Abstract
Pestalotiopsis and related genera, including Neopestalotiopsis and Pseudopestalotiopsis have damaged many plants for many decades; however, there is little available information about the fungi on tropical fruit in Thailand. This study isolated and characterized pestalotioid fungi on tropical fruit, investigated host specificity, and screened whether plant extracts could control the fungi. In total, 53 diseased fruit samples were sampled from eight types of fruit trees (jackfruit, rose apple, mangosteen, plum, snake fruit, rambutan, strawberry, and avocado). Based on morphological characteristics, 44 isolates were classified as belonging to pestalotioid taxa. Of these isolates, seven with distinct characteristics were selected for identification using molecular analysis, and six isolates were identified as Neopestalotiopsis and one as Pseudopestalotiopsis. In the cross-inoculation experiment, the isolates exhibited nonhost specificity and could infect at least two host plants. The isolates were used to screen for a potential biocontrol resource using six crude plant extracts (clove, ginger, lemongrass, mangosteen, roselle, and turmeric). All crude extracts except mangosteen could inhibit the growth of Neopestalotiopsis and Pseudopestalotiopsis. Application of crude plant extracts could be a potential treatment to control these diseases on tropical fruit.
Similar content being viewed by others
Introduction
Pestalotiopsis sensu lato comprises three genera—Neopestalotiopsis, Pseudopestalotiopsis, and Pestalotiopsis sensu stricto—established through phytopathogenic analysis based on the large subunit of the nuclear ribosomal RNA (LSU) region, and asexual morphs1, 2. Asexual morphs in these genera produce acervuli consisting of fusoid or ellipsoid to subcylindrical conidia with five cells, including an apical cell with appendages, a basal cell with an appendage, and three median cells positioned together with distinct colors1, 3.
Neopestalotiopsis, Pseudopestalotiopsis, and Pestalotiopsis infect a wide host range and currently cause problems worldwide4,5,6,7,8,9,10,11,12,13,14,15. The common symptoms of these pathogens include fruit rot, leaf blight, leaf spot, stem rot, grey blight, scabby canker, and post-harvest rot1, 16, 17. Examples of important plant pathogenic species are Pestalotiopsis clavispora, P. furcta, P. versicolor, P. virgatula, Neopestalotiopsis piceana, N. samarangensis, Pseudopestalotiopsis camelliae sinensis, and Ps. indica18,19,20.
Pestalotiopsis sensu lato threaten fruit production in Thailand. Several taxa of Pestalotiopsis, such as P. aeruginea, P. expaillatii, P. palmarum, P. pipericola, and P. theae, cause postharvest fruit rot and leaf blight disease in pear, cacao tree, banana, black pepper, and tea plant21. N. samarangensis causes fruit rot of rose apple18.
Pathogenicity of Pestalotiopsis sensu lato on a host has not been well understood. Plant pathogenic fungi in these genera are generally considered as a secondary pathogen; however, the diseases caused by the fungi have become more severe, such as the recent outbreak of Neopestalotiopsis in strawberries in Florida22. Pestalotiopsis sensu lato from different tissues of the same host exhibited a variation of pathogenicity in different tissues23.
Crude plant extracts can be an alternative to control diseases and can reduce the need to use fungicides. Many plants such as medicinal plants produce antimicrobial compounds and inhibit the growth of plant pathogens24, 25. For example, turmeric plants produce curcumin, eugenol, and eugenyl acetate that are effective against bacteria and fungi26,27,28,29,30.
Pestalotiopsis and its related genera have been problems of tropical fruits in Thailand for decades; however, little knowledge of these fungal pathogens has been available. The aims of the present study were: (1) to explore the pestalotioid fungi causing diseases of tropical fruit in Thailand based on both morphological data and molecular phylogeny using multilocus sequences of the internal transcribed spacer (ITS), the β-tubulin gene, and the translation elongation factor 1 alpha gene (tef1); (2) to study host specificity; and (3) to investigate the effectiveness of plant crude extracts against the mycelial growth of these genera.
Materials and methods
Sample collection
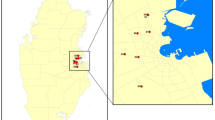
In total, 53 diseased plant samples showing leaf blight and fruit rot symptoms belonging to eight plant species were used, consisting of 5, 5, 13, 5, 6, 8, 6, and 5 samples from jackfruit (Artocarpus heterophyllus Lam.), rose apple (Syzygium samarangense [Blume] Merrill & L. M. Perry), mangosteen (Garcinia mangostana L.), plum (Bouea macrophylla Griffith), snake fruit (Salacca zalacca (Gaertn.) Voss), rambutan (Nephelium lappaceum L.), strawberry (Fragaria × ananassa Duchesne), and avocado (Persea americana Mill.), respectively (Fig. 1, Table 1). The samples were collected from private orchards in Chanthaburi, Phetchaburi, Nakhon Nayok, and Chiang Mai provinces, Thailand with the permission from each orchard owner between February 2017 and January 2018. The samples were recorded, photographed, and brought to the laboratory at the Department of Plant Pathology, Faculty of Agriculture, Kasetsart University (KU), Bangkok, Thailand for mycological isolation and morphological investigation. DNA extraction and PCR amplification were conducted in the National Center for Genetic Engineering and Biotechnology (BIOTEC), Pathum Thani, Thailand.
