Abstract
Chilli (Capsicum annuum L.) is one of the most significant vegetable and spice crop. Wilt caused by Fusarium Sp. has emerged as a serious problem in chilli production. Internal transcribed spacer (ITS) region is widely used as a DNA barcoding marker to characterize the diversity and composition of Fusarium communities. ITS regions are heavily used in both molecular methods and ecological studies of fungi, because of its high degree of interspecific variability, conserved primer sites and multiple copy nature in the genome. In the present study we focused on morphological and molecular characterization of pathogen causing chilli wilt. Chilli plants were collected from four districts of Kashmir valley of Himalayan region. Pathogens were isolated from infected root and stem of the plants. Isolated pathogens were subjected to DNA extraction and PCR amplification. The amplified product was sequenced and three different wilt causing fungal isolates were obtained which are reported in the current investigation. In addition to Fusarium oxysporum and Fusarium solani, a new fungal species was found in association with the chilli wilt in Kashmir valley viz., Fusarium equiseti that has never been reported before from this region. The studies were confirmed by pathogenicity test and re-confirmation by DNA barcoding.
Similar content being viewed by others
Introduction
Chilli (Capsicum annuum L.) is considered as the most important vegetable and spice crop throughout the world which belongs to Genus Capsicum, and family “Solanaceae”1. Chillies have gained their commercial importance in the horticulture industry because of their color and pungency. These are rich source of proteins and vitamins mainly Vitamin A, C and E. Chillies also possess important health benefits due to their antioxidant and anti-inflammatory properties because of the presence of various bioactive molecules like volatile oils, fatty acids, carotenoids and capcaicinoids2,3,4. Chilli is cultivated throughout the world over an area of 1832 thousand hectares producing 2959 thousand tons5. The prime chilli producing countries are China, India, Pakistan, Korea, Indonesia, Turkey and Sri Lanka in Asia; Egypt, Tunisia, Ghana and Nigeria in Africa; Mexico, United States of America in North—Central America; Spain, Yugoslavia, Romania, Italy, Bulgaria and Hungary in Europe and Peru and Argentina in South America6. Among all chilli grown countries, India is the largest chilli producing country followed by China and Pakistan7. The largest producer of Chilli in the world is India with 1400 thousand tonnes of production8, followed by China with 450 thousand tonnes and Mexico with 400 thousand tonnes9,10.
However, the chilli production has been suffering to a great extend because of many bacterial, viral and fungal diseases. More than 40 diseases of chillies are known to be caused by fungi11. One of the important diseases is Chilli wilt to which major loss can be attributed. The common species causing chilli wilt are Phytopthora, verticulum wilt, Rizoctonia root rot and Fusarium wilt12,13. As per the reports Fusarium oxysporum and Fusarium solani are considered the most common fungal species are found associated with Chilli wilt in India14,15 along with Fusarium moniliforme and Fusarium palidoroseum which are reported to be found in some parts of the country16. Total yield loss due to Fusarium wilt has been estimated to be from 10 to 80% in total chilli production on a global level on the basis of selected varieties and climatic conditions17. The disease caused by Fusarium oxysporum and Fusarium solani is a serious complication that reduces growth, fruit yield and quality threatening chilli production18. The common symptoms of fusarium wilt are discoloration and inward rolling of the leaves followed by the wilting of the whole plant which is caused due to the damage in vascular system thus hampering nutrient and water uptake19,20,21. Fusarium equiseti is considered to be a weak pathogen on cereals and is occasionally found to be associated with Fusarium head blight infected kernels22.This species is commonly found in tropical and sub-tropical areas23. The species is a pathogen to varied range of crops and it has been recently reported to be a causal organism of wilt in Capsicum chinense in Mexico24. The pathogenicity of this species is has underestimated though. The species belongs to the F.incarnatum-F. quiseti complex and it is genetically diverse. However, various investigations have been carried out on chilli wilt causing pathogens and several management strategies has been designed for controlling the chilli wilt.
