Abstract
Phylogenetic analyses using mtSSU and nuITS sequences of Buellia violaceofusca (previously placed in Lecanoromycetes), a sterile, sorediate lichen having a trebouxioid photobiont, surprisingly prove that the species is conspecific with Lecanographa amylacea (Arthoniomycetes), a fertile, esorediate species with a trentepohlioid photobiont. These results suggest that L. amylacea and B. violaceofusca are photomorphs of the same mycobiont species, which, depending on the photobiont type, changes the morphology and the reproduction strategy. This is the first example of a lichenized fungus that can select between Trebouxia (Trebouxiophyceae) and trentepohlioid (Ulvophyceae) photobionts. Trebouxia photobionts from the sorediate morphotype belong to at least three different phylogenetic clades, and the results suggest that Lecanographa amylacea can capture the photobiont of other lichens such as Chrysothrix candelaris to form the sorediate morphotype. Phylogenetic analyses based on rbcL DNA data suggest that the trentepohlioid photobiont of L. amylacea is closely related to Trentepohlia isolated from fruticose lichens. The flexibility in the photobiont choice enables L. amylacea to use a larger range of tree hosts. This strategy helps the lichen to withstand changes of environmental conditions, to widen its distribution range and to increase its population size, which is particularly important for the survival of this rare species.
Similar content being viewed by others
Introduction
In lichen symbiosis, the mycobiont associates with a chlorophyll-containing partner, either an eukaryotic alga or a cyanobacterium, or both1. Recent studies revealed even more complex associations with the coexistence of bacteria, endolichenic fungi and basidiomycete yeasts with these primary partners2,3,4. Photobiont flexibility was recognized long ago by the existence of alternative cyanobacterial and green algal morphs (photomorphs or photosymbiodems) of the same mycobiont species5,6. In addition to differences observed in the colour of the thallus caused by the photobiont type, these photosymbiodems also differ in their thallus morphology, sometimes so significantly that, in the past, the chloromorph (with eukaryotic algae) and the cyanomorph (with cyanobacteria) were recognized as separate species, which were even sometimes classified in different genera1,7. Very rarely the photosymbiodems also differ in the additional development of asexual reproductive structures1. When a single fungal partner associates with a green alga and a cyanobacterium simultaneously, the latter photobiont type is located in distinct, usually easily morphologically recognizable areas of the thallus (i.e. cephalodia8), which in some species may enlarge and eventually form independent thalli9.
Switching between closely related eukaryotic green-algal strains is also frequently observed within a single lichen species10,11,12,13,14,15,16,17,18, but is not known to involve changes in the thallus anatomy and morphology. Recent molecular studies also highlight the coexistence of multiple green-algal taxa in a single lichen thallus13,15,19,20. A higher flexibility in photobiont choice is considered as a strategy to survive under new selective pressures and to widen the ecological niche amplitude enabling the occurrence even in nutrient-poor environments1,12,21,22,23,24, and may also drive the evolution of lichens16,25.
Some groups of crustose lichens reproduce mainly vegetatively by the formation of asexual propagules (usually soredia or isidia), whereas the development of ascomata (sexual reproduction) might be more or less limited to occasional events or are completely absent. In particular cases, the ability of the ascomata formation was completely lost in entire genera26,27,28. Determining the systematic position of sterile ascomycetes is difficult or impossible using only morphological or chemical characters of the thallus. In these cases the application of molecular markers and phylogenetic methods allows classification of those sterile samples within higher level taxonomic groups26,27,28.
Sterile lichens are often parts of so-called species pairs. This term, first introduced by du Rietz almost hundred years ago29, states that species pairs are two taxa that are morphologically and chemically very similar but differ in their predominant reproductive modes, i.e. “primary species” is fertile and reproduces sexually (by ascospores formed in apothecia), while “secondary species” is sterile and forms asexual diaspores (by isidia or soredia) (for review see30). Several hypotheses have been proposed to explain the presence of these species pairs and the most widely assumed one suggests that the majority of vegetatively reproducing taxa are members of the species pairs and evolved from the primary sexual taxon29. However, more recently Buschbom & Mueller31 proposed that the founding taxon of the species pair could be a species with either vegetative or sexual reproduction and switches between reproductive modes are triggered through symbiont interactions. Most phylogenetic studies of species pairs reported that neither sexual nor vegetative taxa are monophyletic, but rather intermingled and independent lineages did not correspond to reproductive mode31,32,33,34. However, none of these studies considered the impact of the photobiont type on the lichen reproduction strategy.
The lichen species Buellia violaceofusca G. Thor & Muhr is characterized by a sterile, thin, pale grey to almost white thallus producing dark brownish soralia with a violet tinge (but greenish when abraded), and the lack of detectable substances by thin-layer chromatography. Soralia are scattered, slightly elevated and sometimes confluent. This characteristic corticolous species contains a trebouxioid (green unicellular spherical cells) photobiont and was tentatively assigned to the genus Buellia De Not. due to its similarity with some species of that genus, especially the sorediate B. griseovirens (Turner & Borrer ex Sm.) Almb., and some other genera of Lecanoromycetes35. But because of the absence of apothecia, pycnidia and chemical compounds, the generic position was unclear. In order to verify the generic position of B. violaceofusca, fresh material was collected and sequenced. Here we show (1) that B. violaceofusca is conspecific with Lecanographa amylacea (Ehrh. ex Pers.) Egea & Torrente, (2) L. amylacea represents a first example of a lichen species that can switch between Trebouxia Puymaly and trentepohlioid photobionts, (3) a first example of dimorphism (thallus morphology and type of reproduction) resulting of the interchange between two different eukaryotic green algae taxa for a single mycobiont species, (4) Trebouxia photobionts from the sorediate morphotype known as B. violaceofusca belong to at least three different phylogenetic clades, (5) Lecanographa amylacea can capture the photobiont of other lichens such as Chrysothrix candelaris (L.) J.R.Laundon to form the sorediate morphotype, (6) the trentepohlioid alga of L. amylacea is most closely related to strains isolated from Roccella species, (7) photobiont switching in L. amylacea enables the lichen to widen its distribution range and to increase its population size.
Results
Fungal Phylogeny
The mtSSU dataset consisted of 32 specimens and 590 unambiguously aligned sites. In our mtSSU phylogenetic tree (Fig. 1), the Lecanographaceae are composed of four mainly supported lineages corresponding to the Plectocarpon-Alyxoria and Phacographa-Opegrapha brevis clades and the genera Lecanographa Egea & Torrente and Zwackhia Körb. The five specimens of Buellia violaceofusca are nested in the genus Lecanographa and intermixed with the specimens of Lecanographa amylacea. The nuITS dataset consisted of 10 specimens and 637 unambiguously aligned sites. In our nuITS phylogenetic tree (Fig. 2), the four specimens of Buellia violaceofusca cluster in a polytomy with the three specimens of L. amylacea because the nuITS sequences of these seven specimens are identical. These results prove that B. violaceofusca is conspecific with L. amylacea. Therefore, B. violaceofusca is synonymized with L. amylacea. L. amylacea is morphologically very different from B. violaceofusca notably by a thick, white-powdery thallus often dotted with brown flecks, containing a trentepohlioid photobiont (i.e. filamentous algae containing large amounts of carotenoid pigments, causing the algae to appear yellow orange in color), the absence of soralia and the occasional presence of apothecia. Our results prove thus that B. violaceofusca and L. amylacea represent two photomorphs of the same ascomycete species. These morphotypes are so distinct that they were described as two different species belonging to two different classes (Lecanoromycetes versus Arthoniomycetes).