Symptoms of leaf blight and fruit rot on the original hosts caused by Neopestalotiopsis and Pseudopestalotiopsis. (a, b) Jackfruit leaves (A. heterophyllus); (c, d) rambutan leaves (N. lappaceum); (e, f) plum leaves (B. macrophylla); (g, h) avocado (P. americana); (i, j) strawberry (F. × ananassa); (k, l) leaves of snake fruit (S. zalacca); (m, n) mangosteen (G. mangostana); (o, p) rose apple (S. samarangense).
Fungal isolation and morphological data
The tissue transplantation technique was used to isolate fungal pathogens from the plant samples31. The tissue between healthy and infected parts was cut into 5 mm × 5 mm pieces, dipped in 0.25% sodium hypochlorite for 3 min, rinsed with sterile water, and dried with sterile tissue paper. The samples were placed in sterilized 9 cm Petri dishes, containing potato dextrose agar (PDA; Difco, Oxford, UK), incubated at 25 ± 2 °C for 3 days, and monitored daily. After incubation, the fungal isolates were examined and transferred to new PDA plates for identification. The macroscopic features of the fungi, such as colony color, colony diameter, and the exudates of each isolate, were recorded. Microscopic characteristics such as acervuli and conidial features were examined under a stereomicroscope (Olympus, Tokyo, Japan) and a compound microscope (Carl Zeiss, Jena, Germany) and compared with the features in identification keys and species descriptions1, 8, 9, 18.
DNA extraction, PCR amplification, and sequencing
Total genomic DNA of fungal mycelia grown on PDA at 28 °C for 4 days was extracted using the protocol of Chuaseeharonnachai et al.32 and the CTAB method33. Fragments of the rDNA internal transcribed spacer or ITS1-5.8S-ITS4 region were amplified using the primer pairs ITS1 and ITS4 34; the β-tubulin gene region was amplified using the primer pairs Bt2a and Bt2b 35; and the tef1 gene region was amplified using the primer pairs EF1-526F and EF1-1567R36. Amplification and sequencing reactions, sequences analyses, and consensus of sequences were performed as described by Maharachchikumbura et al.9. The PCR amplicons were visualized by the staining of a 1% agarose electrophoresis gel. Sequencing was performed by Macrogen Inc. (Seoul, Korea). Generated sequences were deposited in NCBI under accession numbers MT628375–MT628381 for ITS sequences, MW776648–MW776654 for β-tubulin sequences, and MW776633–MW776638 for the tef1 gene sequences.
Phylogenetic analyses
The sequences generated in this study were compared with sequences retrieved from the GenBank which were reported1, 37,38,39. ITS, β-tubulin, and tef1 gene sequence data were assembled using the alignment program BioEdit v7.2.540, aligned using an online alignment program MAFFT version 7 (https://mafft.cbrc.jp/alignment/server/41) and edited manually as necessary using the alignment program BioEdit v7.2.540. Phylogenetic tree estimation for each alignment was performed using Maximum Parsimony (MP), Maximum Likelihood (ML), and Bayesian Inference (BI). MP analyses were conducted using PAUP v. 4.0b1042, inferring trees with the heuristic search option with Tree-Bisection Reconnection branch swapping and 1000 random sequence additions. ML and bootstrapping analyses were conducted using RAxML-HPC v.8 on XSEDE43 in the CIPRES Science Gateway (http://www.phylo.org/portal2/44). The ML analysis was performed using the GTRGAMMA model with 1000 bootstrap iterations. The phylogenetic analyses were performed for Bayesian inference using MrBayes v. 3.2.7a45. Markov chain Monte Carlo sampling in MrBayes v.3.2.7a45 was used to determine the posterior probabilities. Four simultaneous Markov chains were run for 5,000,000 generations and trees were sampled every 1000th generation. Support values (ML bootstrap, ML-BS; MP bootstrap, MP-BS; BI posterior probability, BPP) were calculated for all analyses and the tree length (TL), consistency index (CI), retention index (RI), and the rescaled consistency index (RC) for the MP analysis generated under different optimality criteria. Phylogenetic trees were viewed and arranged using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Pathogenicity tests
The fungal isolates were tested for pathogenicity in the original hosts of the pathogens based on the protocol of Phoulivong et al.46. The cross-inoculation experiment was conducted on host and other plants, consisting of mangosteen, avocado, rambutan, snake fruit, plum, strawberry, rose apple, and jackfruit. The selected fruits and leaves were cleaned with tap water and sterilized by dipping them into 70% ethanol for 3 min. The fruit was rinsed three times in sterile water and dried with sterile tissue paper. Each fruit was wounded with a sterile needle. Mycelial agar plugs from 7-day cultures were placed onto the surfaces of the leaf and fruit samples. Each of the treated samples was placed in a separate plastic box that served as a moist chamber and incubated at 25 ± 2 °C. All inoculated fruits and leaves were visually assessed daily and any symptoms were recorded. The fungi were re-isolated, and species comparison was performed to confirm the causal pathogen.