Fusarium species have several morphological characteristics that help these for distinct identification and one of the prominent features is development of various shapes and sizes of macro and micro conidia which are asexual spores. Other structures that they form are called chlamydospores spores25 which ensure the survival of the pathogen in soil and in plant for many years thus making the management and control of the diseases caused by Fusarium species very difficult to cater26,27. These are also identified on the basis of growth rate on agar media and the pigmentations produced by them28. Moreover, the morphological identification can be quite difficult among the Fusarium species29. Identification of the species is done through macro and microscopic analysis but the most reliable form of identification is through the information gathered via nucleotide sequencing from conserved gene regions which include Internal Transcribed Spacer (ITS)30. The sequence information using ITS regions has been immensely used in phylogeny and taxonomy of Fusarium species31 as ITS regions have been known to successfully aid in identification between the species32. ITS is differentiated into two regions ITS1 and ITS2 (genes 18S to 5.8S and 5.8S to 28S respectively)33. There are more than 172,000 fungal ITS sequences present in Genbank34.
A deep comprehension of the populations of pathogens is important as they show variations in pathogenicity, response to management systems, environment and host differences35. Thus population biology of the pathogen needs to be studied with depth. Since, the incidence of wilt is very prevalent in Kashmir so it is the need of an hour to do a detailed investigation of pathogen causing chilli wilt. It has not been yet cleared that among the population of Fusarium species how many are responsible for causing chilli wilt in Kashmir region. In this regard major chilli growing hotspots, pathogens association and prevalence of major pathogen in different areas of Kashmir Himalayas were investigated and morphological and molecular studies were carried out which, for the first time revealed Fusarium equiseti to be one of the causal organisms for chilli wilt in Kashmir along with Fusarium oxysporum, Fusarium Solani.
Materials and methods
Collection, Purification and Maintenance of culture
Survey of vegetable growing areas in four districts of Kashmir valley viz., Srinagar, Budgam, Pulwama and Anantnag was carried out to assess the prevalence of the disease (Table 1).The fungi from the infected samples were isolated using tissue bit technique36. The isolated pathogens were purified using hyphal tip and / or single spore technique37. Pure cultures thus obtained were maintained by repeated sub- culturing at intervals of 45 days for further studies and the sterile cultures were stored at 4 °C in a refrigerator.
Identification and Pathogenicity test
Identification of the pathogen was confirmed on the basis of morphological and the pathological characteristics. Identification was confirmed from division of Plant Pathology, SKUAST-K and also confirmed through DNA barcoding. The pathogen inoculum was multiplied on sand meal agar medium. The medium was prepared by autoclaving 90 g dry sieved sand and 10 g maize meal with 40 ml of distilled water at 1.05 kg cm-2 pressure for half an hour for three consecutive days. The sterilized medium was then inoculated with respective fungi and incubated for three weeks at 25 ± 1 °C with daily shaking of flasks to get uniform growth. The inoculums thus prepared were added to the sterilized sand soil (2:1) potting mixture @ 10 per cent (w/w) by mixing it with upper layer of soil and allowed for 7 days to infest soil38 (Raj and Singh 1973; Najar 2001). Forty five days old chilli seedlings (cv. ‘Shalimar Long’), raised by growing surface sterilized seeds in sterilized sand soil (2:1) potting mixture were gently uprooted and transplanted in the sterilized potting mixture incorporated in sterilized soil served as check. The study on symptom development of chilli wilt disease was undertaken on artificially inoculated potted chilli plants (cv. ‘Shalimar Long’). The chilli seedlings transplanted in infested soil, inoculated with the test pathogen were continuously monitored for symptom development till the plants were completely dead.
Morphological and cultural characteristics of the isolated pathogen(s)
The morphological characteristics of the causal pathogen(s) were studied in-vivo after culturing on artificial medium to identify the associated pathogen(s). The pathogen cultures were grown on potato dextrose agar (PDA) medium and the semi- permanent slides prepared from 10 days old colonies. The important characters studied were:
-
Hyphae, width, septation and colour
-
Conidia, shape, size and colour
-
Conidiophore, shape, size and colour
-
Chlamydospore, shape, size and colour.
