Abstract
While our understanding of the genetics underlying the Brassica-Leptosphaeria pathosystem has advanced greatly in the last decade, differences in molecular responses due to interaction between resistance genes and host genetic background has not been studied. We applied RNAseq technology to monitor the transcriptome profiles of Brassica napus (Bn) lines carrying one of four blackleg R genes (Rlm2, Rlm3, LepR1 & LepR2) in Topas or Westar background, during the early stages of infection by a Leptosphaeria maculans (Lm) isolate carrying the corresponding Avr genes. We observed upregulation of host genes involved in hormone signalling, cell wall thickening, response to chitin and glucosinolate production in all R gene lines at 3 day after inoculation (dai) albeit having higher level of expression in LepR1 and Rlm2 than in Rlm3 and LepR2 lines. Bn-SOBIR1 (Suppressor Of BIR1-1), a receptor like kinase (RLK) that forms complex receptor like proteins (RLPs) was highly expressed in LepR1 and Rlm2 at 3 dai. In contrast Bn-SOBIR1 induction was low in Rlm3 line, which could indicate that Rlm3 may function independent of SOBIR1. Expression of Salicylic acid (SA) related defense was enhanced in LepR1 and Rlm2 at 3 dai. In contrast to SA, expression of Bn genes with homology to PDF1.2, a jasmonic acid (JA) pathway marker, were increased in all Rlm and LepR lines at 6 and 9 dai. Effect of host genetic background on induction of defense, was determined by comparison of LepR1 and LepR2 in Topas vs Westar genotype (i.e. T-LepR1 vs W-LepR1 and T-LepR2 vs W-LepR2). In both cases (regardless of R gene) overall number of defense related genes at the earliest time point (3 dai) was higher in Tops compared to Westar. SA and JA markers genes such as PR1 and PDF1.2 were more induced in Topas compared to Westar introgression lines at this time point. Even in the absence of any R gene, effect of Topas genotype in enhanced defense, was also evident by the induction of PDF1.2 that started at a low level at 3 dai and peaked at 6 and 9 dai, while no induction in Westar genotype was observed at any of these time points. Overall, variation in time and intensity of expression of genes related to defense, was clearly dependent on both R gene and the host genotype.
Similar content being viewed by others
Introduction
Plants are exposed to a myriad of microorganism in their environment. However, natural physical barriers and chemical deterrents guard the plants from the majority of these microbes. The small number of microorganisms that overcome this passive defense still have to evade recognition by the plant cell surface and cytoplasmic receptors that have evolved to perceive conserved structural molecules and virulence factors termed pathogen associated molecular patterns (PAMP) and effectors, respectively1,2,3. Effector-triggered immunity (ETI) is often manifested as a rapid and strong defense response leading to induction of cell death, also known as hypersensitive response (HR), at the site of infection to arrest pathogen growth beyond the point of entry. HR provides early and robust resistance against Leptosphaeria maculans (Lm), the hemibotrophic fungal agent of blackleg disease in canola (oilseed rape, Brassica napus; Bn). The gene expression profile of Bn cotyledons infected with Lm reveals the transition from biotrophy to necrotrophy as infection progresses in a compatible interaction4. Genes related to salicylic acid (SA) pathway are induced at the earlier stages of infection (3 days after inoculation; dai) while expression of jasmonic acid (JA) pathway genes, linked to plant response to necrotrophic pathogens, is up-regulated at the later stages (6–9 dai). Induction of JA defense correlates with the up-regulation of the Lm gene necrosis and ethylene‐inducing peptide‐1 (Nep‐1) ‐like protein, a well-known marker of transition from biotrophy to necrotrophy in fungi4. Becker et al.5 also reported the importance of SA and JA pathways in a Bn line carrying the LepR1 resistance gene in response to a Lm isolate carrying the corresponding AvrLep1 effector. Upon perception of Lm an array of host genes functionally-defined as proteases and protease inhibitors, chitinases, peroxidases, transcription factors (WRKY, AP2/EREBP, MYB), genes related to the production of the secondary metabolites and genes involved in plant cell wall reinforcement were differentially expressed4,5.
Despite being a qualitative trait, the immunity response triggered by R gene Avr gene recognition varies in phenotype; from a highly-localised response seen as minute necrosis, trailing necrosis, no visual symptoms or contained pathogen growth and sporulation6. Both genotype of the host and functional variation of R genes cause variation in the interaction phenotype7. R protein activation and recognition of pathogen effector proteins often requires inter and/or intra-molecular interaction and complex formation with other host proteins8,9. These receptor complexes affect R protein function and consequently plant pathogen interaction phenotype8,9. Natural variation in Arabidopsis has served as a tool to dissect the genetic basis of polymorphism in plant to interaction with pathogens10,11.
Race specific R genes have been widely used in breeding for Brassica napus (canola; oilseed rape) resistance to blackleg disease caused by the ascomycete fungus Leptosphaeria maculans12. Out of nineteen R genes reported from Brassica species, eleven of them (Rlm1, Rlm2, Rlm3, Rlm4, Rlm7, Rlm9, Rlm11, LepR1, LepR2, LepR3 and LepR4) originated from the A genome of Bn and Brassica rapa (Br)12,13. The genetics of the Bn-Ln pathosystem has been greatly advanced by the cloning of Lm Avr genes; AvrLm1, 2, 3, 4–7, 5–9, 6, and 1114,15,16,17,18,19,20,21,22 and the characterisation of the Bn R genes LepR3 and Rlm223,24. Host differential lines are indispensable for genotyping plant pathogen races. In previous studies, Rlm and LepR genes were introgressed into common susceptible Bn doubled-haploid lines Topas (DH16516) or Westar (N-o-1)13,25. This led to generation of seven Topas introgression lines (T-Rlm1, T-Rlm2, T-Rlm3, T-Rlm4, T-LepR1, T-LepR2, T-LepR3), which share 92.9–98.9% of their genomic background with the susceptible parental lines13, and two Westar introgression lines (1065; W-LepR1, 1135; W-LepR2). These introgression lines provide a unique tool to compare the function of different R genes in a common genotype background and also for the dissection of the effect of host genetic background on the defense responses triggered by the same R gene. To gain insight into the molecular mechanism of host genetic background and R gene effect we conducted a comprehensive transcriptome analysis by performing RNAseq (1.5 billion raw reads- 168 samples) on Lm infected cotyledons at 0, 3, 6 and 9 dai. Here we describe the differences in gene expression profiles and defense pathways among these treatments and their correlation with the variation in visual interaction phenotypes.