Preparation of plant extracts
Fresh plants of turmeric (Curcuma longa L. (family Zingiberaceae)), ginger (Zingiber officinale L. (Zingiberaceae)), lemongrass (Cymbopogon citratus L. (Poaceae)), mangosteen (Garcinia mangostana L. (Clusiaceae)), roselle (Hibiscus sabdariffa L. (Malvaceae)), and clove (Syzygium aromaticum L. (Myrtaceae)) were purchased from the Talad Thai market, Khlong Luang district, Pathum Thani province, Thailand. Plants were washed thoroughly under running tap water and soaked in a 2% solution of sodium hypochlorite for 20 min, then rinsed thoroughly with sterilized water, and air-dried at 25 ± 2 °C for 4 h. The dried plant material was milled. To prepare crude extracts, about 500 g of finely ground plant material were submerged in 1000 ml ethanol solvent overnight at 25 ± 2 °C and passed through Whatman filter paper No. 1. The organic phase was evaporated in a rotary vacuum evaporator (BUCHI, Operation Manual Roatavapor® R-210/215, Vacuum Pump V-700/710, Switzerland) under a pressure of 45 mbar at 40 °C, followed by storage at 4 °C until use.
Determination of mycelial inhibition
The modified method of Cho et al.47 was used to determine the effect of the crude extracts on fungal growth. The center of Petri plates was inoculated with a mycelial plug obtained from a colony edge of each cultured fungi at age 7 days. The control was set up using blank agar plug plates (PDA mixed with distilled water). The fungicides (azoxystrobin + tebucolnazole, prochloraz, or captan) were prepared at recommended concentrations of 500, 1000, or 1500 mg/l, respectively. Five replicates of PDA-crude extract per isolate were incubated at 25 ± 2 °C, and radial growth was measured at 7 and 14 days. Inhibition levels (%) were calculated as (X–Y/X) × 100, where X and Y are the diameter of a fungal colony grown in a negative control plate and the diameter of a fungal colony grown in a plate containing a crude plant extract, respectively.
Statistical analysis
The effect of different concentrations of crude extracts on the growth of fungi on PDA media was evaluated based on one-way analysis of variance (ANOVA), and significant differences were determined using Duncan’s multiple range test (P < 0.05). Conidial sizes were evaluated based on ANOVA and Wilcoxon signed-rank tests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors. Plant materials in this study complies with relevant institutional, national, and international guidelines, and legislation.
Results
Morphological and molecular identification
In total, 44 fungi were isolated from eight diseased plant samples, as shown in Fig. 1 and were classified into pestalotioid taxa based on morphological characteristics. Of the isolates, 2, 5, 15, 1, 6, 9, 1, and 5 were derived from jackfruit, rose apple, mangosteen, plum, snake fruit, rambutan, strawberry, and avocado, respectively (Table 1). All isolates were cultivated on PDA and maintained as axenic cultures using the specimen codes PP001–PP044. The most representative characteristic features of colony morphology and the acervuli and conidia were similar in all species. The characteristic lightly concolorous, darkly concolorous, or versicolorous appearance of conidia is considered a key character for identifying fungal species. Of the 44 isolates, 7 distinct morphological characteristics were observed (PP026, PP027, PP035, PP037, PP038, PP039, and PP044), as shown in Fig. 2.
These seven isolates showed different colony patterns on PDA plates, and many had an undulate edge. PP037 produced yellowish aerial mycelia while others displayed whitish mycelia. The colony diameters of PP026, PP027, P035, PP037, PP038, PP039, and PP044 after incubating at 28 °C for 7 days were 76, 81, 83, 83, 70, 65, and 90 mm, respectively. All isolates produced black fruiting bodies or acervuli. Conidia were fusiform with 2–3 apical appendages and 1 basal appendage. Conidial lengths of all isolates varied between about 18 to almost 30 µm (Fig. 3). Conidial widths were about 4.5–7.3 µm. Cells of the conidia and appendages of each isolate were designated as apical appendages, apical cell, three median cells, basal cell, and basal appendage and measured for length accordingly. The median cell of all isolates was approximately similar with lengths of 11.7–18.9 µm. Apical and basal cells were around 2.5–7 µm. The apical appendages of PP035 were 7.7–19.8 µm while the other six isolates were extended to 35.4 µm. Basal appendages ranged from 2 to 11.5 µm.