Phylogenetic relationships among samples within Lecanographaceae based on a data set of mtSSU sequences that resulted from a RAxML analysis. Posterior probabilities obtained from a Bayesian analysis and Maximum Likelihood bootstrap values are shown near the internal branches. Internal branches, considered strongly supported by both analyses, are represented by thicker lines. Collecting numbers of the authors following the species names of the newly generated sequences act as specimen and sequence identifiers, while those retrieved from GenBank are followed by their GenBank number. The samples of Buellia violaceofusca are highlighted.
Phylogenetic relationships among samples within Lecanographa based on a data set of nuITS sequences that resulted from a Bayesian analysis. Posterior probabilities and Maximum Likelihood bootstrap values obtained from a RAxML analysis are shown near the internal branches. Internal branches, considered strongly supported by both analyses, are represented by thicker lines. All sequences were newly generated. Collecting numbers of the authors following the species names act as specimen and sequence identifiers.
Algal phylogenies
The Trebouxia algal dataset consisted of 99 nuITS sequences and 651 unambiguously aligned sites. The Bayesian tree (Harmonic mean was −4936.69) is shown in Fig. 3 with added bootstrap supports from RaxML analysis (ML Optimization Likelihood was −4490.471038). The newly sequenced photobionts revealed in three different clades within Trebouxia spp. The most common photobiont in B. violaceofusca thalli is Trebouxia sp. that groups together with Trebouxia sp. strain TR9 previously reported from Ramalina farinacea (L.) Ach.19. It was found in all Polish specimens (Kukwa 12916, 12998 and 15628) and four specimens from Sweden (i.e. Thor 31255, 31380, 31385 and 31389). A second lineage is related to the T. solaris clade and consists of Trebouxia sp. found in two Swedish specimens (i.e. Thor 31382 and 31385) as well as in four Chrysothrix candelaris specimens (i.e. Thor 31379, 31380, 31382, 31385) growing together with ‘B. violaceofusca’-morphotype. These molecular data strongly support our hypothesis that Lecanographa amylacea can capture the photobiont of Chrysothrix candelaris to form ‘Buellia violaceofusca’ thalli. This possibility was also concluded from the morphological observations of our specimens, because of the presence of Buellia violaceofusca at the margin of Lecanographa amylacea (with trentepohlioid photobiont) thalli in contact with Chrysothrix candelaris. A third lineage consists of two sequences from a specimen of Buellia violaceofusca (Thor 31376) that are nested within the Trebouxia jamesii clade. The latter is a common photobiont reported from e.g. Candelariella vitellina (Hoffm.) Müll. Arg., Lecanora spp., Protoparmelia sp., Ramalina spp., Rhizocarpon geographicum (L.) DC., Rhizoplaca sp.36,37. Interestingly, among the studied samples, we found that two different Trebouxia photobionts may be present in Buellia violaceofusca thalli. In specimen Thor 31385, we detected Trebouxia sp. TR9 and Trebouxia sp. related to T. solaris clade (Fig. 3).
Phylogenetic relationships of Trebouxia photobionts from Lecanographa amylacea and Chrysothrix candelaris based on Bayesian analysis of nuITS dataset. Posterior probabilities and Maximum Likelihood bootstrap values obtained from a RAxML analysis are shown near the internal branches. Newly generated sequences are in bold, with collecting numbers of the authors following the species names. In case of specimens for which more than one sequence was generated the collecting numbers are followed with the number of sequence (i.e. seq. 1 or seq. 2). Specimen of ‘Buellia violaceofusca’ morphotype Thor 31385 is marked with asterisk because two completely different Trebouxia strains were identified in its thallus. GenBank accession numbers of sequences retrieved from GenBank precede the species names. Clades with photobionts from Lecanographa amylacea and Chrysothrix candelaris are highlighted.
The rbcL dataset of trentepohlioid photobionts consisted of 60 sequences and 668 unambiguously aligned sites. The Bayesian tree (Harmonic mean was −9624.90) is shown in Fig. 4 with added bootstrap supports from RaxML analysis (best tree ML = −8571.5689). The four sequenced specimens of Lecanographa amylacea (i.e. Thor 31378, 31379, 31382, 31391) form a lineage closely related to Trentepohlia photobionts isolated from Roccella spp. (JQ617924 from R. linearis; JQ617925 from R. maderensis; JQ617926 from R. tinctoria) collected in Spain and Portugal and reported by Hametner et al.38 as belonging to clade R. However, their proper identification to the species level is not possible due to insufficient data resolution in GenBank for this group of photobionts. The most closely related algae identified to the species level is a free-living Trentepohlia flava (Kützing) De Toni that was isolated from bark39.
Phylogenetic placement of trentepohlioid photobionts from Lecanographa amylacea based on Bayesian analysis of chloroplast rbcL dataset. Posterior probabilities and Maximum Likelihood bootstrap values obtained from a RAxML analysis are shown near the internal branches. Newly generated sequences are in bold, with collecting numbers of the authors following the species names. GenBank accession numbers of sequences retrieved from GenBank precede the species names. Clade with photobionts from Lecanographa amylacea is highlighted.
Taxonomy
Lecanographa amylacea (Ehrh. ex Pers.) Egea & Torrente (Figs 5, 6) Bibliotheca Lichenologica 54: 122 (1994). Bas. Lichen amylaceus Ehrh. ex Pers., Plant. Cryptog. Exsicc. n°303 (1793)/Ann. Bot. (Usteri) 5: 18 (1794). Type: (Germany), Hannoverae, Ehrh. sub Lichen amylaceus 303 (W, lectotype designated by Egea & Torrente40, not seen).
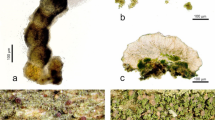
Lecanographa amylacea. (a,b) Thallus of the non-sorediate morphotype of Lecanographa amylacea (a Thor 31378; b, Thor 31379); (c) two apothecia (Thor 31378); (d) trentepohlioid algae (Thor 31382); (e) section of apothecium (Thor 31382); (f) ascospores in water (Thor 31378); (g,h), thallus of the non-sorediate morphotype of Lecanographa amylacea on the right (=white thallus), with the sorediate morphotype (=dark soralia with a greenish to violet tinge) present at the contact with Chrysothrix candelaris (=yellowish sorediate thallus) on the left (Thor 31391).
Syn. nov. Buellia violaceofusca G. Thor & Muhr, Lichenologist 23: 11 (1991). Type: Sweden, Värmland, Visnums Kil par., Nötön Nature reserve, Arskagen, near Lake Vänern, 59°04′N, 14°01′E, c. 50 m, on the northern side of old Quercus robur in deciduous wood, 27 May 1985, L.-E. Muhr 7911 (UPS-holotype!, S-isotype!).