Results
Variation of R gene phenotypic response to Lm infection
We have introduced individual Rlm (resistance to L. maculans) and LepR (Leptosphaeria resistance) genes into the common susceptible Bn cultivar Topas using repeated backcrossing and selfing (BC5S3) that allowed generation of individual Topas lines each harbouring Rlm or LepR gene intervals which was confirmed by genome wide high density SNP profiling of each line, minimizing background effects on R gene performance13. Bn cultivars Topas and Westar being completely susceptible to Lm are routinely used as control in blackleg pathology tests in the greenhouse and field trials. We have previously noted that the cotyledon immune response triggered by R genes in Topas is often more robust compared to the immune response induced by the same R gene in Westar13. To explore the genetics of this variation, we studied the effect of R genes and host genetic background on the global gene expression in response to Lm infection.
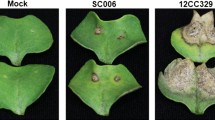
Parallel comparisons were made between Topas introgression resistance lines (T-Rlm2, T-Rlm3, T-LepR1 and T-LepR2) with Bn cv 1065 and 1135 harbouring LepR1 and LepR2 genes, respectively, in Westar background (W-LepR1 and W-LepR2). This allowed to study the effect of plant genotype on the dynamics of host defense against Lm. Plants were inoculated at the seedling stage with the pycnidiospores of Lm isolate 00–100 (Avr phenoytype A2-A3-A5-A6-(8)-A9-(10)-AS-AL1-AL2-(L4))13 and RNAseq was conducted on samples collected at 3, 6 and 9 dai (Supp. Table 1). Concomitantly visual and microscopy phenotypic responses were observed at 3, 6, 9, 12, 15 dai. There were no visible symptoms at 3 dai. At 6 dai a chlorotic ring surrounding the inoculation site became clearly visible which later expanded, leading to the formation of lesion and tissue collapse around the site of inoculation in Topas and Westar while in the Topas introgression lines (T-Rlm2, T-Rlm3, T-LepR1 and T-LepR2) pathogen growth was contained within the inoculation site (Fig. 1). The hypersensitive response (HR) appeared less intense at 6 dai in the Westar introgression lines (W-LepR1 and W-LepR2) compared to T-LepR1 and T-LepR2, judging by the intensity of brown tissue surrounding the inoculation site.
Disease symptoms on inoculated cotyledons of B. napus cv. Topas, Westar and Introgression Lines (ILs) with Leptosphaeria maculans (Lm). (A) Cotyledons of Topas, Westar and ILs photographed at 6, 9, 12 and 15 days after inoculation (dai). (B) Lm hyphae (green) shown by alexa fluor 488 (AF488) conjugate of wheat germ agglutinin (WGA) through field of view (FOV) at lesion on infected cotyledons of Topas, Westar and ILs at 6dai.
RNA sequencing and gene expression profile during infection
RNA reads from three biological replicates (1.5 billion paired-end reads from infected and mock-inoculated cotyledons) were analysed according to the methods described previously4. The number of reads for each sample is presented in the Supp. Table 1. Variability among the samples was determined by conducting principal component analysis (PCA). PCA displayed clear distinction between the transcriptome of introgression lines (ILs) and the susceptible parental lines at different time points (Fig. 2A). PCA distinguished four separate groups. “Group I” consisted of Topas and Westar in which transcript profiles of Lm infected lines at 3 dai grouped under the same cluster as transcript profiles of mock-inoculated Topas and Westar 3, 6 and 9 dai. Transcript profiles of Lm infected T-LepR2, T-Rlm3 and W-LepR1 at 3 dai formed a cluster separated from the transcript profiles of mock-inoculated lines (3, 6 and 9 dai) and Lm infected lines at 6 and 9 dai (Group III). Transcript profile of Lm infected W-LepR2 at 3 dai overlapped with the cluster containing transcript profiles of the mock-inoculated lines at 3, 6 and 9 dai (Group II). Comparison of all the differentially expressed genes (DEG) among the various lines and time points also revealed a delay in response for the two Bn susceptible cultivars (Topas and Westar). Group VI, composed of T-LepR1 and T-Rlm2 indicated a more rapid defense response by LepR1 and Rlm2 evident by clear distinction between transcript profiles of Lm infected lines at (3, 6 and 9 dai) and transcript profiles of mock-inoculated lines at the same time points (Fig. 2A). At 3 dai, fewer genes were differentially expressed in Topas and Westar than in ILs (40 and 22, respectively) while the total number of DEG in the ILs were between 580 and 3669 for the same time point (Fig. 2B).
Global evaluation of RNA-seq and defense response to Leptosphaeria maculans (Lm) in Introgression Lines (ILs) compared to Topas and Westar at 3, 6 and 9dai. (A) PCA displays clear distinction between the transcriptome of resistance lines and wild parents at different time points. (B) Differentially expressed genes (DEGs) obtained at various time points in Topas, Westar and ILs (C) GO term enrichment. Intensity scale: Red represents more up-regulated genes corresponding to GO term in response to Lm. ‘3I/3C’ means ‘3 dai for inoculated with Lm/3 dai for not inoculated control’. ‘6I/6C’ means ‘6 dai for inoculated with Lm/6 dai for not inoculated control’. ‘9I/9C’ means ‘9 dai for inoculated with Lm/9 dai for not inoculated control’.
The most notable and rapid changes in gene expression profile of ILs (3 dai) were changes in expression of genes with known function in plant defense such as response to chitin, oxidative burst, and salicylic acid (SA), jasmonic acid (JA), abscisic acid (ABA), ethylene and glucosinolate pathways. In comparison, for the susceptible Westar and Topas lines, changes in expression of defense pathway-related genes occurred at 6 dai (Fig. 2C). This latent differential gene expression response to infection is depicted in the PCA analysis (Fig. 2A).
Key defense response triggered by R genes
To define common defense pathways triggered by the race-specific resistance genes in Bn in response to Lm we compared the early defense response (3 dai) profile of gene expression across all the ILs. This comparison further validated the key genes involved in seedling resistance to Lm4. The pattern of DEG in all ILs indicated that early detection of Lm by Bn occurs through the activation of cell surface receptors. Examples of these were Rlm2/LepR3 and their paralogues, several Wall-Associated Kinases (WAKs) and Cysteine-rich Receptor-like Kinases (CRKs) containing DUF26 (domain of unknown function 26) as well as BIR1 and SOBIR1 that are components of PAMP/Effector receptor complexes25,26. In this study our data showed strong expression of six copies of Bn SOBIR1 homologues in T-LepR1 and T-Rlm2, peaking at 3 dai, which was also observed in T-LepR2, W-LepR1 and W-LepR2 but to lesser extent, compared to the susceptible controls, Westar and Topas, in which SOBIR1 expression was not induced at 3 dai (Supp. Fig. 1A). Expression of only two out of the six SOBIR1 homologues was induced in T-Rlm3 albeit to a much lower level compared to the other R gene introgression lines (Supp. Fig. 1A). Our data also showed the involvement of some of the CRK family of cell surface receptors in response to Lm at the earlier time points of infection. Among the differentially expressed CRKs with a role in plant defense were homologues of the Arabidopsis genes CRK 4, 5 and 20, which act as HR inducers27,28 (Supp. Fig. 1B) and CRK 2, 10 and 11, involved in chitin-triggered defense response29 (Supp. Fig. 1C).