Conidial sizes of Neopestalotiopsis (PP026, PP027, PP037, PP038, PP039, and PP044) and Pseudopestalotiopsis (PP035). The upper-left corner represents conidial cells that are measured. Box plots are from the measurements of 20 conidia per isolate. Brackets with asterisks denote pairs differing at P < 0.05.
Seven distinct characteristics were considered for molecular analysis using multilocus sequences and six isolates were identified as Neopestalotiopsis spp. and one was Pseudopestalotiopsis sp. For phylogenetic analyses, the alignment for Pseudopestalotiopsis and Neopestalotiopsis consisted of 65 taxa, with Discosia artocreas (CBS124848) as the outgroup taxon. The alignment comprised 1622 characters of which 345 were variable, 752 were constant, and 525 were parsimony-informative characters, including gaps (ITS: 1–430, tef1: 431–892, and β-tubulin: 893–1622). The parsimony analysis resulted in three equally parsimonious trees and the first tree (TL = 2114, CI = 0.597, HI = 0.403, RI = 0.823, and RC = 0.492) was selected, as shown in Fig. 4. Phylogenetic analyses with MP, ML, and BI using the dataset of individual loci were also performed, but no topological phylogenetic conflict was observed (data not shown). Multilocus phylogenetic analyses with our fungal isolates and those for representative Neopestalotiopsis, Pseudopestalotiopsis species retrieved from the NCBI GenBank database were separated into two clades, with an ML bootstrap value of 93%. Six of the isolates (PP026, PP027, PP037, PP038, PP039, and PP044) were clustered with Neopestalotiopsis, while PP035 was sister to Pseudopestalotiopsis isolates (MP-BS = 97%, ML-BS = 96%, and BPP = 1). In this study, the phylogenetic analyses revealed that our representative isolates were supported by moderate to high bootstrap values and were consistent with the morphology-based classification. Based on the morphological and molecular evidence, our results indicated that these seven isolates were candidates for two species, namely Neopestalotiopsis spp. (PP026, PP027, PP037, PP038, PP039, and PP044) and Pseudopestalotiopsis sp. and (PP035).
MP tree based on analysis of combined ITS, β-tubulin, and tef1 dataset. Discosia artocreas (CBS124848) was used as the outgroup taxon, and new isolates are in bold. Bootstrap support values for Maximum Parsimony (MP-BS) and Maximum Likelihood (ML-BS) smaller than 70% (left and middle) are not shown and Bayesian posterior probabilities (BPP) equal to, or higher than 0.95 PP (right) are indicated at the nodes (MP-BS/ML-BS/BPP).
Pathogenicity and cross-inoculation
Since Neopestalotiopsis and Pseudopestalotiopsis are known to have a wide host range, we investigated whether the isolates in this study could cross infect a range of fruit species. Six Neopestalotiopsis (PP026, PP027, PP037, PP038, PP039, and PP044) and one Pseudopestalotiopsis (PP035) were tested to evaluate the effect of cross-inoculation on eight plant species including their original hosts. Light brown lesions surrounded by a dark brown border, which resembled symptoms that occurred in the field, were observed on the leaves and fruits at 3–7 days after inoculation, while these symptoms did not occur in the control inoculated with agar media. The development of a necrotic zone or a lesion of at least 2.0 mm beyond the inoculation site was considered to indicate a positive status of the plant hosts. The results revealed that Neopestalotiopsis PP026 had a wide host range since it caused symptoms on seven fruit species except for avocado, while PP039 and PP044 infected only two fruit species and neither of them was the original host (Table 2). PP027 did not produce symptoms on the original host. Strawberry was highly susceptible to six pestalotioid isolates, except for PP027 (Fig. 5a). Plum ranked second for susceptibility, since it showed symptoms of inoculation with five pestalotioid isolates (Fig. 5b). Conversely, snake fruit displayed inoculation symptoms only with Pseudopestalotiopsis and not with Neopestalotiopsis.
Effect of crude plant extracts and fungicides on mycelial inhibition
Six crude plant extracts from clove, ginger, lemongrass, mangosteen, roselle, and turmeric were studied for their ability to inhibit the growth of Neopestalotiopsis and Pseudopestalotiopsis and compared with fungicide treatments for controls. The results showed that at the highest concentrations (10,000 mg/l), the clove and turmeric crude extracts completely inhibited the growth of all isolates after incubation for 14 days, followed by the effects of ginger, lemongrass, and roselle extracts, whereas the crude extract of mangosteen exhibited comparatively very low activity against all isolates (Fig. 6). Although clove extracts at 1000 mg/l displayed some inhibition, the other crude plant extracts with lower concentrations did not exhibit antifungal activity against the Pseudopestalotiopsis or Neopestalotiopsis species. To compare the effectiveness of crude plant extracts with chemical treatments, three fungicides (azoxystrobin + tebucolnazole, captan, or prochloraz) were tested at the recommended dose. Azoxystrobin + tebucolnazole and prochloraz both showed 100% inhibition of the mycelial growth of all Neopestalotiopsis and Pseudopestalotiopsis isolates, while captan exhibited less effectiveness.