DNA Extraction and PCR Amplification
DNA extraction was carried out using 400 microlitre extraction buffer. This buffer is same as that described by Cenis et al.39. The genomic DNA of fungal isolates was loaded on 0.7% agarose in 1XTAE for 30 min. PCR amplification was carried out using ITS1 andITS2 primer pair for the amplification of all the three isolates. The primers were designed manually using, Oilgocalc, Clustalw software the primer pairs used for amplification were ITS1F Primer (5′CCTGCGGAGGATCATTA 3′), ITS2R Primer (5′TCCTCCGCTTATTGAT3′). PCR amplification for ITS1 5.8S-ITS2 region was carried out in a reaction volume of 25 µl in 0.2 ml PCR tubes. The amplification reaction was carried out in thermo cycler (Applied Biosystems) for 1- 2 h for amplification of ITS1 -ITS2 region followed by gel electrophoresis procedure and compared with 100 bp DNA ladder, respectively (Fig. 1).
Sequencing, Nucleotide Alignment and Phylogenetic Analysis
Amplified PCR products were sequenced at Agri Genome Labs (Infopark Road, Kakkanad, Kerala, India). The same primers utilized for the PCR amplification were used for sequencing as well. PCR products (sequences) were assembled using the DNA Baser V.4 program to produce complete contigs. These were further aligned using the CLUSTAL W method (Bio-Edit). A search of homologous sequences was performed by BLAST analysis at NCBI (http://ncbi.nlm.nih.gov/BLAST). The MEGA7 (Molecular Evolutionary Genomics Analysis Version 7)40 constructed dendrograms from the 10 isolates from the current study and reference strain sequences from GenBank. There were 100 replications for every bootstrap value. For validation of results, an out group fungal pathogen Alternaria was selected.
Results
Diseased plant samples were collected from four districts of Kashmir region of India viz., Srinagar, Budgam, Pulwama and Anantnag on the basis of symptomatology, during 2018–2019. The characteristic symptoms discovered were lesions on the root/stem, brown tube-shaped structure discoloration, yellowing, weakening and death of the plants.
Morphological characterization and identification of the Fusarium solani, Fusarium oxysporum and Fusarium equiseti
Morphological characteristics of the causal pathogens were studied both on host as well as on artificial culture medium to identify the associated pathogen which were studied on PDA medium.
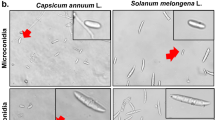
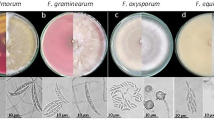
The pure culture of the Fusarium solani initially produced white colonies (Fig. 2a) which gradually turned to light orange coloured at agar base (Fig. 2b) and attained a growth of 90 mm in 10 days of incubation at 25 ± 1 °C. Microscopic observations revealed that mycelium was smooth, branched, cylindrical, septate and 3.50- 5.50 µm in width. Conidiophores were cylindrical, short, simple, septate and measured 88.60–110.50 × 2.50–4.50 µm in size. Microconidia were round to oval in shape, hyaline, 0–1 septate, measuring 7.50–11.00 × 2.80–3.75 µm in size (Fig. 2c) Macroconidia were curved with short apices pedicellate basal cells, hyaline, 3–4 septate, measuring 21.15–32.00 × 3.80–4.75 µm in size (Fig. 2d,e). Clamydospores were intercalary, produced singly or in chains, nearly spherical, hyaline and measured 5.80–9.00 µm in diameter (Fig. 2f). Like Fusarium solani, pure cultures of Fusarium oxysporum produced white colonies at the beginning (Fig. 3a) which later turned to peach brown colour at agar base (Fig. 3b) and attained a growth of 90 mm in 10 days of incubation at 25 ± 1 °C. Mycelium was