Perceived signals of Lm are relayed through the activation of genes encoding for Cyclic Nucleotide-Gated Ion Channels (CNGC) that are likely involved in influx of calcium. CNGC 3, 12 and 19 have been reported to be involved in plant immunity30. Transcripts of CNGC 3, 12 and 19 homologues were enriched in response to Lm at 3 dai in the ILs while these genes were not induced in Topas until 6 dpi (Fig. 3). No induction of CNGC genes was observed for Westar with the exception of CNGC3 (BnaA05g01380D) (Fig. 3). Three genes (BnaA04g09500D, BnaC03g25880D and BnaC06g12520D) annotated as Calcium-binding EF-hand family proteins were also up-regulated at 3 dai.
Comparison of defense pathway genes common among the R genes shows that SA is the main plant hormone involved in the response and is activated in earlier time points (3 dai) in all of the ILs. In total 72 genes showed a strong positive correlation (>0.8) with genes reported to be induced in response to SA (Supp. Fig. 2). The most prominent SA dependent genes were EDS1, FMO1, EDS5, NIM1/NPR1 interacting protein 1 and 2 (NIMIN1 and NIMIN2, Supp. Fig. 3A), PAD4, PR1, PR2, as well as several WRKY transcription factors, in particular WRKY18 and 70 (Supp. Fig. 3B) that are reported to be positive regulators of the SA pathway31. This rapid SA dependent defense response was followed by the induction of antifungal enzymes such as chitinases (CHI) (Supp. Fig. 3C). At later time points (6 and 9 dai), DEG related to JA and auxin (Aux) pathways showed positive and strong correlation although SA pathway was still active at these time points, but to a lesser extent (Supp. Fig. 2).
In addition to Flavin-containing Monooxygenase (FMO), up-regulation of genes such as ALD1 (BnaA03g38440D and BnaC0345280D, infected vs mock; RPKM = 120 vs 0 at 3 dai) and amino acid transporter Lys/His transporter 1 and 7 (BnC01g03360D and BnaAnn11580D, infected vs mock; RPKM = 113 vs 0 at 3 dai) indicated the early potentiation and systemic spread of defense signals, prompting the systemic acquired resistance (SAR) response32,33,34. Arabidopsis ALD1 (agd2-like Defense Response Protein 1) mutants are reported to be impaired in SAR, susceptible to bacteria pathogens and compromised in defense against P. syringae35. Two BAP2 (BON1 Associated Proteins) genes (BnaA03g56970D and BnaC03g25440D) were up-regulated at 3 dai (70 RPKM in ILs compared to 0 to 0.2 RPKM in all mock control at 3, 6 and 9 dai). The expression level of BAP2 in infected tissue decreased at 6 and 9 dai to 10–20 RPKM. BAP2 is a negative regulator of cell death and its over-expression, along with BON1, prevents cell death36.
We predicted gene networks associated with response to fungi, response to chitin, response to salicylic acid, defense response by callose deposition and transcriptional regulatory elements involved in response to Lm based on co-expressed genes (Fig. 4A–F). Among the differentially expressed transcription factors with a role in plant defense were WRKY, AP2/EREBP and MYB. Expression of several WRKY transcription factors were up-regulated in the ILs. Transcripts of WRKY 70, 51, 50, 33, 18 associated with response to fungi and MYB51, MYB2 and WRKY 62, 53 related to salicylic acid response were enriched by 3 dai in the ILs. Among several WRKY genes, WRKY33 and MYB51 (predicted transcriptional regulator for glucosinolates) showed the highest expression difference to mock in the ILs (Fig. 4G).
Predicted networks these are involved in early response (3 dai) to Leptosphaeria maculans (Lm) in Introgression Lines (ILs). A predicted network that is involved in (A) response to fungus, (B) response to salicylic acid, (C) chitin response, (D) immune effector process, (E) defense response by cell wall thickening or callose deposition, (F) hypersensitive response at 3dai. TFs are highlighted in yellow. (G) Heatmaps present the difference in expression of B. napus genes with homology to WRKY33 and MYB51 in Lm-inoculated compared with mock-treated cotyledons of Topas, Westar and ILs at 3.6 and 9 dai. The colors correspond to log 2 RPKM (Infected – Uninfected) ranging from red (high) to green (low). Euclidean distance for the distances measure and complete linkage for clusters linkage criteria were selected.
Genetic background and R gene effect on the host defense response
Clustering of ILs and their respective susceptible lines, based on the number of differentially expressed genes, provided an overview and comparison of the dynamics of defense in each of these lines. Little change in gene expression was detected in the two susceptible lines Topas and Westar at the earliest time point (3 dai, Fig. 2B). The most enhanced gene expression (in terms of the total number of DEG) occurred in T-LepR1 followed by T-Rlm2, T-LepR2 and W-LepR1 (Fig. 2B). In W-LepR2 and T-Rlm3, the total number of DEG was significantly reduced showing a transcriptome profiles between the susceptible and T-LepR1, T-Rlm2. Genes related to proteins targeted to the membrane, defense against fungi, response to chitin, Reactive Oxygen Species (ROS), Systemic Acquired Resistance (SAR), SA, JA and genes involved in hypersensitive response, glucosinolate pathway and callose deposition were most prevalent in ILs (particularly in T-LepR1 and T-Rlm2) at 3 dai (Fig. 2C). In comparison, in Topas and Westar enrichment for genes related to these pathways occurred at 6 dai (Fig. 2C). All of these genes continued to be differentially expressed in T-LepR1, T-LepR2 and T-Rlm2 until 9 dai (Fig. 2C). At 3 dai, overexpression of marker genes related to SA (PR1, WRK70, ICS1) and JA (PDF1) were noticed in all ILs, however it was again most enhanced in T-LepR1 and T-Rlm2 (Supp. Fig. 3B and Fig. 5). Up-regulation of these marker genes occurred at 6 and 9 dai in all other R lines as well as Topas (Supp. Fig. 3B and Fig. 5). Induction of SA marker genes in Westar started at 6 and 9 dai however with much lower expression level as compared to Topas (Supp. Fig. 3B). Interestingly PDF1, the marker for JA pathway, was not upregulated in Westar at any of these time points (Fig. 5).