Effect of six crude plant extracts (clove, ginger, lemongrass, mangosteen, roselle, and turmeric) at concentrations of 10, 100, 1,000, and 10,000 mg/L on mycelial growth inhibition of Neopestalotiopsis (PP026, PP027, PP037, PP038, PP039, and PP044) and Pseudopestalotiopsis (PP035) compared to inhibition of azoxystrobin + tebucolnazole, captan, or prochloraz fungicides. Same lowercase letters represent no significant difference (P < 0.05).
Discussion
The genus Pestalotiopsis was previously separated from the genus Pestalotia and classified in the Ascomycota, Coleomycetes, as proposed by Barr48. In the present study, a new re-classification of Pestalotiopsis was performed including two other genera. The genus Neopestalotiopsis spp. is characterized by versicolorous median cells, Pestalotiopsis spp. is characterized by lightly pigmented concolorous median cells, and Pseudopestalotiopsis spp. are characterized by darkly concolorous median cells1.
Neopestalotiopsis and Pseudopestalotiopsis are not highly host-specific, and these taxa may have the ability to infect a range of hosts49, 50. In this study, all tested isolates could infect the surface tissue of many plants based on the wounding inoculation method, which avoids any interference caused by host penetration resistance. Typical symptoms were observed including the browning of foliage, leaf blight, fruit rot, and the presence of black acervuli on diseased tissue. However, some fungal species failed to infect other inoculated hosts that might have been caused by environmental conditions, especially the temperature of the room used to incubate the inoculated fruits, which might have been too high for these genera exhibiting an optimum temperature of 22–28 °C 51 and optimal relative humidity ranging from 65 to 100%49.
Examination of the efficacy of the six plant extracts against Neopestalotiopsis and Pseudopestalotiopsis species showed that crude extracts from clove and turmeric had the highest antifungal efficacy against all the tested isolates. The antimicrobial properties of clove and turmeric have been well established, especially against bacterial and fungal pathogens. These two species produce active agents such as curcumin, eugenol, and eugenyl acetate, which are effective against bacteria and phytopathogenic fungi28, 29. Roselle, lemongrass, and ginger extracts are also reported to possess antibacterial and antifungal properties against pathogens26, 27, 30, while mangosteen are not effective. Although xanthones in mangosteen affect Alternaria solani, Aspergillus flavus, and Fusarium spp.52, mangosteen extracts display comparatively very low activity against Colletotrichum gloeosporioides53.
Crude plant extracts are comparable to fungicides to control Neopestalotiopsis and Pseudopestalotiopsis. Clove and turmeric extracts on the inhibition of mycelial growth of pestalotioid fungi are equivalent to the efficiency of azoxystrobin + tebucolnazole or prochloraz, which had 100% inhibition. Similarly, azoxystrobin and prochloraz can inhibit P. microspora, which causes leaf spot disease of Photinia fraseri in China54. The present study showed that captan inhibited 20–80% of the growth of pestalotioid fungi. Similarly, Esiegbuya et al.55 reported the effect of captan at 250 and 500 ppm on the in vitro mycelial growth of P. clavispora; however, the fungicide did not completely inhibit the pathogen.
Therefore, it was assumed that the antifungal properties of these plant extracts may be principally due to the presence of unknown compounds that could not be analyzed and identified in the present study. Hence, further study should investigate the identification of the active compounds of plant extracts that control all pathogens, and the tested plants may be used for developing new fungicides that are safe for consumers and farmers and are effective in the field.
Data availability
Sequence data that support the findings of this study have been deposited in GenBank with the accession numbers: MT628375–MT628381, MW776648–MW776654, and MW776633–MW776638.
References
Maharachchikumbura, S. S. N., Hyde, K. D., Groenewald, J. Z., Xu, J. & Crous, P. W. Pestalotiopsis revisited. Stud. Mycol. 79, 121–186. https://doi.org/10.1016/j.simyco.2014.09.005 (2014).
Nozawa, S., Ando, K., Phay, N. & Watanabe, K. Pseudopestalotiopsis dawaina sp. nov. and Ps. kawthaungina sp. nov.: Two new species from Myanmar. Mycol. Prog. 17, 865–870. https://doi.org/10.1007/s11557-018-1398-1 (2018).