Sequenced specimens of esorediate morphotype of Lecanographa amylacea. Sweden: Västmanland, Kungsör parish, 3 km NNW Kungsör town, Jägaråsen, E of dirt road, 20 m, 59°27.036′N, 16°04.613′E, on dying, large Quercus robur in open, grazed deciduous forest, with Buellia violaceofusca, 25 July 2015, Thor 31379 (UPS); ibidem, 59°26.808′N, 16°04.754′E, on large Quercus robur in open, grazed deciduous forest, with Buellia violaceofusca, Thor 31382 (UPS); ibidem, Rytterne parish, 1.2 km SW Tidö Castle, 2 m S the dirt road, 59°30.073′N, 16°27.558′E, on large, exposed Quercus robur, Thor 31391 (UPS).
Sequenced specimens of sorediate morphotype of Lecanographa amylacea (originally identified as Buellia violaceofusca). Belgium: Ardennes, Wellin, à 4 km au sud de Chanly, ruisseau le Glan, 260 m, sur Fraxinus en forêt de feuillus, 29 September 2007, Ertz 10169 (BR); ibidem, St-Hubert, vallée de la Masblette, en amont du Pont Mauricy, 320 m, forêt de feuillus mixte de fond de vallée, sur un gros tronc de Fraxinus, 13 February 2011, Ertz 16062 (BR). Poland: Równina Bielska, Białowieża Primeval forest, Białowieża National Park, forest section no256, plot F07, Querco-Piceetum, on Quercus robur, May 2015, Kukwa 15628 & Łubek (BR, UGDA); plot A11, Circaeo-Alnetum, on bark of fallen Fraxinus excelsior, August 2014, Kukwa 12916 & Łubek (UGDA); plot M10, Circaeo-Alnetum, on bark of snag of dead Fraxinus excelsior, October 2014, Kukwa 12998 & Łubek (UGDA). Sweden: Uppland, S of Uppsala, Alsike parish, 300 m NW of Krusenberg mansion, 15 m, 59°44′13.62′′N, 17°38′37.51′′E, dense, grazed deciduous forest, on old large Quercus robur, 27 June 2015, Thor 31376 (BR, UPS); Västmanland, Kungsör parish, 3 km NNW Kungsör town, Jägaråsen, E of dirt road, 20 m, 59°26.879′N, 16°04.711′E, on large Quercus robur in open, grazed deciduous forest, 25 July 2015, Thor 31380 (UPS).
Additional sequenced specimens for the photobionts of Lecanographa amylacea (both morphotypes) and/or associated Chrysothrix candelaris. Sweden: Södermanland, Hölö parish, 1 km NW Tullgarn Castle, E of dirt road, 20 m, 58°57.467′N, 17°34.463′E, on one of two nearby (5 m) very large Quercus robur in open, grazed, deciduous forest, 25 July 2015, Thor 31378 (UPS). Västmanland, Kungsör parish, 3 km NNW Kungsör town, Jägaråsen, E of dirt road, 20 m, 59°26.808′N, 16°04.754′E, on large Quercus robur in open, grazed deciduous forest, 25 July 2015, Thor 31382 (UPS); ibidem, 59°26.810′N, 16°04.779′E, Thor 31385 (UPS); Rytterne parish, 620 m SW Tidö Castle, 3 m N the dirt road, 25 m, 59°30.119′N, 16°28.176′E, on large, dead Quercus robur with almost no bark, 25 July 2015, Thor 31389 (UPS); Gotland, Fide parish, 1.2 km NNW Fide church, W of the road, the wooded meadow Fide annex, 10 m, 57°04.993′N, 18°18.588′E, on Quercus robur 287 cm in circumference, 21 May 2015, Thor 31255 & Knutsson (UPS).
Discussion
Here we report Lecanographa amylacea as a first example of a lichen that uses either Trebouxia (Trebouxiophyceae) or trentepohlioid algae (Ulvophyceae) as primary photobiont. This is also the first example of a lichen with two very distinct photomorphs, both containing distantly related green algae. Indeed, none of the studies that previously recorded green algae switch within single lichen species suggested an impact on the thallus morphology (see introduction). The high degree of dimorphism involving alterations in the thallus anatomy and morphology was so far only known from lichens in which the same mycobiont associates with either a green alga or a cyanobacterium (e.g. Dendriscocaulon–Lobaria5,7,8). Moreover, photobiont switching between different species of green algae so far mainly concerned lichens belonging to the class Lecanoromycetes. One exception is Coniocarpon cinnabarinum DC., which belongs to the Arthoniomycetes and associates with the trentepohlialean algae Printzina lagenifera (Hildebrand) R.H. Thompson & Wujek and one of its close relative, probably belonging to the same genus18. However, no impact on the thallus appearance has ever been recorded before when the closely related green algae exchange in the same lichen species.
The two types of photobionts (trentepohlioid versus Trebouxia) with which Lecanographa amylacea associates are obviously responsible for the strong dimorphism observed. Once L. amylacea has gained one of the Trebouxia photobionts, the lichen species can disseminate by means of soredia that propagate jointly both symbionts and may form independent sorediate thalli previously recognized as Buellia violaceofusca. The Trebouxia photobiont species might be maintained in this sterile sorediate photomorph as this reproduction strategy should allow preservation of the relationship among symbionts. However, maintenance of the symbiotic associations seems to be rather an option than a strict consequence of joint symbiont dispersal in lichens41 and horizontal transmissions with other lichen species are possible as proved e.g. for the genus Lepraria Ach., which disperses solely by soredia-like granules14.
In addition to the alteration of morphology, the type of photobiont might influence the sexuality of the mycobiont. The Trebouxia-morphotype of L. amylacea is not known to form ascomata, contrary to its trentepohlioid morphotype, which is on the other hand never sorediate. When L. amylacea is fertile, ascospores, which disseminate only the fungal partner, could switch photobionts by the capture of Trebouxia algae from other lichens or from free-living, non-lichenized Trebouxia (despite these latter appear to be less frequently encountered), leading to the subsequent development into the sorediate morphotype. Buschbom & Mueller31 hypothesized that switches between reproductive modes are triggered through symbiont interactions. They suggested that the asexual reproductive mode is advantageous when the relationship between the mycobiont and the photobiont is optimal in a given environment allowing the rapid dispersion of both partners: in this situation, it is advantageous for the mycobiont not to switch algae. In disadvantageous environmental situations, sexual reproduction of the mycobiont is preferred because this would increase the chance to “escape” from its current partner. The sexual reproduction allows the mycobiont to acquire new partners as well as generates variability through recombination, with the development of new genotypes that may be better adapted to local and changing conditions. Similarly, Ellis & Coppins42 suggested that the asexually reproducing crustose lichens are better adapted to stable habitats. However, it is unclear if this might also be the case for the asexual/sexual morphotypes of L. amylacea. Studies at local and global scales should be performed to better understand which might be the combination of environmental factors that drive the asexual/sexual reproduction in L. amylacea and those responsible of the photobiont switches.