recorded to be smooth, cylindrical, septate and branched with 3.00- 4.80 µm in width. Conidiophores were measured to be 87.40–112.50 × 2.50–5.00 µm in size. Microconidia were ellipsoidal to cylindrical, straight or curved in shape, born on short philaids, hyaline, 0–1 septate and measuring 6.80–16.00 × 3.50–4.00 µm in size (Fig. 3c). Macroconidia were fusiform, pointed at ends, pedicellate basal cells, hyaline, 2–4 septate and measuring 32.50–44.00 × 4.00–5.50 µm in size (Fig. 3d,e). Clamydospores were produced terminally, singly or in chains, nearly spherical, hyaline and measured 5.00–11.00 µm in diameter (Fig. 3f). in the case of Fusarium equiseti pure culture produced white colonies (Fig. 4a) later turning to peach orange colour at agar base (Fig. 4b) with a growth of 90 mm in 10 days of incubation at 25 ± 1 °C. Microscopic observations revealed that mycelium was smooth, branched, cylindrical, septate and 3.25- 4.80 µm in width. Conidiophores were cylindrical, short, simple, septate and measured 72.50–106.30 × 3.00–4.5.0 µm in size. Microconidia were oval, hyaline, 0–1 septate and measuring 12.00–16.00 × 3.20–4.50 µm in size (Fig. 4c). Macroconidia were curved with tapered and elongated apical cell, prominent foot cell, hyaline, 2–5 septate and measuring 24.00–40.00 × 4.00–6.00 µm in size (Fig. 4d,e). Clamydospores were produced intercalary, singly or in chains, nearly spherical, hyaline and measured 5.00- 11.00 µm in diameter. Morphological characters of Fusarium solani, Fusarium oxysporum and Fusarium equiseti are represented in Tables 2, 3 and 4 respectively.
Pathogenicity and Symptomatology
The Pathogenicity of isolated fungi was established by confirming Koch’s postulates on potted chilli plants cv. ‘Shalimar Long’ (Figs. 5, 6). The initial disease symptoms in case of Fusarium equiseti inoculated seedlings developed after 10- 12 days of inoculation and in case of Fusarium solani and Fusarium oxysporum the initial symptoms were observed after 19–20 days of inoculation. The pathogens were re-isolated from the inoculated seedlings and compared with their original inoculated cultures. The re-isolated pathogens completely resembled the original inoculated fungi in their morphological, cultural and pathogenic characteristics, so satisfied Koch’s postulates. Pathogen inoculated plants developed initial symptoms as light green to yellowish discoloration of leaves (Fig. 7) followed by their shriveling, drooping and ultimately wilting leading to death of the whole plant. When the collar region of the plant was cut vertically, the vascular bundles showed brownish discoloration. Comparative microscopic results depicting Conidia, Chlamydospore and Mycelium of the three fungi show clear difference (Fig. 8).
Molecular Characterization and phylogeny
After sequencing the PCR product and analyzing with BLASTn, all sequences showed 98%-100% sequence homology with GenBank sequences. Sequences were submitted to NCBI GenBank and accession numbers were obtained (Table 1). Phylogenetic analysis revealed that our isolates clustered along with other submitted Fusarium isolates from GenBank. The sequences of Fusarium isolates formed same cluster with Fusarium equiseti, Fusarium solani and Fusarium oxysporum, but in separate subclusters. Alternaria formed a different cluster (outgroup) in the dendrogram (Fig. 9). We observed that using ITS region of the 10 isolates collected from different locations of J&K showed sequence homology with isolates reported from different regions particularly isolates of India, China, Egypt and Thailand.