Kang, J. C., Hyde, K. D. & Kong, R. Y. Studies on amphisphaeriales: The amphisphaeriaceae (sensu stricto). Mycol. Res. 103, 53–64. https://doi.org/10.1017/S0953756298006650 (1999).
Amrutha, P. & Vijayaraghavan, R. Evaluation of fungicides and biocontrol agents against Neopestalotiopsis clavispora causing leaf blight of strawberry (Fragaria × ananassa Duch). Int. J. Curr. Microbiol. App. Sci. 7, 622–628. https://doi.org/10.20546/ijcmas.2018.708.067 (2018).
Guo, J. W. et al. First report of pseudostem black spot caused by Pestalotiopsis microspora on Tsao-ko in Yunnan, China. Plant Dis. 100, 1021. https://doi.org/10.1094/PDIS-08-15-0920-PDN (2016).
Ismail, S. I., Zulperi, D., Norddin, S. & Ahmad-Hamdani, S. First report of Neopestalotiopsis saprophytica causing leaf spot of oil palm (Elaeis guineensis) in Malaysia. Plant Dis. 101, 1821. https://doi.org/10.1094/PDIS-02-17-0271-PDN (2017).
Li, L., Pan, H., Chen, M. Y. & Zhong, C. H. First report of Pestalotiopsis microspora causing postharvest rot of kiwifruit in Hubei province, China. Plant Dis. 100, 2161. https://doi.org/10.1094/PDIS-01-16-0059-PDN (2016).
Lu, L. M. et al. Identification of Pestalotiopsis clavispora causing brown leaf spot on Chinese bayberry in China. Can. J. Plant Pathol. 37, 397–402. https://doi.org/10.1080/07060661.2015.1065001 (2015).
Maharachchikumbura, S. S. N. et al. A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Divers. 56, 95–129. https://doi.org/10.1007/s13225-012-0198-1 (2012).
Palou, L., Montesinos-Herrero, C., Guardado, A. & Taberner, V. First report of Pestalotiopsis clavispora causing postharvest fruit rot of loquat in Spain. J. Plant Pathol. 95, S4.69-S4.77. https://doi.org/10.4454/JPP.V95I4.005 (2013).
Shen, H. F. et al. First report of Pestalotiopsis microspora causing leaf spot of oil palm (Elaeis guineensis) in China. Plant Dis. 98, 1429. https://doi.org/10.1094/PDIS-02-14-0163-PDN (2014).
Shi, H. J. et al. First report of Pestalotiopsis microspora as a causal agent of black spot of pecan (Carya illinoinensis) in China. Plant Dis. 99, 1276. https://doi.org/10.1094/PDIS-01-15-0079-PDN (2015).
Silva, A. C. et al. Pestalotiopsis pini sp. nov., an emerging pathogen on stone pine (Pinus pinea L.). Forests 11, 805. https://doi.org/10.3390/f11080805 (2020).
Gerardo-Lugo, S. S. et al. Characterization of Neopestalotiopsis species associated with mango grey leaf spot disease in Sinaloa, Mexico. Pathogens 9, 788. https://doi.org/10.3390/pathogens9100788 (2020).
Valencia, A. L., Torres, R. & Latorre, B. A. First report of Pestalotiopsis clavispora and Pestalotiopsis spp. causing postharvest stem end rot of avocado in Chile. Plant Dis. 95, 492–492. https://doi.org/10.1094/PDIS-11-10-0844 (2011).
Feng, Y. R., Liu, B. S. & Sun, B. B. First report of leaf blotch caused by Pestalotiopsis clavispora on Rosa chinensis in China. Plant Dis. 98, 1009. https://doi.org/10.1094/PDIS-01-14-0036-PDN (2014).
Yasuda, F., Kobayashi, T., Watanabe, H. & Izawa, H. Addition of Pestalotiopsis spp. to leaf spot pathogens of Japanese persimmon. J. Gen. Plant Pathol. 69, 29–32. https://doi.org/10.1007/s10327-002-0011-1 (2003).
Maharachchikumbura, S. S. N., Guo, L. D., Chukeatirote, E., McKenzie, E. H. C. & Hyde, K. D. A destructive new disease of Syzygium samarangense in Thailand caused by the new species Pestalotiopsis samarangensis. Trop. Plant Pathol. 38, 227–235. https://doi.org/10.1590/S1982-56762013005000002 (2013).
Norphanphoun, C. et al. Morphological and phylogenetic characterization of novel pestalotioid species associated with mangroves in Thailand. Mycosphere 10, 531–578. https://doi.org/10.5943/mycosphere/10/1/9 (2019).
Suwannarach, N., Kumla, J., Bussaban, B. & Lumyong, S. New report of leaf blight disease on eucalyptus (Eucalyptus camaldulensis) caused by Pestalotiopsis virgatula in Thailand. Can. J. Plant Pathol. 34, 306–309. https://doi.org/10.1080/07060661.2012.680501 (2012).