A higher flexibility in the photobiont choice is considered as a strategy to increase ecological tolerance, enabling the lichen to withstand changes of environmental conditions (e.g. shifts in light or humidity regimes), to colonize diverse microhabitats, and to widen its distribution range8,12,16,21,22. As a consequence, it increases the population size of the fungal partner15, which is particularly important for the survival of a rare species such as L. amylacea. Since its description from Sweden in 1991, ‘Buellia violaceofusca’ (=the Trebouxia-morphotype of L. amylacea) has been recorded from several European countries43,44. This morphotype is even present in regions where the trentepohlioid morphotype has never been recorded (e.g. Belgium or Białowieża Forest in Poland) suggesting that the Trebouxia photobionts may offer higher fitness in some habitats or in climatically different regions. The Trebouxia morphotype might also be more easily dispersed with successful new colonization as soredia are produced in large quantities and disseminate the fungal and algal symbionts together. The trentepohlioid morphotype of L. amylacea appears to be exclusively confined to old Quercus trees40,45,46,47,48, while the Trebouxia-morphotype can sometimes colonize other phorophytes, mainly Fraxinus43,47,48,49 and Acer44,50 in addition to Quercus. The use of a larger range of tree hosts is an obvious advantage that might explain the success of the Trebouxia-morphotype in some regions. In addition, the Trebouxia-morphotype has the capacity to colonize much younger Quercus trees than the trentepohlioid morphotype, the latter only occurring on Quercus over 200 years old with > 60 mm bark crevices as shown in Sweden45,51,52. As a consequence, the Trebouxia-morphotype has a wider ecological niche on Quercus trees compared to the trentepohlioid morphotype, with a much higher number of Quercus trees being thus suitable for colonization by the Trebouxia-morphotype51,52. Despite the ability to switch photobionts, L. amylacea (including ‘B. violaceofusca’) is rare throughout its distributional range, being often known from small populations (on a single or a few trees). With other epiphytic species occurring on old Quercus trees in semi-open landscapes, it disappears notably because of shading by developing secondary woodland52, but the Trebouxia-morphotype of L. amylacea appears to have the capacity to survive in more shaded conditions, and might thus replace the trentepohlioid morphotype in more dense forests. Studies on the forest types and of the environmental parameters, particularly light and humidity, which favour each morphotypes should be performed to better understand their distribution.
The identification of trentepohlioid algae in lichens are difficult on the basis of morphological features even at the generic level, especially as sexual stages of algae are usually suppressed in lichens18. As a consequence, we used molecular methods to investigate the identity of the trentepohlioid alga of L. amylacea (Fig. 4). Unfortunately, the taxonomic coverage in GenBank for this group of algae is still very limited and uneven, and many deposited sequences are from samples identified to the genus level or above, with many genera being polyphyletic. Brooks et al.53 summarized the most recent molecular studies and showed five well-supported lineages within Trentepohliales, of which none correspond to the morphologically defined genera. Lichenized trentepohlioid photobionts were found within most of these clades. It is estimated that approximately 23% of lichen-forming fungi associate with trentepohlioid algae54. In our study, the photobiont of Lecanographa amylacea was found to be closely related to Trentepohlia spp. isolated from different Roccella species (Fig. 4), which were shown to belong to clade R closely related to strains representing e.g. Printzina bosseae (De Wildeman) R.H.Thompson & Wujek, Trentepohlia annulata F. Brand, T. iolithus (L.) Wallroth18, and T. flava (Kützing) De Toni (Fig. 4). Because molecular studies only started to elucidate the identity and diversity of trentepohlialean photobionts involved in lichen symbiosis18,38,54, more work is needed to investigate the selectivity and specificity (as defined by Yahr et al.12, selectivity denotes the frequency of association among partners, whereas specificity denotes the phylogenetic range of associated partners) with this type of photosynthetic partner. Although previous studies suggested that at least some bark-inhabiting lichens may switch their trentepohlialean photobionts18,54, none of these studies showed photobiont switch from coccoid green algae to trentepohlioid photobiont.
The most common trebouxioid photobiont detected in sorediate morphotype of L. amylacea thalli is Trebouxia sp. TR9 previously reported only from Ramalina farinacea19. In one of these thalli, the same photobiont as in Chrysothrix candelaris was detected together with Trebouxia sp. TR9 (Thor 31385; Fig. 3). Such multiple algal genotypes have been previously found in a single thallus of different species13,15,20, which may be beneficial in terms of lichen’s ability to respond to environmental changes or to occupy diverse microenvironments22. Moreover, we also found the Trebouxia-morphotype of L. amylacea to contain the same photobiont as found in the neighboring thallus of C. candelaris (Thor 31382; Fig. 3). This suggests that Lecanographa amylacea may obtain photobionts from different lichens, one of those being probably Chrysothrix candelaris. In the specimen Thor 31382, ‘B. violaceofusca’-like soralia were present along the margins of the trentepohlioid morphotype of L. amylacea thalli contiguous to those of Chrysothrix candelaris, without borders between them (see also Fig. 6g,h for specimen Thor 31391). It suggests that the growing thallus of L. amylacea is able to capture the photobiont from C. candelaris by entering its neighboring thalli (so called ‘horizontal transmission’ allowing acquisition of new symbionts). Moreover, thalli of C. candelaris often become paler and appear to be killed in close vicinity of the ‘B. violaceofusca’-morphotype thalli. Phenomenon in which the mycobiont captures the photobiont of other lichen species has been already described long ago (10for Diploschistes muscorum (Scop.) R. Sant. in the Ostropales in Lecanoromycetes). However, it is known that photobionts with identical nuITS sequences are present in lichens growing within the same lichen community, without necessarily implying that the photobiont was taken over from other lichens, as free-living Trebouxia can also be captured55. A study of the photobionts of entire lichen communities as well as their free-living algae will be needed to identify the different photobiont sources that might explain the existence of the multiple trebouxioid-algal taxa detected in the ‘B. violaceofusca’-morphotype.
Composite or joint thalli of the two morphotypes of L. amylacea have not been reported so far thus explaining why connection between the two previously recognized taxa, B. violaceofusca and L. amylacea, has never been suggested before, nor a possible placement of the former one in the Arthoniomycetes. Both morphotypes have even been used as different indicator species of old trees in several ecological studies51,52,56.
Lecanographa amylacea belongs to the Lecanographaceae, a family of species that are either lichenized with a trentepohlioid photobiont or that are lichenicolous57,58. The presence of a morph with Trebouxia algae in this family is therefore surprising. Interestingly, Lecanographa insolita Lendemer & K. Knudsen was also recently described as having a trebouxioid photobiont59. However, this lichen has never been found fertile and has not been sequenced, so the species was only tentatively assigned to the Lecanographaceae on basis of secondary chemistry and of the anatomy of immature ascomata observed in only one specimen. Its taxonomic affiliation should be verified by sequencing to determine whether it is a second example of trebouxioid photomorph in the family. Species of Arthoniales having unicellular green algae are otherwise only known in the Arthoniaceae (e.g. Arthonia phlyctiformis Nyl.) and the Chrysotricaceae (e.g. Chrysothrix candelaris)58. Molecular studies are needed to determine the identity of the green unicellular algae living in symbiosis with Arthoniales, our study being the first step in this field. L. amylacea represents an interesting model to explore the evolutionary mechanisms that generate fungal specificity and selectivity in the Arthoniomycetes and lichens in general.