Discussion
Chilli is a widely grown crop all over the world for its flavor and colour; and is believed to have many health beneficial properties like anti-inflammatory and antioxidant potential41. However, from past few years the production of chilli has been hampered due to many prevailing abiotic and biotic stresses which include many bacterial and fungal diseases42. Among the fungal diseases most dominating is Fusarium species to which 50–80% losses in the production are attributed43. The pathogens causing wilt in chilli in the regions of Kashmir valley were isolated and later confirmed as Fusarium oxysporum, Fusarium solani and Fusarium equiseti based on the morpho-cultural characteristics as mentioned by Booth et al.44 as well as by sequencing results. The disease was prevalent in all the surveyed locations with varied levels of incidence which ranged from 22 to 55 per cent. The disease incidence was highest in district Budgam (52.10%) followed by Pulwama (48.70%) and Srinagar (35.20%). However, the incidence was least (26.70%) in district Anantnag. Fusarium equiseti has been reported to cause wilt in cauliflower in china and wilt in cumin reported from parts of India45 and has not been yet reported to cause chilli wilt in Himalayan region of Kashmir valley. On the basis of morpho-cultural characteristics, pathogenicity and comparison with authentic description given by Booth44, Domsch et al.46, Brayford47 and Zhu et al.48 the pathogens were identified as Fusarium Oxysporum Schlecht., Fusarium equiseti (Corda.) Sacc. and Fusarium solani.
All the three isolates viz., F. oxysporum, F. solani, F. equiseti isolates evaluated within the present study were infective in nature with slight variation in virulence. The appearance of the specific symptom/disease severity varied depending on the pathogenicity or virulence level of specific isolate of fungus. The fungal pathogens isolated from wilted chilli plants, when artificially inoculated through rhizosphere inoculation technique on potted chilli plants exhibited typical disease symptoms. Koch’s postulates were confirmed by re-isolating the pathogen from the artificially inoculated and infected plants. The plants developed initial symptoms on the second week of inoculation. The initial symptoms showed light green to yellowish discoloration of leaves followed by their shriveling, drooping and finally death of whole plant at fourth week of inoculation. The dried leaves remained clinged on the wilted plants. When the collar region of the plant was cut vertically, the vascular bundles showed brownish discoloration. The initial symptoms were first recorded in F.equiseti so, this strain of pathogen was more virulent as compared to other pathogens which were artificially inoculated (F.oxysporum, F.solani). F.oxysporum has also been reported from various regions of Kashmir valley and from many parts of India49 F.solani has been also reported from many parts of India50,51. F. equiseti was shown to be prevelent in five different locations in Kashmir valley.
In the present studies, the internal transcribed spacer (ITS) amplification with genera and/or species specific ITS primers, clearly differentiated all the three isolates of fungi into F. oxysporum, F. solani, F. equiseti. In a recent study PCR when carried out on Fusarium found in Capsicum species using ITS regions, the sequences successfully resulted in identification of five Fusarium species as F. solani, F. oxysporum, F. equiseti, F. incarnatum, F. chlamydosporum showing dominance in F. solani and F. equiseti52. Based on amplification and size of the amplicons in different isolates, amplified with ITS primer combinations viz., ITS1F and ITS4Rdirected the amplification of ~ 500 bp ITS-rRNA uniform amplicons in all the isolates of F. oxysporum , F. solani, F. equiseti. These ITS primer mixtures, which targeted and amplified a specific gene/region within the internal transcribed spacer (ITS) regions, are used for straight forward and early identification of fungal species viz., F. oxysporum, F. solani, F. equiseti from the infected plant tissues. The PCR products of ITS1-5.8S-ITS2 gene were sequenced and the pairwise and multiple alignment of pathogens identified through sequencing was carried out by Clustal Omega and later were published in Gene bank. (https://www.ebi.ac.uk/Tools/msa/clustalo/). While comparing the query sequence of ITS1 5.8S-ITS2 with the database sequence. Tree based method (Neighbour joining., NJ) were applied to test the efficacy of twelve DNA barcodes candidates for the identification of fungal species. Sequences were assembled, edited and multiple sequence alignments were performed using the ClustalW tool from MEGA 7.0 and a NJ tree, which was recommended as the standard barcoding method which was adopted and constructed using MEGA 7.0 software. In the present investigation three query sequences of different fungal species which were identified through sequencing were clustered together according to their similarity with their respective hits viz., Fusarium equiseti, Fusarium solani, Fusarium solani showing closeness with each other also.
