L | TrL | 36.86 ± 0.18 Y |
| 14 | Anadenanthera peregrina | F | B | ApB | 54.00 ± 0.07 M | 37 | Mimosa pudica | F | L | MpF | 27.01 ± 0.07 Z |
| 15 | Bowdichia virgilioides | F | L | BvL | 52.35 ± 0.17 N | 38 | Cedrela odorata | Ml | L | CodL | 24.02 ± 0.03 AA |
| 16 | Copaifera officinalis | F | B | CoB | 52.25 ± 0.15 N | 39 | Senna viarum | F | L | SvL | 22.04 ± 0.15 AB |
| 17 | Hymenaea courbaril | F | L | HcL | 52.21 ± 0.19 N | 40 | Tithonia diversifolia | A | F | TdF | 19.43 ± 0.31 AC |
| 18 | Croton colombianus | E | L | CcL | 51.96 ± 0.23 N | 41 | Virola elongata | My | L | VeL | 16.29 ± 0.04 AC |
| 19 | Mimosa colombiana | F | A | McA | 51.74 ± 0.16 N | 42 | Hymenaea courbaril | F | S | HcS | 14.28 ± 0.18 AE |
| 20 | Baccharis macrantha | A | F | BmF | 51.71 ± 0.11 NO | 43 | Cedrela odorata | Ml | B | CodB | 14.23 ± 0.09 AE |
| 21 | Miconia resima | Mt | L | MrL | 51.68 ± 0.11 NO | 44 | Tithonia diversifolia | A | S | TdS | 12.93 ± 0.18 AF |
| 22 | Nectandra amazonum | L | L | NamL | 51.64 ± 0.05 NO | - | Dithane | - | - | 99.1 ± 0.06 | |
| 23 | Cotoneaster pannosus | Ro | Fr | CpFr | 51.02 ± 0.08 OP | - | Prochloraz | - | - | 94.4 ± 0.36 |
| Plant | Plant Part a | Code b | CGI c (%) |
|---|---|---|---|
| Piper elongatum | L | PeL | 83.52 ± 0.78 A |
| Tithonia diversifolia | S | TdS | 73.95 ± 0.18 B |
| Croton hibiscifolius | S | ChS | 47.60 ± 0.11 C |
| Piper elongatum | S | PeS | 24.01 ± 0.15 D |
| Piper aduncum | L | PaL | 21.13 ± 0.02 E |
| Tithonia diversifolia | F | TdF | 15.36 ± 0.06 F |
| Dithane | - | - | 97.36 ± 0.02 |
| Prochloraz | - | - | 53.62 ± 0.26 |
| Piperaduncin C (1) | Asebogenin (2) | (−)-Methyllinderatin (3) | |
|---|---|---|---|
| MGI IC50 (µM) a | 38.2 (31.5–44.8) | 25.6 (23.5–29.3) | 689.2 (623.8–744.9) |
| CGI IC50 (µM) b | >1000 | >1000 | 22.3 (17.8–29.7) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cárdenas-Laverde, D.; Barbosa-Cornelio, R.; Coy-Barrera, E. Antifungal Activity against Fusarium oxysporum of Botanical End-Products: An Integration of Chemical Composition and Antifungal Activity Datasets to Identify Antifungal Bioactives. Plants 2021, 10, 2563. https://doi.org/10.3390/plants10122563
Cárdenas-Laverde D, Barbosa-Cornelio R, Coy-Barrera E. Antifungal Activity against Fusarium oxysporum of Botanical End-Products: An Integration of Chemical Composition and Antifungal Activity Datasets to Identify Antifungal Bioactives. Plants. 2021; 10(12):2563. https://doi.org/10.3390/plants10122563
Chicago/Turabian StyleCárdenas-Laverde, Diego, Ricardo Barbosa-Cornelio, and Ericsson Coy-Barrera. 2021. "Antifungal Activity against Fusarium oxysporum of Botanical End-Products: An Integration of Chemical Composition and Antifungal Activity Datasets to Identify Antifungal Bioactives" Plants 10, no. 12: 2563. https://doi.org/10.3390/plants10122563