Methods
Amplification and sequencing
Well-preserved and freshly collected specimens lacking any visible symptoms of fungal infection were used for DNA isolation. Genomic DNA was isolated from fresh specimens using the CTAB extraction protocol (for the specimens of Lecanographa atropunctata Sparrius, Saipunk. & Wolseley, L. cf. farinosa (Hepp) Egea & Torrente, L. hypothallina (Zahlbr.) Egea & Torrente and L. uniseptata Ertz, van den Boom, Tehler & Degreef) or modified CTAB protocol60 (for the specimens of Buellia violaceofusca Kukwa 12916, 12998). Hand-made sections of the hymenium (e.g. Lecanographa amylacea, Thor 31382), or the thallus (e.g. L. amylacea, Thor 31379, 31391), as well as soredia (specimens of Buellia violaceofusca and Chrysothrix candelaris) were also used for direct PCR as described in Ertz et al.61. For amplification of Trebouxia photobionts one or two reactions from the same thallus were performed.
Amplification reactions were prepared for a 50 µl final volume containing 5 µl 10 × DreamTaq Buffer (Thermo Scientific), 1.25 µl of each of the 20 µM primers, 5 µl of 2.5 mg mL−1 bovin serum albumin (Thermo Scientific), 4 µl of 2.5 mM each dNTPs (Thermo Scientific), 1.25 U DreamTaq DNA polymerase (Thermo Scientific), and 1 µl of template genomic DNA or, in case of direct PCRs, lichen fragments. For amplification of algal DNA (Trebouxia and trentepohlioid photobionts) StartWarm HS-PCR Mix (A&A Biotechnology) was used. A targeted fragment of about 0.8 kb of the mtSSU rDNA was amplified using primers mrSSU1 and mrSSU3R62 and a fragment of about 0.6 kb of the nuITS rDNA (ITS1 + 5.8 S + ITS4) using primers ITS1F and ITS463. We also amplified a fragment of algal nuITS rDNA (ITS1 + 5.8 S + ITS4) using Trebouxia-specific primers AL1500bf 64 and ITS4M13 and a fragment of trentepohlioid chloroplast rbcL gene using the following primers a-ch-rbcL-203–5′MPN and a-ch-rbcL-991–3′-MPN54. The yield of the PCRs was verified by running the products on a 1% agarose gel using ethidium bromide. PCR products were purified using Clean-Up Concentrator (A&A Biotechnology). Both strands were sequenced by Macrogen® using amplification primers. Sequence fragments were assembled with Sequencher version 5.3 (Gene Codes Corporation, Ann Arbor, Michigan). Sequences were subjected to ‘megablast’ searches to verify their closest relatives and to detect potential contaminations (no contaminations were found).
Phylogenetic analyses of fungi
Because the first mtSSU sequences obtained from Buellia violaceofusca from Belgium and Poland were identical based on BLASTN search to the single mtSSU sequence of Lecanographa amylacea present in GenBank (viz. KF707650), more samples of the two lichens were collected in Sweden and sequenced. In addition, mtSSU and nuITS sequences were also generated for four more species of Lecanographa. A total of thirteen new mtSSU sequences for this study were obtained for placing the newly sequenced samples in a phylogeny of the family (Table 1). Members representing all genera currently accepted in the Lecanographaceae and for which a mtSSU sequence was available, were selected from Ertz & Tehler57, Frisch et al.58 and Van den Broeck et al.65. In order to test if B. violaceofusca is conspecific with L. amylacea, we also sequenced a more variable locus i.e. 10 nuITS (ITS1 + 5.8 S + ITS4; Table 1). Opegrapha vulgata (Ach.) Ach. was chosen as the outgroup species for the mtSSU tree and Lecanographa atropunctata for the nuITS tree. The sequences were aligned using MAFFT v6.814b66 within Geneious and improved manually using Mesquite 3.04. No regions were excluded from the nuITS dataset. Ambiguous regions corresponding to a total of 371 aligned sites were delimited manually following Lutzoni et al.67 and excluded from the mtSSU dataset. These regions were mainly due to a long insertion in the sequence of O. celtidicola (positions 197 to 475 of the sequence EU704066), and to the regions corresponding to positions 410 to 441 and 677 to 687 in the sequence KJ851024 of Zwackhia sorediifera and positions 597 to 644 in the sequence EU704080 of O. vulgata.
Bayesian analyses were carried out on the fungal mtSSU and the nuITS datasets separately using the Metropolis-coupled Markov chain Monte Carlo (MCMCMC) method in MrBayes v. 3.2.6 on the CIPRES Web Portal68. Best-fit evolutionary models were estimated using Akaike Information Criterion (AIC69) as implemented in jModelTest2 2.1.670. The TVM + I + G and the HKY + G models were selected respectively for the mtSSU and the nuITS datasets. For each of them, two parallel MCMCMC runs were performed, each using four independent chains and 20 million generations, sampling trees every 1000th generation. Tracer v. 1.671 was used to ensure that stationarity was reached by plotting the log-likelihood values of the sample points against generation time. Convergence between runs was also verified using the PSRF (Potential Scale Reduction Factor), where all values were equal or close to 1.000. Posterior probabilities (PP) were determined by calculating a majority-rule consensus tree generated from the 30002 post-burnin trees of the 40002 trees sampled by the two MCMCMC runs using the sumt option of MrBayes. In addition, a Maximum Likelihood (ML) analysis was performed using RAxML-HPC2 v.8.2.1072 with 1000 ML bootstrap iterations (ML-BS) and the GTRGAMMA model.
The RAxML tree did not contradict the Bayesian tree topology for the strongly supported branches. Therefore, only the RAxML tree is shown for the mtSSU dataset with the bootstrap support values and the posterior probabilities of the Bayesian analysis added near the internal branches (Fig. 1), whereas only the Bayesian tree is shown for the nuITS dataset with the posterior probabilities and the bootstrap support values of the RAxML analysis added near the internal branches (Fig. 2). ML-BS ≥ 70 and PP ≥ 0.95 were considered to be significant. Phylogenetic trees were visualized using FigTree v. 1.4.273.
Phylogenetic analyses of trebouxioid algae
In case of algal nuITS rDNA region, the newly obtained sequences were aligned together with Trebouxia sequences generated in the study of Voytsekhovich & Beck36 which are deposited in GenBank as PopSet 940816236 and additionally with best-hits from BLAST searches of newly sequenced photobionts, i.e. Trebouxia sp. TR9 (KU716051) and Trebouxia sp. P-400-IaSc (AM159210), using MAFFT on XSEDE (7.305) on the CIPRES Web Portal68. It was followed with a selection of ambiguous positions using Gblocks 0.91b74,75 using less stringent settings (i.e. allow smaller final blocks, gap positions within the final blocks and less strict flanking positions). The excluded regions were the terminal ends corresponding to positions 1–2632 and 3300–7640 of the sequence KU716051 (Trebouxia sp. TR9) and positions 1–689 of the sequence AM159210 (Trebouxia sp. P-400-IaSc). Moreover, the excluded regions were mainly due to insertions, i.e. in Trebouxia gelatinosa (e.g. positions 57–253 of the sequence KT819972), T. arboricola and T. crenulata (e.g. positions 158–189 of the sequence KT819921), T. gigantea and T. incrustata (e.g. positions 458–543 of the sequence KT819922), T. arboricola, T. crenulata, T. decolorans, T. solaris and Trebouxia sp. P-400-IaSc (e.g. positions 581–599 of the sequence KT819921).