(A) An overview of MPK-VQ-JA signaling in Brassica napus and Leptosphaeria maculans (Lm) interaction. Identification of networks involved in earlier response to blackleg in ILs provided the evidence that WRKY33 is involved in response to Lm. Accumulation of transcripts associated with WRKY33, VQ16 and VQ23 homologues were enriched by 3 dai in resistance lines. RNAseq analysis revealed all 3 copies of VQ16 is suppressed after invasion by pycnidiospores of Leptosphaeria maculans in Topas and Westar, consequently one copy of PDF1.2 is highly suppressed in susceptible lines whilst it is highly accumulated in resistance lines. (B) Heatmaps present the difference in expression of B. napus genes with homology to JA (PDF1.2) in Lm-inoculated compared with mock-treated cotyledons of Topas, Westar and resistance lines at 3, 6, 9 dai. The colors correspond to log 2 RPKM (Infected – Uninfected) ranging from red (high) to green (low). Euclidean distance for the distances measure and complete linkage for clusters linkage criteria were selected.
A search of the KEGG database using DEG from T-LepR1, T-LepR2, T-Rlm2 and T-Rlm3 at 3 dai was performed. Pathways related to metabolic processes involved in energy production, such as glycolysis and the pentose phosphate pathway, TCA cycle, mitochondrial electron transport, ATP biosynthesis, and biosynthesis of some amino acids such as lysine and methionine, whose catabolism leads to energy production37, as well as biosynthesis of glutamic acid, arginine, serine, and glycine, which are associated with photorespiration38 were highly activated (based on the number of DEG associated with these pathways) in T-LepR1 and T-Rlm2 as compared to T-LepR2 and T-Rlm3.
Pathogens rely on host-derived nutrients for their growth. Balance of influx and efflux of nutrients between the source and sink tissue either promotes or limits pathogen growth39. We searched for the DEGs encoding nutrient transporters and noticed that nitrate, sucrose and amino acid transporters were the most abundant among the DEG in T-Rlm2 and T-LepR1 followed by T-LepR2 and T-Rlm3. In addition, there was a noticeable reduction in the number of nutrient transporters in W-LepR1 and W-LepR2 compared to T-LepR1 and T-LepR2, respectively. A search of plant defense-related GO terms revealed saturation for T-Rlm2 and T-LepR1 followed by T-LepR2 and T-Rlm3 and a significant reduction in W-LepR1 and W-LepR2. The most notable defense-related GO terms were related to cell wall modification (e.g. callose deposition, pectin metabolism), HR response, oxidative burst in response to defense, SA and glucosinolate biosynthesis, SA, JA, ABA, and SAR mediated signaling pathways, defense response to pathogens, response to chitin and response to toxins (Fig. 2C). Judging by the phenotypic response (Fig. 1A) Rlm2 and LepR1 display a more robust and rapid resistance response than Rlm3 and LepR2. Similarly as described above, pattern of DEG indicated a rapid response in defense and coordinated induction of pathways that provide energy and nutrient during the induction of defense in T- Rlm2 and T- LepR1 compared to T-LepR2 and T-Rlm3. This pattern of gene regulation occurred at 6 dai in Rlm3 and LepR1 lines. Likewise, effect of host genetic background on defense response initiated by the same R gene was evident from the prevalence of DEG related to biotic stress in T-LepR1 and T-LepR2 compared to W-LepR1 and W-LepR2, respectively.
Comparison of defense response in Topas and Westar with and without R genes
We have previously observed that some R genes in the Topas background show an enhanced resistance response compared to the level of resistance when the same R gene is expressed in Westar13. Also, development of lesions in Topas at the earlier stages of infection is slower compared to Westar, although at the late phases of infection (10–14 dai) development of lesion in both cultivars is comparable. HORMONOMETER software40 was used to catalogue hormone defense networks in response to Lm infection in which the DEGs as the input query genes were compared against a database of Arabidopsis genes expressed in response to the application of different hormones. Comparison between the gene expression profile of Westar and Topas at 3 dai revealed a positive correlation between DEG related to SA and JA pathways in Topas at 3 dai while expression of genes related to these pathways showed a negative correlation for the same time point in Westar, indicating the importance of SA and JA in boosting the defence response in Topas (Supp. Fig. 4). A network of differentially expressed transcription factors generated by SeqEnrich41 showed the enrichment for phytoalexin camalexin in Topas vs Westar at 3 dai (Supp. Fig. 5A,B). Among the genes with confirmed role in defense, expressed in Topas but absent from Westar at 3 dai, were genes with homology to the Arabidopsis Extensin (AT1G21310), a hydroxyproline-rich glycoprotein (HRGPs), with reported roles in plant defense through strengthening of the plant cell wall42,43. High expression of several chitinase genes at 3 dai was detected in Topas (Supp. Fig. 3C). Another DEG in Topas with antimicrobial activity was a GDSL LIPASE reported to function in disrupting fungal spores as well as inducing SAR44. Induction of defense pathways in Topas and Westar were further examined when the same R gene was present in both backgrounds. Comparison of DEG in T-LepR1 and W-LepR1 and similarly between T-LepR2 and W-LepR2 provided further insight into molecular mechanisms of enhanced resistance in Topas vs Westar. Although presence of LepR1 and LepR2 in Westar triggered the induction of the main defense hormones, notably SA and JA as well as auxin, Et and brassinosteroid pathways, the overall number of DEG related to these pathways were higher in T-LepR1 and T-LepR2 compared to W-LepR1 and T-LepR2, respectively (Supp. Fig. 4). A search of the KEGG database with DEG in T-LepR1 and W-LepR1 revealed that a serine/threonine kinase (OX1) is expressed at 3 dai in T-LepR1 but not in W-LepR1. Ox1 is required for basal resistance and activation of MPK3 and MPK445. Genes related to the glucosinolate pathway were induced in T-LepR1 at 3 dai but not in W-LepR1. The brassinazole resistance (BZR1) gene (BnaA06g13460D) was suppressed in T-LepR1. It has been shown that silencing BZR1 improved tobacco plants’ resistance to tobacco mosaic virus46. Activation of BZR1 resulted in impaired PAMP-triggered ROS production and enhanced susceptibility to adapted and non-adapted strains of the bacterium Pseudomonas syringae47.