Conclusion
In the present study, three isolates of fungi were collected and isolated from diseased root/stem samples of chilli the pathogenicity was demonstrated on very vulnerable chilli plants. The pathogenicity of the contagious spp. isolates was confirmed based on the capacity of each isolate to cause ailment and appearance of the particular side effects viz., yellowing, wilting or plant death. Morphological characteristics of the causal pathogen were studied both on host as well as on artificial culture medium to identify the associated pathogen. Gene sequencing studies of ITS1-5.8S-ITS2 gene were elucidated and all the sequences were submitted in GenBank (NCBI). The major chilli growing hotspots of Kashmir Valley were investigated in the current study. Among many pathogens F. oxysporum and F. solani and Fusarium equiseti were found to be prevalent in these areas. Fusarium equiseti incidence is reported for first time in Kashmir valley.
Change history
22 April 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41598-021-88867-4
References
Muthukumar, T. & Sathya, R. Endorhizal fungal association and colonization patterns in solanaceae. Pol. Bot. J. 62(2), 287–299 (2017).
Zhuang, Y. et al. Bioactive characteristics and antioxidant activities of nine peppers. J. Funct. Foods 4(1), 331–338 (2012).
Wahba, N. M., Ahmed, A. S. & Ebraheim, Z. Z. Antimicrobial effects of pepper, parsley, and dill and their roles in the microbiological quality enhancement of traditional Egyptian Kareish cheese. Foodborne Pathog. Dis. 7(4), 411–418 (2010).
Palevitch, D. & Craker, L. Nutritional and medical importance of red pepper (Capsicum spp). J. Herbs Spices Med. Plants 3(2), 55–83 (1996).
Thomas, S., Kumar, D., and Ahmad, A. Marketing of Green Chilli in Kaushambi District of Uttar Pradesh, India.
Sridhar, K., Rajesh, V. & Omprakash, S. A critical review on agronomic management of pests and diseases in chilli. Int. J. Plant Anim. Environ. Sci. 4, 284–289 (2014).
Alston, J. M., Beddow, J. M. & Pardey, P. G. Agricultural research, productivity, and food prices in the long run. Science 325(5945), 1209–1210 (2009).
Subbiah, A. & JeyAkumAr, S. Production and marketing of chillies. Facts You 29(6), 1–19 (2009).
Farooqi, A. A., Sreeramu, B. & Srinivasappa, K. Cultivation of Spice Crops (Universities Press, Hyderabad, 2005).
Reddy, R. J. Problems of red chilli growers and cold storage units—a case study of Guntur district. Int. J. Res. Soc. Sci. 2(4), 684–697 (2012).
Rangaswami, G. Diseases of Crop Plant in India 570 (Printice-Hall of India Private Ltd., New Delhi, India, 1979).
Sanogo, S. Chile pepper and the threat of wilt diseases. Plant Health Prog. 4(1), 23 (2003).
Singh, J. et al. Screening of chili cultivars against fusarium wilt of chilli (Capsicum annuum L.). Int. J. Agric. Sci. Res. 7(1), 235–240 (2017).
Khan, K.A., et al., Chilli Wilt Disease: A Serious problem in Chilli cultivation in India.
Tembhurne, B. et al. Molecular characterization and screening for Fusarium (Fusarium solani) resistance in Chilli (Capsicum annuum L.) genotypes. Int. J. Curr. Microbiol. Appl. Sci. 6(9), 1585–1597 (2017).
Naik, M. Wilt of chilli with special reference to cultural, morphological, molecular characterization and pathogenic variability of Fusarium isolates of India. in Proc. Midterm Review Meeting of the Project held at Indian Institute of Vegetable Research, Varanasi. 2006.