Bayesian analyses were carried out for algal nuITS dataset using the Metropolis-coupled Markov chain Monte Carlo (MCMCMC) method in MrBayes v. 3.2.6 on the CIPRES Web Portal68. GTR + I + G best-fit evolutionary model was selected based on Akaike Information Criterion (AIC69) as implemented in MrModelTest276. Two parallel MCMCMC runs were performed, each using four independent chains and 20 million generations, sampling trees every 1000th. Tracer v. 1.671 was used to ensure that stationarity was reached by plotting the log-likelihood values of the sample points against generation time. Convergence between runs was also verified using the PSRF (Potential Scale Reduction Factor), where all values were equal or close to 1.000. Posterior probabilities (PP) were determined by calculating a majority-rule consensus tree generated from the 60002 post-burnin trees of the 80002 trees sampled by the two MCMCMC runs using the sumt option of MrBayes. In addition, a Maximum Likelihood (ML) analysis was performed using RAxML-HPC2 v.8.2.1072 with 1000 ML bootstrap iterations (ML-BS) and the GTRGAMMA model.
The RAxML tree did not contradict the Bayesian tree topology for the strongly supported branches, therefore, only the Bayesian tree is shown for algal nuITS rDNA dataset with the bootstrap support values and posterior probabilities of the Bayesian analysis added near the branches (Fig. 3). ML-BS ≥ 70 and PP ≥ 0.9 were considered to be significant and are shown near the branches.
Phylogenetic analyses of trentepohlioid algae
For the trentepohlioid rbcL gene analysis, the newly generated sequences were aligned together with all sequences of Trentepohliales (200 records) available in GenBank using Seaview software77,78 employing muscle option and excluding ambiguous positions in Gblocks 0.91b74,75 using less stringent settings (i.e. allow smaller final blocks, gap positions within the final blocks and less strict flanking positions). The preliminary Bayesian analysis was carried out using the Metropolis-coupled Markov chain Monte Carlo (MCMCMC) method in MrBayes v. 3.2.6 on the CIPRES Web Portal68. Based on the tree obtained, we decided to reduce our dataset to 60 sequences using selected representatives (one or two sequences) of all main lineages. Moreover, sequences that were significantly shorter were also excluded from analyses. Terminal ends of the alignment corresponding to positions 1–321 and 990–1427 in the sequence KM464712 of Trentepohlia annulata were excluded from the alignment.
Bayesian analyses were carried out for trentepohlioid rbcL dataset using the Metropolis-coupled Markov chain Monte Carlo (MCMCMC) method in MrBayes v. 3.2.6 on the CIPRES Web Portal68. GTR + I + G best-fit evolutionary model was selected based on Akaike Information Criterion (AIC69) as implemented in MrModelTest276. Two parallel MCMCMC runs were performed, each using four independent chains and 10 million generations, sampling trees every 1000th. Tracer v. 1.671 was used to ensure that stationarity was reached by plotting the log-likelihood values of the sample points against generation time. Convergence between runs was also verified using the PSRF (Potential Scale Reduction Factor), where all values were equal or close to 1.000. Posterior probabilities (PP) were determined by calculating a majority-rule consensus tree generated from the 15002 post-burnin trees of the 20002 trees sampled by the two MCMCMC runs using the sumt option of MrBayes. In addition, a Maximum Likelihood (ML) analysis was performed using RAxML-HPC2 v.8.2.1072 with 1000 ML bootstrap iterations (ML-BS) and the GTRGAMMAI model.
The RAxML tree did not contradict the Bayesian tree topology for the strongly supported branches, therefore, only the Bayesian tree is shown for trentepohlioid rbcL dataset with the bootstrap support values and posterior probabilities of the Bayesian analysis added near the branches (Fig. 4). ML-BS ≥ 70 and PP ≥ 0.9 were considered to be significant and are shown near the branches.
Phylogenetic trees were visualized using FigTree v. 1.4.273.
The DNA sequences generated during the current study are available in the GenBank repository (https://www.ncbi.nlm.nih.gov; see Table 1) and from the corresponding author on reasonable request.
References
Moncada, B., Coca, L. F. & Lücking, R. Neotropical members of Sticta (lichenized Ascomycota: Lobariaceae) forming photosymbiodemes, with the description of seven new species. Bryologist 116, 169–200 (2013).
Chagnon, P. L., U’Ren, J. M., Miadlikowska, J., Lutzoni, F. & Arnold, A. E. Interaction type influences ecological network structure more than local abiotic conditions: evidence from endophytic and endolichenic fungi at a continental scale. Oecologia 180, 181–191, https://doi.org/10.1007/s00442-015-3457-5 (2016).
Aschenbrenner, I. A., Cernava, T., Berg, G. & Grube, M. Understanding microbial multi-species symbioses. Front. Microbiol. 7, 180, https://doi.org/10.3389/fmicb.2016.00180 (2016).
Spribille, T. et al. Basidiomycete yeasts in the cortex of ascomycete macrolichens. Science 353, 488–492, https://doi.org/10.1126/science.aaf8287 (2016).
Dughi, R. Etude comparée du Dendriscocaulon bolacinum Nyl. et de la céphalodie fruticuleuse du Ricasolia amplissima (Scop.) Leight. B. Soc. Bot. Fr. 83, 671–693 (1936).
Armaleo, D. & Clerc, P. Lichen chimeras: DNA analysis suggests that one fungus forms two morphotypes. Exp. Mycol. 15, 1–10 (1991).
Magain, N., Goffinet, B. & Sérusiaux, E. Further photomorphs in the lichen family Lobariaceae from Reunion (Mascarene archipelago) with notes on the phylogeny of Dendriscocaulon cyanomorphs. Bryologist 115, 243–254 (2012).
James, P. W. & Henssen, A. The morphological and taxonomic significance of cephalodia in Lichenology : Progress and Problems (eds D. H. Brown, D. L. Hawksworth, & R. H. Bailey) 27–77 (Academic Press, 1976).
Miadlikowska, J. The blue-green morphotype of Peltigera venosa in Poland. Graphis Scripta 9, 21–22 (1998).
Friedl, T. Thallus development and phycobionts of the parasitic lichen Diploschistes muscorum. Lichenologist 19, 183–191 (1987).
Piercey-Normore, M. D. & DePriest, P. T. Algal switching among lichen symbioses. Am. J. Bot. 88, 1490–1498 (2001).
Yahr, R., Vilgalys, R. & Depriest, P. T. Strong fungal specificity and selectivity for algal symbionts in Florida scrub Cladonia lichens. Mol. Ecol. 13, 3367–3378, https://doi.org/10.1111/j.1365-294X.2004.02350.x (2004).