Discussion
Plant response to pathogen infection leads to significant changes in the plant’s transcript profile. Despite overlaps and commonality between responses of different plant species to various pathogens, variation in plant phenotypic and molecular interactions to pathogens also occurs due to differences in R genes and host genetic backgrounds. Natural variation in Arabidopsis has been exploited to capture genetics for defense polymorphism against many oomycete, fungal and bacterial pathogens48. For some pathogens such as Lm, genetics and genomics of the defense response needs to be investigated using its natural host, as Arabidopsis does not provide an ideal model system49. Here we have taken advantage of several well-defined introgression lines each harboring individual R genes against blackleg pathogen in the Bn cv Topas, a common susceptible genotype. We monitored phenotypic interaction and changes in gene expression profile due to immunity response triggered by the race-specific resistance genes Rlm2, Rlm3, LepR1 and LepR2 in two susceptible Bn genetic background, Topas and Westar.
Comparing the transcript profiles of Topas and Westar and also W-LepR1 with T-LepR1 and W-LepR2 with T-LepR2 revealed a delay in defense response in Westar compared to Topas. While at the earliest time point i.e. 3 dai, changes in a limited number of defense-related genes were noticed in Topas, the transcript profile of infected Westar plants was the same as Westar mock inoculated controls. Similarly, judging by the defense related GO terms, immunity triggered by LepR1 and LepR2 was less intense in the Westar background compared to the immunity response induced by these same genes in Topas. Early induction of genes related to cell wall strengthening and production of antifungal compounds such as chitinases and lipases occurred in Topas but was not observed in Westar. The GO enrichment of camalexin in Topas early in infection was observed as compared to Westar (Supp. Fig. 5A,B). An Arabidopsis Phytoalexin Deficient 3 (pad3) mutant50 that is defective in camalexin production has been reported to be more susceptible to Lm infection51. The combined effect of R gene and host genetic background in generating a stronger defense response was also evident by expansion of DEG for SA, JA and Et pathways in T-LepR1, T-LepR2 compared to W-LepR1 and W-LepR2, respectively. Clustering of R gene ILs confirmed the boosting effect of the Topas genetic background on the level of gene expression at 3 dai for LepR1 in Topas compared to LepR1 in Westar. Differentially expressed genes in Topas vs Westar background could prove to be useful as markers to select the most suitable genotype as a recipient parent when developing Bn cultivars with resistance to Lm. The effect of R genes on spontaneity and robustness of defense response was measured based on the number and scope of defense related genes by comparing differentially expressed genes in T-Rlm2, T-Rlm3, T-LepR1 and T-LepR2. Based on the microscopy observation of the strength of the interaction phenotype and the genes’ ability to limit the pathogen growth over the course of infection, these R genes could be ordered as T-LepR1/T-Rlm2; T-LepR2 and finally T-Rlm3 from the most robust to a weaker defense response. As shown in the heat map of DEG at 3 dai (Supp. Fig. 3C), in T-LepR1, T-Rlm2 and to certain extent in T-LepR2, a strong induction of chitin-responsive genes, a known PAMP, and induction of genes related to callose deposition, up-regulation of ROS related genes, induction of SA and JA and to a lesser extent glucosinolate pathways, regulation of hypersensitive response and induction of downstream MAP kinases were the most prominent and well-documented indicators of plant immunity responses. It was only at 6 dai that these pathways were up-regulated to the same extent in T-LepR2 however, in the case of T-Rlm3, expression of defense related pathways was significantly less than in the other Topas introgression lines.
PAMP triggered immunity (PTI) provides basal defense upon detection of conserved pathogen molecules while effector triggered immunity (ETI) provides a rapid and strong defence in response to pathogen virulence (effector) genes. We investigated the PTI and ETI defense against Lm by comparing the DEG with the Arabidopsis genes associated with PTI and ETI as reported by Dong et al.52. This comparison revealed strong induction of PTI and ETI related genes in T-LepR1 and T-Rlm2 (Supp. Fig. 6A). It has also been reported that Non-race Specific Disease Resistance 1 (NDR1) is a conserved downstream regulator of R signalling53. Interaction of NDR1 and RIN4 results in transduction of extracellular pathogen-derived signals53. The difference in expression of Bn genes with homology to NDR1 related to ETI54 in Lm-inoculated compared with mock-treated cotyledons of Topas, Westar and the ILs revealed activation of these genes in incompatible hosts at 3 dai, with comparatively higher activity in T-Rlm2, T-LepR1 and T-LepR2 (Supp. Fig. 6B).
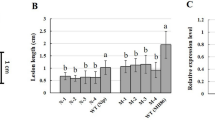
Among several WRKY transcription factors, WRKY11 and WRKY17 have both been reported to be involved in JA–dependent defense response55. Our result showed up-regulation of both in the ILs as compared to Topas and Westar. The WRKY33 transcription factor is reported to be important for plant resistance to hemibiotrophic and necrotrophic pathogens and to be involved in response to chitin, production of secondary metabolic and the phytoalexin biosynthetic pathway (Fig. 4A,C). Bimolecular fluorescence complementation previously revealed that WRKY33 interacts with nuclear-encoded SIGMA FACTOR BINDING PROTEIN 1 (SIB1) and SIB256. Both SIB1 (VQ16) and SIB2 (VQ23) contain a short VQ motif that is important for interaction with WRKY33. Transcripts associated with WRKY33, VQ16 and VQ23 homologues were enriched at 3 dai in the ILs (Fig. 5). Comparative transcriptomic analysis identified all 3 copies of VQ16 were suppressed in the Lm infected Topas and Westar plants. Figure 5 presents a model describing the possible role of VQ proteins and WRKY genes in the induction and suppression of defense against Lm. Quantification of expression of VQ16, WRKY33 and PDF1.2 were conducted by Droplet Digital PCR (ddPCR) which confirmed the RNAseq expression data (Supp. Fig. 7).
While monitoring the receptor complex-associated SOBIR1 homologues, it was noted that these were strongly expressed in T-LepR1 and T-Rlm2, with expression peaking early in the time course (3 dai). It was previously reported that the Arabidopsis thaliana LRR-receptor-like kinase (LRR-RLK) suppressor of Bir-1(AtSOBIR1) interacts with LRR-RLPs resistance genes57. We have previously demonstrated that SOBIR1 binds with both Rlm2 and its allelic variant LepR3, which are membrane-bound receptor-like proteins (RLPs)23,24. This result suggests that SOBIR1 is also required for successful LepR1 defense response, and that LepR1 may also encode an RLP. Conversely, very low expression of the SOBIR1 homologues was observed during the Rlm3-induced defence response, which could indicate that Rlm3 may function independent of SOBIR1.