Loganathan, M., et al. Morphological, cultural and molecular characterizations of Fusarium wilt infecting tomato and chilli. in National Symposium on Abiotic and Biotic Stress Management in Vegetable Crops. Indian Society of Vegetable Science, IIVR. 2013.
Prasannath, K., Mahendran, S., and Karunakaran, S. Control of Fusarium wilt of chilli (Capsicum annuum L.) by crude plant extracts2011.
MacHardy, W., M. WE, and B. CH, Vascular wilt Fusaria: infection and pathogenesis. 1981.
Rivelli, V., A wilt of pepper incited by Fusarium oxysporum f. spp. capsici f. spp. nov, 1989, MS Thesis, Louisiana State University, Baton Rouge.
Morid, B., Hajmansoor, S. & Kakvan, N. Screening of resistance genes to Fusarium root rot and Fusarium wilt diseases in tomato (Lycopersicon esculentum) cultivars using RAPD and CAPs markers. Eur. J. Exp. Biol. 2(4), 931–939 (2012).
Goswami, R. S. & Kistler, H. C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5(6), 515–525 (2004).
Goswami, R. S., Dong, Y. & Punja, Z. K. Host range and mycotoxin production by Fusarium equiseti isolates originating from ginseng fields1. Can. J. Plant Path. 30(1), 155–160 (2008).
Mejía-Bautista, M. A. et al. Actividad in vitro de Bacillus spp. en la inhibición de crecimiento micelial de Fusarium equiseti Y Fusarium solani aislado de chile habanero (Capsicum chinense Jacq.). Agrociencia 50(8), 1123–1135 (2016).
Teixeira, L.M., Caracterização de isolados de Fusarium oxysporum e resistência de genótipos de Passiflora à fusariose. 2015.
Raghu, S., Benagi, V. & Nargund, V. Cultural, morphological and pathogenic variability among the isolates of Fusarium solani causing wilt disease of Chilli (Capsicum annuum L.). J. Pure Appl. Microbiol. Shahjahanabad 10(1), 599–604 (2016).
El Kichaoui, A., et al., Development of Beauveria bassiana-Based Bio-Fungicide Against Fusarium Wilt Pathogens for Capsicum Annuum, a Promising Approach Toward Vital Biocontrol Industry in Gaza Strip. IUG Journal of Natural Studies, 2017.
Leslie, J. F. & Summerell, B. A. The Fusarium Laboratory Manual (Wiley, New York, 2008).
Lievens, B., Rep, M. & Thomma, B. P. Recent developments in the molecular discrimination of formae speciales of Fusarium oxysporum. Pest Manag. Sci. Formerly Pest. Sci. 64(8), 781–788 (2008).
Singha, I. M. et al. Identification and characterization of Fusarium sp. using ITS and RAPD causing fusarium wilt of tomato isolated from Assam, North East India. J. Genet. Eng. Biotechnol. 14(1), 99–105 (2016).
Menezes, J. P. et al. Variabilidade genética na região its do rDNA de isolados de Trichoderma spp. (Biocontrolador) e Fusarium oxysporum f. sp. chrysanthemi. Ciência e Agrotecnologia 34(1), 132–139 (2010).
Chen, C. A. et al. Secondary structure and phylogenetic utility of the ribosomal internal transcribed spacer 2 (ITS2) in scleractinian corals. Zool. Stud. 43(4), 759–771 (2004).
Hillis, D. M. & Dixon, M. T. Ribosomal DNA: molecular evolution and phylogenetic inference. Q. Rev. Biol. 66(4), 411–453 (1991).
Schoch, C. L. et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. 109(16), 6241–6246 (2012).
McDonald, B. A. & Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40(1), 349–379 (2002).
Ferniah, R. S. et al. Characterization and pathogenicity of Fusarium oxysporum as the causal agent of Fusarium wilt in chili (Capsicum annuum L.). Microbiol. Indones. 8(3), 5 (2014).