Plant Protection Research and Development Office, Department of Agriculture (DOA). Host Index of Plant Diseases in Thailand. 266 (Bangkok, 2016).
Baggio, J. S. et al. Outbreak of leaf spot and fruit rot in Florida strawberry caused by Neopestalotiopsis spp. Plant Dis. 105, 305–315. https://doi.org/10.1094/PDIS-06-20-1290-RE (2021).
Nozawa, S. et al. Infection sources of Pestalotiopsis sensu lato related to loquat fruit rot in Nagasaki Prefecture, Japan. J. Gen. Plant Pathol. 86, 173–179. https://doi.org/10.1007/s10327-020-00908-4 (2020).
Neela, F. A., Sonia, I. A. & Shamsi, S. Antifungal activity of selected medicinal plant extract on Fusarium oxysporum Schlecht: The causal agent of fusarium wilt disease in tomato. Am. J. Plant Sci. 5, 2665–2671. https://doi.org/10.4236/ajps.2014.518281 (2014).
Pretorius, J. C., Magama, S., Zietsman, P. C. & van Wyk, B. E. Growth inhibition of plant pathogenic bacteria and fungi by extracts from selected South African plant species. S. Afr. J. Bot. 69, 186–192. https://doi.org/10.1016/S0254-6299(15)30344-6 (2003).
Agarwal, M., Walia, S., Dhingra, S. & Khambay, B. P. S. Insect growth inhibition, antifeedant and antifungal activity of compounds isolated/derived from Zingiber officinale Roscoe (ginger) rhizomes. Pest Manag. Sci. 57, 289–300. https://doi.org/10.1002/ps.263 (2001).
Barman, H., Roy, A. & Das, S. K. Evaluation of plant products and antagonistic microbes against grey blight (Pestalotiopsis theae), a devastating pathogen of tea. Afr. J. Microbiol. Res. 9, 1263–1267. https://doi.org/10.5897/AJMR2015.7391 (2015).
Duduk, N. et al. Antifungal activity of three essential oils against Colletotrichum acutatum, the causal agent of strawberry anthracnose. J. Essent. Oil-Bear. Plants 18, 529–537. https://doi.org/10.1080/0972060X.2015.1004120 (2015).
Kim, M. K., Choi, G. J. & Lee, H. S. Fungicidal property of Curcuma longa L. rhizome-derived curcumin against phytopathogenic fungi in a greenhouse. J. Agric. Food Chem. 51, 1578–1581. https://doi.org/10.1021/jf0210369 (2003).
Saha, D., Dasgupta, S. & Saha, A. Antifungal activity of some plant extracts against fungal pathogens of tea (Camellia sinensis). Pharm. Biol. 43, 87–91. https://doi.org/10.1080/13880200590903426 (2005).
Agrios, G. N. Plant Pathology 5th edn. (Academic Press, 2005).
Chuaseeharonnachai, C. et al. Synnematotriadelphia gen. nov. (S. stilboidea comb. nov. and S. synnematofera comb. nov.) and Triadelphia hexaformispora sp. nov. in the family Triadelphiaceae. Mycol. Prog. 19, 127–137. https://doi.org/10.1007/s11557-019-01547-6 (2020).
Liu, F., Hou, L., Raza, M. & Cai, L. Pestalotiopsis and allied genera from Camellia, with description of 11 new species from China. Sci. Rep. 7, 866. https://doi.org/10.1038/s41598-017-00972-5 (2017).
White, T. J., Bruns, T., Lee, S. & Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols Guide Methods Appl. https://doi.org/10.1016/b978-0-12-372180-8.50042-1 (1990).
Glass, N. L. & Donaldson, G. C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61, 1323–1330 (1995).
Rehner, S. A. Primers for Elongation Factor 1-alpha (EF1-alpha) (accessed 02 Nov 2021); http://ocid.nacse.org/research/deephyphae/EF1primer.pdf (2001).
Jayawardena, R. S. et al. One stop shop III: Taxonomic update with molecular phylogeny for important phytopathogenic genera: 51–75 (2019). Fungal Divers. 98, 77–160. https://doi.org/10.1007/s13225-019-00433-6 (2019).
Maharachchikumbura, S. S. N., Guo, L. D., Liu, Z. Y. & Hyde, K. D. Pseudopestalotiopsis ignota and Ps camelliae spp. Nov. associated with grey blight disease of tea in China. Mycol. Prog. 15, 22. https://doi.org/10.1007/s11557-016-1162-3 (2016).
Ran, S. F. et al. Two new records in Pestalotiopsidaceae associated with Orchidaceae disease in Guangxi Province, China. Mycosphere 8, 121–130. https://doi.org/10.5943/mycosphere/8/1/11 (2017).