Our data showed a repression of cytokinin (CK) responsive genes during early infection (3 dai) of Lm in Topas and resistance lines while the induction of CK was observed in Westar (Supp. Fig. 4). CK level in Bn cotyledons increases upon Lm infection58. The role of CK has been previously reported in various pathosystems. Some pathogenic fungi such as Magnaporthe oryzae, Cladosporium fulvum, Ustilago maydis or Claviceps purpurea produce CK to compromise the host defense59,60,61. A search for the genes involved in the CK pathway identified isopentenyltransferase (IPT) and adenosine kinase (AK) in the genome of Lm58. Previous observation that the level of CK was elevated in the Lm infected tissues needs to be further explored to determine the origin of CK (pathogen or host) and its importance in defense against Lm.
Previous Quantitative trait locus (QTL) studies suggested a role for cysteine-rich protein kinase genes in quantitative resistance to blackleg disease in Brassica napus62. Association of CRK11 within a functional network of plant immunity related genes (based on DEG at 3 dai) further supports its importance in defence against Lm (Supp. Fig. 8). In the case of quantitative resistance to pathogens, additive effect of genes and the combined interaction of host genotype background and environment account for variation in phenotypes. However, immunity response triggered by single R genes is generally thought to be less variable. By taking advantage of R gene introgression lines described previously13 we show that, despite the involvement of the same pathogen effector and plant R proteins in triggering the initial immunity response, there are clear differences in the dynamics of defense related gene expression and this is influenced by the host genetic background. Robust immune response and arrest of the pathogen at the site of penetration is highly desirable when developing resistant cultivars. Furthering our understanding of how R genes interact with host genotype background will help ensure selection of the best germplasm for robust expression of resistance.
Method and Materials
Plant growth condition and pathogen inoculation
Plant material comprised canola introgression lines (ILs) containing (i) LepR1, LepR2, Rlm2 and Rlm3, in Topas background (T-LepR1, T-LepR2, T-Rlm2, T-Rlm3), (ii) LepR1 and LepR2, in Westar-N-o-1 background (W-LepR1, W-LepR2), and Brassica napus cv. Topas (Topas DH16516) and Westar N-o-1, without known resistance genes13. Plants were grown in the growth chamber at 20 °C, 16 h light, with the light intensity c. 450 μmol m−2 s−1 at the bench level, and 18 °C, 8 h dark. For fungal inoculation, a small wound was made in the center of each cotyledon lobe (four wounds per plant) of 7 day-old seedlings and 10 μL of 2 × 107 spores/mL suspension was applied to each wound. In each of three experimental replications, half the one week-old plants from each IL, Topas and Westar (60 plants per line per condition) were inoculated with pycnidiospores of Lm isolate 00–100. The remaining seedlings were “mock” inoculated in the same manner with H2O.
RNA isolation and RNA sequencing
RNA was prepared from three biological replicates of infected and mock-inoculated cotyledons. Twelve discs per sample were collected using a standard hole punch and pooled, with each disc centred on the initial wound site. Samples were snap frozen in liquid nitrogen for later extraction. For RNA isolation, samples were ground in liquid nitrogen, then extracted with TRIzol LS reagent (Invitrogen) and purified by applying to the Ambion mini RNA kit following the manufacturer’s instructions. RNA was DNAase treated, quantified by Qubit fluorometer (Invitrogen) and checked for quality by Agilent Bioanalyzer 2100 (Agilent Technologies). Only samples with RNA integrity numbers above 8.0 were used for sequencing. Sequence reads (100 bp paired-end) were generated with Illumina TruSeq- high output version 3 chemistry on a HiSeq 2500 (Illumina, Inc.) at NRC-Plant Biotechnology Institute (NRC-PBI), Saskatoon, Saskatchewan, Canada.
Read mapping and analysis of RNA-Seq data
Transcriptomic analyses were carried out based on the methods described previously4. In total, 1.5 billion raw reads were analyzed for 168 samples in this study. Reads were trimmed, adaptor sequences were removed using Trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic) and then mapped to the B. napus (Genome Resources – Genoscope)63 and Lm19 genomes using CLC Genomics Workbench (QIAGEN, Aarhus, Denmark). Transcript abundance was measured as log 2 RPKM (Reads Per Kilobase of exon per Million mapped reads). Genes with a false discovery rate (FDR-BH) less than 0.05 were considered differentially expressed (DEG) using the empirical analysis of DGE Tool, which implements edgeR analysis64 and DESeq2 package65. Principal component analysis (PCA) was performed to assess the variability among samples.
Droplet Digital PCR (ddPCR) analysis
Quantification of expression of WRKY33, PDF1.2 and VQ16 were conducted by Droplet Digital PCR (ddPCR) according to the methods described previously66. RNA of Lm infected and mock inoculated Topas and Westar (from Group I), W-LepR1 (from Group II), T-LepR2 (from Group III) and T-LepR1 (from Group VI) at 3, 6, 9 dai were adjusted to 1 μg of RNA, and cDNA from three biological replicates was synthesized using an iScript™ Advanced cDNA Synthesis Kit according to the manufacturer’s protocol (Bio‐RAD). All primers and probes (Supp. Table 2) were designed using “Quest tool” complimented by IDT (https://www.idtdna.com/PrimerQuest/Home/Index) and “BnActin” was used as reference. ddPCR was performed using a QX100 Droplet Digital PCR (ddPCR™) System – Bio-Rad. The Bio-Rad QuantaSoft™ Analysis Pro (QuantaSoft AP) software was used to calculate the ratio signal of assay/Actin. In order to get RNAseq and ddPCR result comparable, heat map was generated based on: Log2 (Assay/Actin) (mean of replicates): inoculated with Lm/not inoculated control and Log2 RPKM (mean of replicates): inoculated with Lm/not inoculated control (Supp. Fig. 7).
Functional classification based on Blast2Go and KEGG
Annotation of DEG was performed by Blast2Go-pro67 and then KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis was done using KEGG function of Blast2Go-pro software to assign predicted pathways for DEG. HORMONOMETER40 was used to evaluate the transcriptome response through the perspective of similar events that happened upon hormonal activation in Arabidopsis.
Functional network analysis
The putative A. thaliana orthologs of Bn-DEGs were identified and then considered for a functional network analysis. An integrated pathway and interaction data based on co-expressed, co-localized genes and gene ontology information was thus identified using GeneMANIA, which was implemented through Cytoscape. Transcriptional landscape was also generated using SeqEnrich41.
Confocal microscopy: observation of Lm hyphae
To visualize Lm hyphae, the area 1 cm around the wound that was made in the center of each cotyledon of 7 day-old seedlings of ILs and susceptible lines was excised at 6 dai. The samples were soaked in a staining solution containing WGA-AlexaFluor488 (Invitrogen) at room temperature for 10 min. Observations with a TCS-SP5 confocal laser-scanning microscope were done at objectives of 20x. WGA–Alexa Fluor 488 was detected with a 488 nm excitation and 500–540 nm emission wavelength. Raw images were first deconvolved using AutoQuant X3 software (10 iterations) and then were imported into Imaris 7.4.1 to remove background noise.