Pathak, V.N., Essentials of plant pathology. Essentials of plant pathology., 1972.
Davey, C. & Papavizas, G. Comparison of methods for isolating Rhizoctonia from soil. Can. J. Microbiol. 8(6), 847–853 (1962).
Cenis, J. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 20(9), 2380 (1992).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33(7), 1870–1874 (2016).
Rahman, M., Rahman, S., and Akter, A. Comparative performance of some insecticides and botanicals against chilli fruit borer (Helicoverpa armigera). J. Exp. Sci. 2011.
Abdel-Fattah, G. & Al-Amri, S. Induced systemic resistance in tomato plants against Fusarium oxysporum f. sp. lycopersici by different kinds of compost. Afr. J. Biotechnol. 11(61), 12454–12463 (2012).
Madhavi, M. et al. Integrated Management of Wilt of Chilli Incited by. Indian J. Plant Prot. 34(2), 225–228 (2006).
Booth, C., The genus fusarium. The genus Fusarium., 1971.
Ramchandra, S. S. & Bhatt, P. Morphological and molecular identification of Fusarium isolated from cumin wilt. Int. J. Plant Protec 4(2), 359–362 (2011).
Domsch, K. H., Gams, W. & Anderson, T.-H. Compendium of Soil Fungi Vol. 1 (Academic Press, London, 1980).
Brayford, D. & Samuels, G. J. Some didymosporous species of Nectria with nonmicroconidial Cylindrocarpon anamorphs. Mycologia 85(4), 612–637 (1993).
Zhu, Z. et al. Identification and characterization of Fusarium species associated with wilt of Eleocharis dulcis (Chinese water chestnut) in China. Plant Dis. 98(7), 977–987 (2014).
Rajeswari, P. and Kannabiran, B. In Vitro Effects of Antagonistic Microorganisms on Fusarium oxysporum [Schlecht. Emend. Synd & Hans] Infecting Arachis hypogaea L. J. Phytol. (2011).
Naik, M., Rani, G. D. & Madhukar, H. Identification of resistant sources against wilt of chilli (Capsicum annuum L) caused by Fusarium solani (Mart.) Sacc. J. Mycopathol. Res. 46(1), 93–96 (2008).
Maruti, T. et al. Reaction of chilli (Capsicum annuum L.) genotypes and hybrids against Fusarium wilt (Fusarium solani). J. Spices Arom. Crops 23(2), 186–191 (2014).
dos Anjos, I. V. et al. Molecular characterization of isolates of Fusarium spp. associated with wilt in Capsicum spp. J. Agric. Sci. 11(6), 328 (2019).
Funding
The project was initially submitted to DBT, Govt of India for Funding under grant application no. BT/PR30182/AGIII/103/1084/2018 but was rejected. Seeing the importance of the problem caused by Chilli wilt throughout India and the huge economic loss incurred by India every year due to this disease, the project was carried out and the cost incurred in it was paid from the salary of the corresponding author.
Author information
Authors and Affiliations
Contributions
K.Z.M.: conceptualization, data curation, K.Z.M., S.M., N.A., A.H.: formal analysis, investigation, A.H., N.A.K., R.S.R., M.A.M.: methodology, visualization, writing—original draft, writing—review and editing. K.Z.M.: funding acquisition, project administration, resources, software, supervision, validation, writing—review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained errors in Figures 3, 4 and 6.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hami, A., Rasool, R.S., Khan, N.A. et al. Morpho-molecular identification and first report of Fusarium equiseti in causing chilli wilt from Kashmir (Northern Himalayas). Sci Rep 11, 3610 (2021). https://doi.org/10.1038/s41598-021-82854-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-82854-5
This article is cited by
-
Identification of fungal species associated with chilli fruit disease in North-Central Vietnam
Journal of Plant Pathology (2023)
-
Occurrence and characterization of Alternaria alternata causing leaf spot of soybean (Glycine max) in Pakistan
Journal of Plant Pathology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.