Hall, T. BioEdit: Biological Sequence Alignment Editor for Win95; Ibis Biosciences: Carlsbad, CA, USA, 2007.
Katoh, K., Rozewicki, J. & Yamada, K. D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. https://doi.org/10.1093/bib/bbx108 (2019).
Swofford, D. L. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer Associates, Sunderland (2002).
Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics https://doi.org/10.1093/bioinformatics/btu033 (2014).
Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop (GCE). IEEE, New Orleans, LA, 1–8. https://doi.org/10.1109/GCE.2010.5676129 (2010).
Ronquist, F., Teslenko, M. & Vander, M. P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. https://doi.org/10.1093/sysbio/sys029 (2012).
Phoulivong, S., McKenzie, E. H. C. & Hyde, K. D. Cross infection of Colletotrichum species; a case study with tropical fruits. Curr. Res. Environ. Appl. Mycol. 2, 99–111. https://doi.org/10.5943/cream/2/2/2 (2012).
Cho, S. M., Gu, Y. S. & Kim, S. B. Extracting optimization and physical properties of yellowfin tuna (Thunnus albacares) skin gelatin compared to mammalian gelatins. Food Hydrocoll. 19, 221–229. https://doi.org/10.1016/j.foodhyd.2004.05.005 (2005).
Barr, M. E. Pestalosphaeria, a new genus in the Amphisphaeriaceae. Mycologia 67, 187–194 (1975).
El-Gali, Z. I. Effect of some ecological factors on growth of Pestalotiopsis spp. isolated from mastic shrubs leaves. J. Adv. Bot. Zool. 5, 1–5 (2017).
Hopkins, K. E. & McQuilken, M. P. Characteristics of Pestalotiopsis associated with hardy ornamental plants in the UK. Eur. J. Plant Pathol. 106, 77–85 (2000).
Keith, L. M., Velasquez, M. E. & Zee, F. T. Identification and characterization of Pestalotiopsis spp. causing scab disease of guava, Psidium guajava, Hawaii. Plant Dis. 90, 16–23. https://doi.org/10.1094/PD-90-0016 (2006).
Ovalle-Magallanes, B., Eugenio-Pérez, D. & Pedraza-Chaverri, J. Medicinal properties of mangosteen (Garcinia mangostana L.): A comprehensive update. Food Chem. Toxicol. 109, 102–122. https://doi.org/10.1016/j.fct.2017.08.021 (2017).
Prasothong, N., Plainsiricha, M., Bussaman, P., Luckantinvong, V. & Wongsawas, M. Effect of mangosteen (Garcinia mangostana L.) peel extract on anthracnose disease (Colletotrichum gloeosporioides Penz.) of mango fruit cv. Nam Dok Mai. in The 7th National Agricultural System Conference, Faculty of Technology, Mahasarakham University (2011).
Bin, G., Xingping, L., Chao, X. & Hongyan, Z. Chemical control against Pestalotiopsis microspora in the leaves of Photinia fraseri. J. Northw. For. Univ. 28, 131–135 (2013).
Esiegbuya, O. D. et al. In-vitro evaluation of some selected fungicides against Pestalotiopsis clavispora and Pseudocochliobolus eragrotidis isolated from Vitellaria paradoxa seedlings. J. Agric. Vet. Sci. 7, 80–85 (2014).
Acknowledgements
The authors thank Dr. Jamrearn Buaruang from Ramkhamhaeng University for his support in the preparation of crude plant extract. Dr. Jennifer Luangsa-ard, Dr. Theerayut Toojinda, and Dr. Wonnop Visessanguan provided support with fungal molecular work at BIOTEC. The authors also thank the Kasetsart University Research and Development Institute for the writing assistance.
Author information
Authors and Affiliations
Contributions
The first author (A.D.) carried out all the experiments as part of her MS thesis. O.P. and N.B. were supervisor and co-supervisor, respectively, for this thesis. W.L. and S.N. helped A.D. during the thesis with molecular work and in writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Darapanit, A., Boonyuen, N., Leesutthiphonchai, W. et al. Identification, pathogenicity and effects of plant extracts on Neopestalotiopsis and Pseudopestalotiopsis causing fruit diseases. Sci Rep 11, 22606 (2021). https://doi.org/10.1038/s41598-021-02113-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-02113-5
This article is cited by
-
Antifungal activity of essential oil-encapsulated lipid nanoemulsions against Neopestalotiopsis rosae causing leaf spot on strawberry
Journal of Plant Diseases and Protection (2023)
-
Characterization and identification of fungicide insensitive Pestalotiopsis-like species pathogenic to tea crop in India
World Journal of Microbiology and Biotechnology (2023)
-
Neopestalotiopsis clavispora: a causal agent of twig dieback on highbush blueberries in Serbia
Journal of Plant Diseases and Protection (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.