References
Jones, J. D. G. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Thomma, B. P. H. J., Nürnberger, T. & Joosten, M. H. A. J. The Plant Cell-Of PAMPs and Effectors: The Blurred PTI-ETI Dichotomy. Plant Cell 23, 4–15 (2011).
Varden, F. A. et al. Taking the stage: effectors in the spotlight. Current Opinion in Plant Biology 38, 25–33 (2017).
Haddadi, P., Ma, L., Wang, H. & Borhan, M. H. Genome-wide transcriptomic analyses provide insights into the lifestyle transition and effector repertoire of Leptosphaeria maculans during the colonization of Brassica napus seedlings. Mol Plant Pathol 17 (2016).
Becker, G. B. et al. Transcriptome analysis of the Brassica napus–Leptosphaeria maculans pathosystem identifies receptor, signaling and structural genes underlying plant resistance. The Plant Journal 90, 573–586 (2017).
Jia, Y., McAdams, S. A., Bryan, G. T., Hershey, H. P. & Valent, B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014 (2000).
Frank, S. A. Specificity versus detectable polymorphism in host—parasite genetics. Proc. R. Soc. Lond. B 254, 191–197 (1993).
Yang, S. et al. Genome-wide investigation on the genetic variations of rice disease resistance genes. Plant Mol Biol 62, 181–193 (2004).
Harris, C. J., Slootweg, E. J., Goverse, A. & Baulcombe, D. C. Stepwise artificial evolution of a plant disease resistance gene. Proc Natl Acad Sci USA 110, 21189–21194 (2013).
Holub, E. B. The arms race is ancient history in Arabidopsis, the wildflower. Nat Rev Genet 2, 516–527 (2001).
Holub, E. B. Natural history of Arabidopsis thaliana and oomycete symbioses. European Journal of Plant Pathology 122, 91–109 (2008).
Raman, H., Raman, R. & Larkan, N. In Plant Breeding from Laboratories to Fields Ch. Chapter 4 (2013).
Larkan, N. J., Yu, F., Lydiate, D. J., Rimmer, S. R. & Borhan, M. H. Single R Gene Introgression Lines for Accurate Dissection of the Brassica - Leptosphaeria Pathosystem. Front Plant Sci 7, 1771 (2016).
Gout, L. et al. Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol Microbiol 60, 67–80 (2006).
Fudal, I. et al. Heterochromatin-Like Regions as Ecological Niches for Avirulence Genes in the Leptosphaeria maculans Genome: Map-Based Cloning of AvrLm6. MPMI 20, 459–470 (2007).
Parlange, F. et al. Leptosphaeria maculans avirulence gene AvrLm4-7 confers a dual recognition specificity by the Rlm4 and Rlm7 resistance genes of oilseed rape, and circumvents Rlm4-mediated recognition through a single amino acid change. Mol Microbiol 71, 851–863 (2009).
Balesdent, M. H. et al. The dispensable chromosome of Leptosphaeria maculans shelters an effector gene conferring avirulence towards Brassica rapa. New Phytol 198, 887–898 (2013).
Van de Wouw, A. P., Lowe, R. G., Elliott, C. E., Dubois, D. J. & Howlett, B. J. An avirulence gene, AvrLmJ1, from the blackleg fungus, Leptosphaeria maculans, confers avirulence to Brassica juncea cultivars. Mol Plant Pathol 15, 523–530 (2014).
Ghanbarnia, K. et al. Rapid identification of the Leptosphaeria maculans avirulence gene AvrLm2 using an intraspecific comparative genomics approach. Mol Plant Pathol 16, 699–709 (2015).
Ghanbarnia, K. et al. Leptosphaeria maculans AvrLm9: a new player in the game of hide and seek with AvrLm4-7. Mol Plant Pathol 19, 1754–1764 (2018).
Plissonneau, C. et al. A game of hide and seek between avirulence genes AvrLm4-7 and AvrLm3 in Leptosphaeria maculans. New Phytol 209, 1613–1624 (2016).
Plissonneau, C., Rouxel, T., Chevre, A. M., Van De Wouw, A. P. & Balesdent, M. H. One gene-one name: the AvrLmJ1 avirulence gene of Leptosphaeria maculans is AvrLm5. Mol Plant Pathol 19, 1012–1016 (2018).
Larkan, N. J. et al. The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol 197 (2013).
Larkan, N. J., Ma, L. & Borhan, M. H. The Brassica napus receptor-like protein RLM2 is encoded by a second allele of the LepR3/Rlm2 blackleg resistance locus. Plant Biotechnol J 13, 983–992 (2015).
Yu, F., Lydiate, D. J., Gugel, R. K., Sharpe, A. G. & Rimmer, S. R. Introgression of Brassica rapa subsp. sylvestris blackleg resistance into B. napus. Mol. Breed 30, 1495–1506 (2012).
Ma, L. & Borhan, M. H. The receptor-like kinase SOBIR1 interacts with Brassica napus LepR3 and is required for Leptosphaeria maculans AvrLm1-triggered immunity. Front Plant Sci 6, 933 (2015).
Chen, K., Du, L. & Chen, Z. Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Molecular Biology 53, 61–74 (2003).
Chen, K., Fan, B., Du, L. & Chen, Z. Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Molecular Biology 56, 271–283 (2004).
Bourdais, G. et al. Large-Scale Phenomics Identifies Primary and Fine-Tuning Roles for CRKs in Responses Related to Oxidative Stress. PLoS Genet 11 (2015).
Moeder, W., Urquhart, W., Ung, H. & Yoshioka, K. The role of cyclic nucleotide-gated ion channels in plant immunity. Mol Plant 4, 442–452 (2011).
Chen, C. & Chen, Z. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol 129, 706–716 (2002).
Cecchini, N. M., Jung, H. W., Engle, N. L., Tschaplinski, T. J. & Greenberg, J. T. ALD1 Regulates Basal Immune Components and Early Inducible Defense Responses in Arabidopsis. Mol Plant Microbe Interact 28, 455–466 (2015).
Yang, H. & Ludewig, U. Lysine catabolism, amino acid transport, and systemic acquired resistance: what is the link? Plant Signal Behav 9, e28933 (2014).
Yang, H., Postel, S., Kemmerling, B. & Ludewig, U. W. E. Altered growth and improved resistance of Arabidopsis against Pseudomonas syringae by overexpression of the basic amino acid transporter AtCAT1. Plant, Cell & Environment 37, 1404–1414 (2014).