Guzow-Krzemińska, B. Photobiont flexibility in the lichen Protoparmeliopsis muralis as revealed by ITS rDNA analyses. Lichenologist 38, 469–476 (2006).
Nelsen, M. P. & Gargas, A. Dissociation and horizontal transmission of codispersing lichen symbionts in the genus Lepraria (Lecanorales: Stereocaulaceae). New Phytol. 177, 264–275, https://doi.org/10.1111/j.1469-8137.2007.02241.x (2008).
Muggia, L., Perez-Ortega, S., Kopun, T., Zellnig, G. & Grube, M. Photobiont selectivity leads to ecological tolerance and evolutionary divergence in a polymorphic complex of lichenized fungi. Ann. Bot. 114, 463–475, https://doi.org/10.1093/aob/mcu146 (2014).
Fernandez-Mendoza, F. et al. Population structure of mycobionts and photobionts of the widespread lichen Cetraria aculeata. Mol. Ecol. 20, 1208–1232, https://doi.org/10.1111/j.1365-294X.2010.04993.x (2011).
Leavitt, S. D., Nelsen, M. P., Lumbsch, H. T., Johnson, L. A. & St Clair, L. L. Symbiont flexibility in subalpine rock shield lichen communities in the Southwestern USA. Bryologist 116, 149–161 (2013).
Hametner, C., Stocker-Wörgötter, E. & Grube, M. New insights into diversity and selectivity of trentepohlialean lichen photobionts from the extratropics. Symbiosis 63, 31–40 (2014).
del Campo, E. M. et al. South European populations of Ramalina farinacea (L.) Ach. share different Trebouxia algae. Bibl. Lichenolog. 105, 247–256 (2010).
Moya, P., Molins, A., Martinez-Alberola, F., Muggia, L. & Barreno, E. Unexpected associated microalgal diversity in the lichen Ramalina farinacea is uncovered by pyrosequencing analyses. PloS One 12, e0175091, https://doi.org/10.1371/journal.pone.0175091 (2017).
Demmig-Adams, B. et al. Differences in the susceptibility to light stress in two lichens forming a phycosymbiodeme, one parter possessing and one lacking the xanthophyll cycle. Oecologia 84, 451–456 (1990).
Piercey-Normore, M. D. The lichen-forming ascomycete Evernia mesomorpha associates with multiple genotypes of Trebouxia jamesii. New Phytol. 169, 331–344, https://doi.org/10.1111/j.1469-8137.2005.01576.x (2006).
Peksa, O. & Škaloud, P. Do photobionts influence the ecology of lichens? A case study of environmental preferences in symbiotic green alga Asterochloris (Trebouxiophyceae). Mol. Ecol. 20, 3936–3948 (2011).
Williams, L. et al. Lichen acclimation to changing environments: Photobiont switching vs. climate-specific uniqueness in Psora decipiens. Ecol. Evol. 7, 2560–2574, https://doi.org/10.1002/ece3.2809 (2017).
Magain, N. & Serusiaux, E. Do photobiont switch and cephalodia emancipation act as evolutionary drivers in the lichen symbiosis? A case study in the Pannariaceae (Peltigerales). PloS One 9, e89876, https://doi.org/10.1371/journal.pone.0089876 (2014).
Arup, U. & Grube, M. Where does Lecanora demissa (Ascomycota, Lecanorales) belong? Lichenologist 31, 419–430 (1999).
Ekman, S. & Tønsberg, T. Most species of Lepraria and Leproloma form a monophyletic group closely related to Stereocaulon. Mycol. Res. 106, 1262–1276 (2002).
Lendemer, J. C. & Hodkinson, B. P. A radical shift in the taxonomy of Lepraria s.l.: molecular and morphological studies shed new light on the evolution of asexuality and lichen growth form diversification. Mycologia 105, 994–1018 (2013).
Poelt, J. D. K. Der Artenpaare bei den Flechten. Vorträge aus dem Gesamtgebiet der Botanik 4, 187–198 (1970).
Crespo, A. & Pérez-Ortega, S. Cryptic species and species pairs in lichens: A discussion on the relationship between molecular phylogenies and morphological characters. An. Jardín Bot. Madrid 66S1, 71–81 (2009).
Buschbom, J. & Mueller, G. M. Testing “species pair” hypotheses: evolutionary processes in the lichen-forming species complex Porpidia flavocoerulescens and Porpidia melinodes. Mol. Biol. Evol. 23, 574–586, https://doi.org/10.1093/molbev/msj063 (2006).
Lohtander, K., Myllys, L., Sundin, R., Källersjö, M. & Tehler, A. The species pair concept in the lichen Dendrographa leucophaea (Arthoniales): analyses based on ITS sequences. Bryologist 101, 404–411 (1998).
Myllys, L., Lohtander, K. & Tehler, A. β-Tubulin, ITS and group I intron sequences challenge the species pair concept in Physcia aipolia and P. caesia. Mycologia 93, 335–343 (2011).
Tehler, A., Ertz, D. & Irestedt, M. The genus Dirina (Roccellaceae, Arthoniales) revisited. Lichenologist 45, 427–476 (2013).
Thor, G. & Muhr, L.-E. Buellia violaceofusca, a new lichen from Sweden. Lichenologist 23, 11–13 (1991).
Voytsekhovich, A. & Beck, A. Lichen photobionts of the rocky outcrops of Karadag massif (Crimean Peninsula). Symbiosis 68, 9–24 (2016).
Singh, G. et al. Fungal-algal association patterns in lichen symbiosis linked to macroclimate. New Phytol. 214, 317–329, https://doi.org/10.1111/nph.14366 (2017).
Hametner, C., Stocker-Wörgötter, E., Rindi, F. & Grube, M. Phylogenetic position and morphology of lichenized Trentepohliales (Ulvophyceae, Chlorophyta) from selected species of Graphidaceae. Phycol. Res. 62, 170–186 (2014).
Rindi, F., Lam, D. W. & Lopez-Bautista, J. M. Phylogenetic relationships and species circumscription in Trentepohlia and Printzina (Trentepohliales, Chlorophyta). Mol. Phylogenet. Evol. 52, 329–339 (2009).
Egea, J. M. & Torrente, P. El género de hongos liquenizados Lecanactis (Ascomycotina). Bibl. Lichenolog. 54, 1–205 (1994).
Wornik, S. & Grube, M. Joint dispersal does not imply maintenance of partnerships in lichen symbioses. Microbial Ecol. 59, 150–157 (2009).
Ellis, C. & Coppins, B. J. Reproductive strategy and the compositional dynamics of crustose lichen communities on aspen (Populus tremula L.) in Scotland. Lichenologist 39, 377–391 (2007).
Schultz, M., Wirth, V. & Feuerer, T. Erstfunde von Flechten und lichenicolen Pilzen in Deutschland. Herzogia 20, 329–334 (2007).
Šoun, J., Vondrák, J. & Bouda, F. Rare and little known species in the Třebíč Region and its surroundings. Bryonora 56, 1–23 (2015).
Ranius, T., Johansson, P., Niclas, B. & Niklasson, M. The influence of tree age and microhabitat quality on the occurrence of crustose lichens associated with old oaks. J. Veg. Sci. 19, 653–662 (2008).
Almborn, O. Distribution and ecology of some south Scandinavian lichens. Vol. 1, 1-254 (C.W.K. Gleerup, 1948).
Coppins, B. J., Scheidegger, C. & Aptroot, A. Buellia De Not. In The lichens of Great Britain and Ireland (eds C. W. Smith et al.) 228–238 (The Natural History Museum, London, 2009).
Wirth, V., Hauck, M. & Schultz, M. Die Flechten Deutschlands. Vol. 2 (Eugen Ulmer KG, Stuttgart, 2013).
Diederich, P. et al. New or interesting lichens and lichenicolous fungi from Belgium, Luxembourg and northern France. XII. Bull. Soc. Nat. luxem. 110, 75–92 (2009).
Türk, R. & Berger, F. Neue und seltene Flechten sowie lichenicole Pilze aus den Ostalpen III. Linzer Biol. Beitr. 31, 929–953 (1999).
Johansson, V., Ranius, T. & Snäll, T. Epiphyte metapopulation dynamics are explained by species traits, connectivity, and patch dynamics. Ecology 93, 235–241 (2012).
Johansson, V., Ranius, T. & Snäll, T. Development of secondary woodland decreases epiphyte metapopulation sizes in wooded grasslands. Biol. Conserv. 172, 49–55 (2014).
Brooks, F. E., Rindi, F., Suto, Y., Ohtani, S. & Green, M. The Trentepohliales (Ulvophyceae, Chlorophyta): an unusual algal order and its novel plant pathogen. Cephaleuros. Plant Dis. 99, 740–753 (2015).
Nelsen, M. P., Rivas Plata, E., Andrew, C. J., Lücking, R. & Lumbsch, H. T. Phylogenetic diversity of trentepohlialean algae associated with lichen-forming fungi. J. Phycol. 47, 282–290 (2011).
Beck, A., Friedl, T. & Rambold, G. Selectivity of photobiont choice in a defined lichen community: inferences from cultural and molecular studies. New Phytol. 139, 709–720 (1998).
Ranius, T., Eliasson, P. & Johansson, P. Large-scale occurrence patterns of red-listed lichen and fungi on old oaks are influenced both by current and historical habitat density. Biodivers. Conserv. 17, 2371–2381 (2008).
Ertz, D. & Tehler, A. The phylogeny of Arthoniales (Pezizomycotina) inferred from nucLSU and RPB2 sequences. Fungal Divers. 49, 47–71 (2011).
Frisch, A., Thor, G., Ertz, D. & Grube, M. The Arthonialean challenge: Restructuring Arthoniaceae. Taxon 63, 727–744 (2014).
Lendemer, J. C. & Knudsen, K. Lecanographa insolita, an unusual new species of Roccellaceae from western North America. The Bryologist 113, 350–355 (2010).
Guzow-Krzemińska, B. & Węgrzyn, G. Potential use of restriction analysis of PCR-amplified DNA fragments in taxonomy of lichens. Mycotaxon 76, 305–313 (2000).
Ertz, D. et al. A large-scale phylogenetic revision of Roccellaceae (Arthoniales) reveals eight new genera. Fungal Divers. 70, 31–53 (2015).
Zoller, S., Scheidegger, C. & Sperisen, C. PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. Lichenologist 31, 511–516 (1999).
White, T. J., Bruns, T. D., Lee, S. B. & Taylor, J. W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics in PCR protocols—a guide to methods and applications (eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, & T. J. White) 315–322 (Academic, 1990).
Helms, G., Friedl, T., Rambold, G. & Mayrhofer, H. Identification of photobionts from the lichen family Physciaceae using algal-specific ITS rDNA sequences. Lichenologist 33, 73–86 (2001).
Van den Broeck, D. et al. H eterocyphelium leucampyx (Arthoniales, Ascomycota): another orphaned mazaediate lichen finds its way home. Lichenologist 49, 333–345 (2017).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Lutzoni, F., Wagner, P., Reeb, V. & Zoller, S. Integrating ambiguously aligned regions of DNA sequences in phylogenetic analyses without violating positional homology. Systematic Biol. 49, 628–651 (2000).
Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees in Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November, 20101–8 (2010).
Akaike, H. Information theory and an extension of the maximum likelihood principle in Proceedings of the 2nd International Symposium on Information Theory (eds B. N. Petrov & F. Csaki) 267–281 (Akademiai Kiado, 1973).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772, https://doi.org/10.1038/nmeth.2109 (2012).
Rambaut, A. & Drummond, A. J. Tracer v1.6 http://beast.bio.ed.ac.uk/ (2007).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313, https://doi.org/10.1093/bioinformatics/btu033 (2014).
Rambaut, A. FigTreev1.4. 2 http://tree.bio.ed.ac.uk/software/figtree/ (2012).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000).
Dereeper, A. et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic acids Res. 36, W465–469, https://doi.org/10.1093/nar/gkn180 (2008).
Nylander, J. A. A. MrModeltest 2.0. Program distributed by the author (Evolutionary Biology Centre, 2004).
Galtier, N., Gouy, M. & Gautier, C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12, 543–548 (1996).
Gouy, M., Guindon, S. & Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224, https://doi.org/10.1093/molbev/msp259 (2010).
Acknowledgements
We thank very warmly Cyrille Gerstmans, Myriam Dehaan and Wim Baert for their help with the molecular work and other technical parts of the manuscript. We also thank Dr Małgorzata Kapusta (Department of Plant Cytology and Embryology, University of Gdańsk) for help with photographing photobionts. The research leading to these results has received financial support from the Polish-Norwegian Research Programme operated by the National Centre for Research and Development under the Norwegian Financial Mechanism 2009–2014 in the frame of Project Contract No Pol-Nor/196829/87/2013.
Author information
Authors and Affiliations
Contributions
D.E. sampling, molecular work, phylogenetic analyses of the mycobiont and redaction of the manuscript. M.K. sampling and redaction of the manuscript. B.G.K. molecular work, phylogenetic analyses of the photobionts and redaction of the manuscript. A.L. and G.T. sampling. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ertz, D., Guzow-Krzemińska, B., Thor, G. et al. Photobiont switching causes changes in the reproduction strategy and phenotypic dimorphism in the Arthoniomycetes. Sci Rep 8, 4952 (2018). https://doi.org/10.1038/s41598-018-23219-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-23219-3
This article is cited by
-
On Holobionts, Holospecies, and Holoniches: the Role of Microbial Symbioses in Ecology and Evolution
Microbial Ecology (2023)
-
New lineages of photobionts in Bolivian lichens expand our knowledge on habitat preferences and distribution of Asterochloris algae
Scientific Reports (2021)
-
Morpho-molecular characterization of rock-inhabiting lichen Dermatocarpon miniatum (Verrucariaceae, Ascomycota) and its symbiont in Indian Himalayas
The Nucleus (2021)
-
Species delimitation in the cyanolichen genus Rostania
BMC Evolutionary Biology (2020)
-
Genome-scale data reveal the role of hybridization in lichen-forming fungi
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.