Song, J. T., Lu, H., Mcdowell, J. M. & Greenberg, J. T. A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J 40, 200–12 (2004).
Yang, H., Yang, S., Li, Y. & Hua, J. The Arabidopsis BAP1 and BAP2 genes are general inhibitors of programmed cell death. Plant Physiol 145, 135–146 (2007).
Tabe, L. & Higgins, T. J. V. Engineering plant protein composition for improved nutrition. Trends in Plant Science 3, 282–286 (1998).
Less, H., Angelovici, R., Tzin, V. & Galili, G. Coordinated gene networks regulating Arabidopsis plant metabolism in response to various stresses and nutritional cues. Plant Cell 23, 1264–1271 (2011).
Fatima, U. & Senthil-Kumar, M. Plant and pathogen nutrient acquisition strategies. Front Plant Sci 6, 750 (2015).
Volodarsky, D., Leviatan, N., Otcheretianski, A. & Fluhr, R. HORMONOMETER: a tool for discerning transcript signatures of hormone action in the Arabidopsis transcriptome. Plant Physiol 150, 1796–1805 (2009).
Becker, M. G., Walker, P. L., Pulgar-Vidal, N. C. & Belmonte, M. F. SeqEnrich: A tool to predict transcription factor networks from co-expressed Arabidopsis and Brassica napus gene sets. PLoS One 12, e0178256 (2017).
Roberts, K. & Shirsat, A. H. Increased extensin levels in Arabidopsis affect inflorescence stem thickening and height. Journal of Experimental Botany 57, 537–545 (2006).
Wei, G. & Shirsat, A. H. Extensin over-expression in Arabidopsis limits pathogen invasiveness. Molecular Plant Pathology 7, 579–592 (2006).
Oh, I. S. et al. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 17, 2832–2847, d (2005).
Rentel, M. et al. OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsi. Nature 427 (2004).
Deng, X. G. et al. Role of brassinosteroid signaling in modulating Tobacco mosaic virus resistance in Nicotiana benthamiana. Sci Rep 6, 20579 (2016).
Fan, M. et al. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell 26, 828–841 (2014).
Holub, E. B. Natural variation in innate immunity of a pioneer species. Curr Opin Plant Biol 10, 415–424 (2007).
Šašek, V. et al. Recognition of avirulence gene AvrLm1 from hemibiotrophic ascomycete Leptosphaeria maculans triggers salicylic acid and ethylene signaling in Brassica napus. Mol. Plant–Microbe Interact 25, 1238–1250 (2012).
Zhou, N., Tootle, T. L. & Glazebrook, J. Arabidopsis PAD3, a Gene Required for Camalexin Biosynthesis, Encodes a Putative Cytochrome P450 Monooxygenase. Plant Cell 11, 2419–2428 (1999).
Persson, M., Staal, J., Oide, S. & Dixelius, C. Layers of defense responses to Leptosphaeria maculans below the RLM1- and camalexin-dependent resistances. New Phytol 182, 470–482 (2009).
Dong, X., Jiang, Z., Peng, Y. L. & Zhang, Z. Revealing shared and distinct gene network organization in Arabidopsis immune responses by integrative analysis. Plant Physiol 167, 1186–1203 (2015).
Selote, D., Shine, M. B., Robin, G. P. & Kachroo, A. Soybean NDR1-like proteins bind pathogen effectors and regulate resistance signaling. New Phytol 202, 485–498 (2014).
Zhang, J. et al. Effector-triggered and pathogen-associated molecular pattern–triggered immunity differentially contribute to basal resistance to Pseudomonas syringae. MPMI 23, 940–948 (2010).
Journot-Catalino, N., Somssich, I. E., Roby, D. & Kroj, T. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18, 3289–3302 (2006).
Lai, Z. et al. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23, 3824–3841 (2011).
Liebrand, T. W. et al. Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci 110, 10010–10015 (2013).
Trdá, L. et al. Cytokinin metabolism of pathogenic fungus Leptosphaeria maculans involves isopentenyltransferase, adenosine kinase and cytokinin oxidase/dehydrogenase. Front Microb 8, 1374 (2016).
Chanclud, E. & Morel, J. B. Plant hormones: a fungal point of view. Mol Plant Pathol 17 (2016).
Hinsch, J. et al. De novo biosynthesis of cytokinins in the biotrophic fungus claviceps purpurea. Environmental Microb 17, 2935–2951 (2015).
Bruce, S. A., Saville, B. J. & Emery, R. J. N.Ustilago maydis produces cytokinins and abscisic acid for potential regulation of tumor gormation in maize. J Plant Growth Regul 30, 51–63 (2011).
Larkan, J. et al. Multi-environment QTL studies suggest a role for cysteine-rich protein kinase genes in quantitative resistance to blackleg disease in Brassica napus. BMC Plant Biology 16, 1–16 (2016).
Chalhoub, B. et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014).
Taylor, S. C., Laperriere, G. & Germain, H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Scientific Reports 7, 2409 (2017).
Conesa, A. & Gotz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 619832 (2008).
Acknowledgements
The authors would like to thank Elena Beynon and Colin Kindrachuck for assisting with RNA extraction and Cathy Coutu and Jayvee Sadia for help with confocal microscopy and ddPCR, respectively. Funding for the project was provided by SaskCanola and AAFC through the Growing Forward 2 (GF2) initiative.
Author information
Authors and Affiliations
Contributions
P.H., N.J.L. and M.H.B. designed the experiment. P.H. and N.J.L. performed the experiments. P.H., N.J.L. and M.H.B. wrote the main manuscript text. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Haddadi, P., Larkan, N.J. & Borhan, M.H. Dissecting R gene and host genetic background effect on the Brassica napus defense response to Leptosphaeria maculans. Sci Rep 9, 6947 (2019). https://doi.org/10.1038/s41598-019-43419-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-43419-9
This article is cited by
-
Transcriptomics of temperature-sensitive R gene-mediated resistance identifies a WAKL10 protein interaction network
Scientific Reports (2024)
-
Status and advances in mining for blackleg (Leptosphaeria maculans) quantitative resistance (QR) in oilseed rape (Brassica napus)
Theoretical and Applied Genetics (2021)
-
Candidate Rlm6 resistance genes against Leptosphaeria. maculans identified through a genome-wide association study in Brassica juncea (L.) Czern
Theoretical and Applied Genetics (2021)
-
In silico characterization and expression of disease-resistance-related genes within the collinear region of Brassica napus blackleg resistant locus LepR1′ in B. oleracea
Journal of General Plant Pathology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.